Commensal Fecal Microbiota Profiles Associated with Initial Stages of Intestinal Mucosa Damage: A Pilot Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Volunteers
2.2. Fecal Sample Processing
2.3. DNA Extraction and Microbiota Metataxonomic Analyses
2.4. Fecal Short-Chain Fatty Acid Concentrations
2.5. Statistical Analyses
3. Results
3.1. General Characteristics of the Sample Population
3.2. Fecal Microbiota Profiles
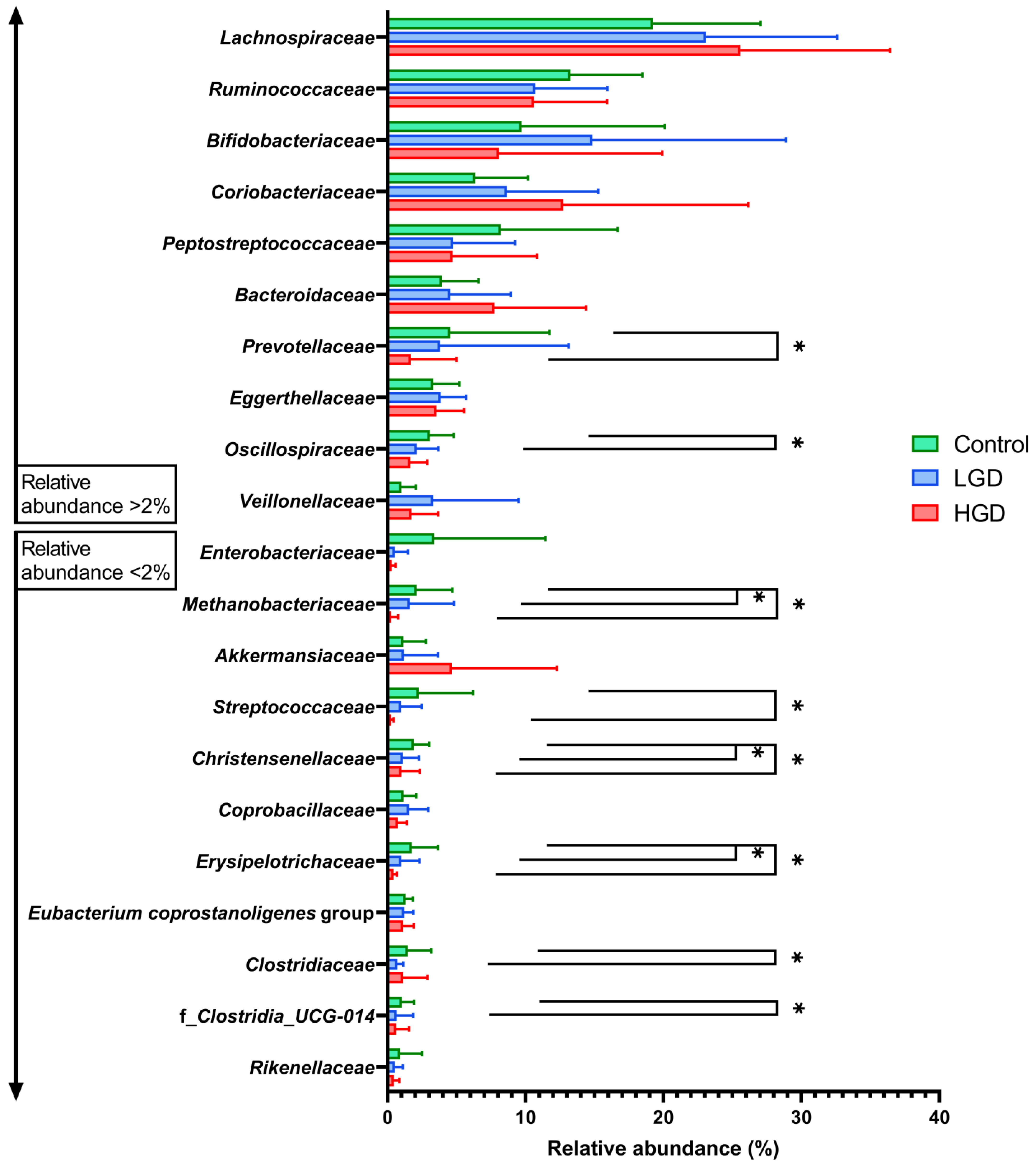
3.2.1. Microbiota Comparison between Control and Polyps Groups
3.2.2. Microbiota Comparison between Individuals with Hyperplastic Polyps and Conventional Adenomas
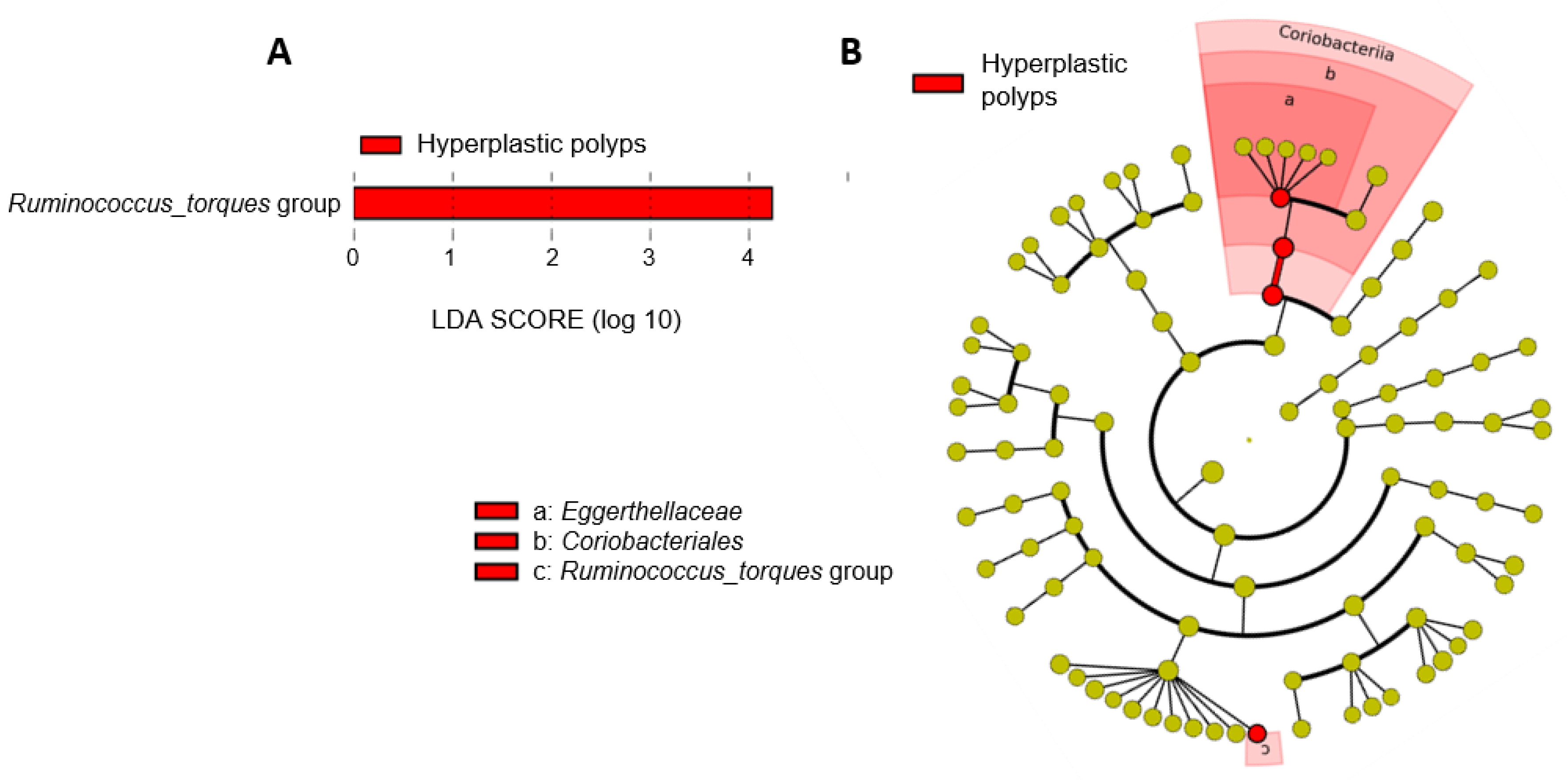
3.2.3. Shifts in the Microbiota of Individuals with Hyperplastic Polyps
3.2.4. Shifts in the Microbiota of Individuals with Conventional Adenomas
3.3. Fecal Short-Chain Fatty Acids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Laconi, E.; Marongiu, F.; DeGregori, J. Cancer as a Disease of Old Age: Changing Mutational and Microenvironmental Landscapes. Br. J. Cancer 2020, 122, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.H.; Goel, A.; Chung, D.C. Pathways of Colorectal Carcinogenesis. Gastroenterology 2020, 158, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Jakszyn, P.; Cayssials, V.; Buckland, G.; Perez-Cornago, A.; Weiderpass, E.; Boeing, H.; Bergmann, M.M.; Vulcan, A.; Ohlsson, B.; Masala, G.; et al. Inflammatory Potential of the Diet and Risk of Colorectal Cancer in the European Prospective Investigation into Cancer and Nutrition Study. Int. J. Cancer 2020, 147, 1027–1039. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of Colorectal Cancer: Incidence, Mortality, Survival, and Risk Factors. Gastroenterol. Rev. Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Powers, S.; Zhu, W.; Hannun, Y.A. Substantial Contribution of Extrinsic Risk Factors to Cancer Development. Nature 2015, 529, 43–47. [Google Scholar] [CrossRef]
- Snover, D.C. Diagnostic and Reporting Issues of Preneoplastic Polyps of the Large Intestine with Early Carcinoma. Ann. Diagn. Pathol. 2019, 39, 1–14. [Google Scholar] [CrossRef]
- Vennelaganti, S.; Cuatrecasas, M.; Vennalaganti, P.; Kennedy, K.F.; Srinivasan, S.; Patil, D.T.; Plesec, T.; Lanas, A.; Hörndler, C.; Andraws, N.; et al. Interobserver Agreement Among Pathologists in the Differentiation of Sessile Serrated from Hyperplastic Polyps. Gastroenterology 2021, 160, 452–454. [Google Scholar] [CrossRef]
- Sanabria, M.C.; Umaña, A.; Serrano, M.L.; Sánchez, M.; Mesa, J.; Hernández, G.A. Vías de Carcinogénesis Colorrectal y Sus Implicaciones Clínicas. [Pathways of Colorectal Carcinogenesis and Their Clinical Implications]. Rev. Colomb. Cancerol. 2012, 16, 170–181. [Google Scholar] [CrossRef]
- Rex, D.K.; Ahnen, D.J.; Baron, J.A.; Batts, K.P.; Burke, C.A.; Burt, R.W.; Goldblum, J.R.; Guillem, J.G.; Kahi, C.J.; Kalady, M.F.; et al. Serrated Lesions of the Colorectum: Review and Recommendations from an Expert Panel. Am. J. Gastroenterol. 2012, 107, 1315–1329. [Google Scholar] [CrossRef]
- Chang, J.J.; Chien, C.H.; Chen, S.W.; Chen, L.W.; Liu, C.J.; Yen, C.L. Long Term Outcomes of Colon Polyps with High Grade Dysplasia Following Endoscopic Resection. BMC Gastroenterol. 2020, 20, 376. [Google Scholar] [CrossRef] [PubMed]
- Cronin, P.; Joyce, S.A.; O’toole, P.W.; O’connor, E.M. Dietary Fibre Modulates the Gut Microbiota. Nutrients 2021, 13, 1655. [Google Scholar] [CrossRef]
- O’Keefe, S.J.D. Diet, Microorganisms and Their Metabolites, and Colon Cancer. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 691–706. [Google Scholar] [CrossRef] [PubMed]
- Portune, K.J.; Beaumont, M.; Davila, A.-M.; Tomé, D.; Blachier, F.; Sanz, Y. Gut Microbiota Role in Dietary Protein Metabolism and Health-Related Outcomes: The Two Sides of the Coin. Trends Food Sci. Technol. 2016, 57, 213–232. [Google Scholar] [CrossRef]
- Nogacka, A.M.; Gómez-Martín, M.; Suárez, A.; González-Bernardo, O.; de los Reyes-Gavilán, C.G.; González, S. Xenobiotics Formed during Food Processing: Their Relation with the Intestinal Microbiota and Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 2051. [Google Scholar] [CrossRef] [PubMed]
- Tjalsma, H.; Boleij, A.; Marchesi, J.R.; Dutilh, B.E. A Bacterial Driver–Passenger Model for Colorectal Cancer: Beyond the Usual Suspects. Nat. Rev. Microbiol. 2012, 10, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.A.; Dominianni, C.; Shapiro, J.A.; Church, T.R.; Wu, J.; Miller, G.; Yuen, E.; Freiman, H.; Lustbader, I.; Salik, J.; et al. The Gut Microbiota in Conventional and Serrated Precursors of Colorectal Cancer. Microbiome 2016, 4, 69. [Google Scholar] [CrossRef]
- Rezasoltani, S.; Ghanbari, R.; Looha, M.A.; Mojarad, E.N.; Yadegar, A.; Stewart, D.; Aghdaei, H.A.; Zali, M.R. Expression of Main Toll-Like Receptors in Patients with Different Types of Colorectal Polyps and Their Relationship with Gut Microbiota. Int. J. Mol. Sci. 2020, 21, 8968. [Google Scholar] [CrossRef]
- Ruiz-Saavedra, S.; Zapico, A.; del Rey, C.G.; Gonzalez, C.; Suárez, A.; Díaz, Y.; de los Reyes-Gavilán, C.G.; González, S. Dietary Xenobiotics Derived from Food Processing: Association with Fecal Mutagenicity and Gut Mucosal Damage. Nutrients 2022, 14, 3482. [Google Scholar] [CrossRef]
- International Human Microbiome Standards (IHMS). IHMS_SOP 06 V2: Standard Operating Procedure for Fecal Samples DNA Extraction, Protocol Q. Available online: https://brd.nci.nih.gov/brd/sop/download-pdf/2105 (accessed on 27 October 2023).
- Nogacka, A.M.; Salazar, N.; Arboleya, S.; Ruas-Madiedo, P.; Mancabelli, L.; Suarez, A.; Martinez-Faedo, C.; Ventura, M.; Tochio, T.; Hirano, K.; et al. In Vitro Evaluation of Different Prebiotics on the Modulation of Gut Microbiota Composition and Function in Morbid Obese and Normal-Weight Subjects. Int. J. Mol. Sci. 2020, 21, 906. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Yachida, S.; Mizutani, S.; Shiroma, H.; Shiba, S.; Nakajima, T.; Sakamoto, T.; Watanabe, H.; Masuda, K.; Nishimoto, Y.; Kubo, M.; et al. Metagenomic and Metabolomic Analyses Reveal Distinct Stage-Specific Phenotypes of the Gut Microbiota in Colorectal Cancer. Nat. Med. 2019, 25, 968–976. [Google Scholar] [CrossRef] [PubMed]
- Aprile, F.; Bruno, G.; Palma, R.; Mascellino, M.T.; Panetta, C.; Scalese, G.; Oliva, A.; Severi, C.; Pontone, S. Microbiota Alterations in Precancerous Colon Lesions: A Systematic Review. Cancers 2021, 13, 3061. [Google Scholar] [CrossRef]
- Yang, Y.; Misra, B.B.; Liang, L.; Bi, D.; Weng, W.; Wu, W.; Cai, S.; Qin, H.; Goel, A.; Li, X.; et al. Integrated Microbiome and Metabolome Analysis Reveals a Novel Interplay between Commensal Bacteria and Metabolites in Colorectal Cancer. Theranostics 2019, 9, 4101–4114. [Google Scholar] [CrossRef] [PubMed]
- Kong, C.; Liang, L.; Liu, G.; Du, L.; Yang, Y.; Liu, J.; Shi, D.; Li, X.; Ma, Y. Integrated Metagenomic and Metabolomic Analysis Reveals Distinct Gut-Microbiome-Derived Phenotypes in Early-Onset Colorectal Cancer. Gut 2023, 72, 1129–1142. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Li, Q.; Tang, Z.; Yan, L.; Zhang, L.; Zheng, Q.; Zeng, X.; Chen, G.; Yue, H.; Li, J.; et al. Alterations of the Gut Microbiome and Fecal Metabolome in Colorectal Cancer: Implication of Intestinal Metabolism for Tumorigenesis. Front. Physiol. 2022, 13, 854545. [Google Scholar] [CrossRef] [PubMed]
- Waters, J.L.; Ley, R.E. The Human Gut Bacteria Christensenellaceae Are Widespread, Heritable, and Associated with Health. BMC Biol. 2019, 17, 83. [Google Scholar] [CrossRef]
- Chen, J.; Vitetta, L. Inflammation-Modulating Effect of Butyrate in the Prevention of Colon Cancer by Dietary Fiber. Clin. Color. Cancer 2018, 17, e541–e544. [Google Scholar] [CrossRef]
- Gheorghe, A.S.; Negru, Ș.M.; Preda, M.; Mihăilă, R.I.; Komporaly, I.A.; Dumitrescu, E.A.; Lungulescu, C.V.; Kajanto, L.A.; Georgescu, B.; Radu, E.A.; et al. Biochemical and Metabolical Pathways Associated with Microbiota-Derived Butyrate in Colorectal Cancer and Omega-3 Fatty Acids Implications: A Narrative Review. Nutrients 2022, 14, 1152. [Google Scholar] [CrossRef]
- Qi, Z.; Zhibo, Z.; Jing, Z.; Zhanbo, Q.; Shugao, H.; Weili, J.; Jiang, L.; Shuwen, H. Prediction Model of Poorly Differentiated Colorectal Cancer (CRC) Based on Gut Bacteria. BMC Microbiol. 2022, 22, 312. [Google Scholar] [CrossRef]
- Fan, J.Q.; Zhao, W.F.; Lu, Q.W.; Zha, F.R.; Lv, L.B.; Ye, G.L.; Gao, H.L. Fecal Microbial Biomarkers Combined with Multi-target Stool DNA Test Improve Diagnostic Accuracy for Colorectal Cancer. World J. Gastrointest. Oncol. 2023, 15, 1424–1435. [Google Scholar] [CrossRef] [PubMed]
- Sereme, Y.; Mezouar, S.; Grine, G.; Mege, J.L.; Drancourt, M.; Corbeau, P.; Vitte, J. Methanogenic Archaea: Emerging Partners in the Field of Allergic Diseases. Clin. Rev. Allergy Immunol. 2019, 57, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Guindo, C.O.; Drancourt, M.; Grine, G. Digestive Tract Methanodrome: Physiological Roles of Human Microbiota-Associated Methanogens. Microb. Pathog. 2020, 149, 104425. [Google Scholar] [CrossRef] [PubMed]
- Engels, C.; Ruscheweyh, H.J.; Beerenwinkel, N.; Lacroix, C.; Schwab, C. The Common Gut Microbe Eubacterium hallii also Contributes to Intestinal Propionate Formation. Front. Microbiol. 2016, 7, 713. [Google Scholar] [CrossRef] [PubMed]
- Fekry, M.I.; Engels, C.; Zhang, J.; Schwab, C.; Lacroix, C.; Sturla, S.J.; Chassard, C. The Strict Anaerobic Gut Microbe Eubacterium hallii Transforms the Carcinogenic Dietary Heterocyclic Amine 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP). Environ. Microbiol. Rep. 2016, 8, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Li, B.; Tu, C.; Chen, X.; Guo, M. Correlations between Intestinal Microbiota and Clinical Characteristics in Colorectal Adenoma/Carcinoma. Biomed. Res. Int. 2022, 2022, 3140070. [Google Scholar] [CrossRef] [PubMed]
- Mori, G.; Rampelli, S.; Orena, B.S.; Rengucci, C.; De Maio, G.; Barbieri, G.; Passardi, A.; Casadei Gardini, A.; Frassineti, G.L.; Gaiarsa, S.; et al. Shifts of Faecal Microbiota During Sporadic Colorectal Carcinogenesis. Sci. Rep. 2018, 8, 10329. [Google Scholar] [CrossRef] [PubMed]
- Kaakoush, N.O. Insights into the Role of Erysipelotrichaceae in the Human Host. Front. Cell. Infect. Microbiol. 2015, 5, 84. [Google Scholar] [CrossRef]
- Yoon, H.; Kim, N.; Park, J.H.; Kim, Y.S.; Lee, J.; Kim, H.W.; Choi, Y.J.; Shin, C.M.; Park, Y.S.; Lee, D.H.; et al. Comparisons of Gut Microbiota Among Healthy Control, Patients with Conventional Adenoma, Sessile Serrated Adenoma, and Colorectal Cancer. J. Cancer Prev. 2017, 22, 108–114. [Google Scholar] [CrossRef]
- Zheng, J.; Hoffman, K.L.; Chen, J.-S.; Shivappa, N.; Sood, A.; Browman, G.J.; Dirba, D.D.; Hanash, S.; Wei, P.; Hebert, J.R.; et al. Dietary Inflammatory Potential in Relation to the Gut Microbiome: Results from a Cross-Sectional Study. Br. J. Nutr. 2020, 124, 931–942. [Google Scholar] [CrossRef]
- Yan, H.; Qin, Q.; Chen, J.; Yan, S.; Li, T.; Gao, X.; Yang, Y.; Li, A.; Ding, S. Gut Microbiome Alterations in Patients with Visceral Obesity Based on Quantitative Computed Tomography. Front. Cell. Infect. Microbiol. 2022, 11, 823262. [Google Scholar] [CrossRef]
- Dong, Y.; Xu, R.; Chen, X.; Yang, C.; Jiang, F.; Shen, Y.; Li, Q.; Fang, F.; Li, Y.; Shen, X. Characterization of Gut Microbiota in Adults with Coronary Atherosclerosis. PeerJ 2023, 11, e15245. [Google Scholar] [CrossRef] [PubMed]
- Cai, P.; Xiong, J.; Sha, H.; Dai, X.; Lu, J. Tumor Bacterial Markers Diagnose the Initiation and Four Stages of Colorectal Cancer. Front. Cell. Infect. Microbiol. 2023, 13, 1123544. [Google Scholar] [CrossRef] [PubMed]
- Ahmad Kendong, S.M.; Raja Ali, R.A.; Nawawi, K.N.M.; Ahmad, H.F.; Mokhtar, N.M. Gut Dysbiosis and Intestinal Barrier Dysfunction: Potential Explanation for Early-Onset Colorectal Cancer. Front. Cell. Infect. Microbiol. 2021, 11, 1244. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Q.; Du, H.; Cheng, X.; Cheng, X.; Tang, Y.; Pan, L.; Wang, Q.; Lin, J. Characteristics of Fecal Gut Microbiota in Patients with Colorectal Cancer at Different Stages and Different Sites. Oncol. Lett. 2019, 18, 4834–4844. [Google Scholar] [CrossRef]
- DeDecker, L.; Coppedge, B.; Avelar-Barragan, J.; Karnes, W.; Whiteson, K. Microbiome Distinctions between the CRC Carcinogenic Pathways. Gut Microbes 2021, 13, 1854641. [Google Scholar] [CrossRef]
- Mailhe, M.; Ricaboni, D.; Vitton, V.; Gonzalez, J.-M.; Bachar, D.; Dubourg, G.; Cadoret, F.; Robert, C.; Delerce, J.; Levasseur, A.; et al. Repertoire of the Gut Microbiota from Stomach to Colon Using Culturomics and Next-Generation Sequencing. BMC Microbiol. 2018, 18, 157. [Google Scholar] [CrossRef]
- Saresella, M.; Marventano, I.; Barone, M.; La Rosa, F.; Piancone, F.; Mendozzi, L.; d’Arma, A.; Rossi, V.; Pugnetti, L.; Roda, G.; et al. Alterations in Circulating Fatty Acid Are Associated with Gut Microbiota Dysbiosis and Inflammation in Multiple Sclerosis. Front. Immunol. 2020, 11, 1390. [Google Scholar] [CrossRef]
- Zapico, A.; Arboleya, S.; Ruiz-Saavedra, S.; Gómez-Martín, M.; Salazar, N.; Nogacka, A.M.; Gueimonde, M.; de los Reyes-Gavilán, C.G.; González, S. Dietary Xenobiotics, (Poly)Phenols and Fibers: Exploring Associations with Gut Microbiota in Socially Vulnerable Individuals. Front. Nutr. 2022, 9, 1000829. [Google Scholar] [CrossRef]




| Variables | Control (n = 20) | Polyps (n = 34) |
|---|---|---|
| Male gender | 6 (30.0%) | 19 (55.9%) |
| Age (years) | 60 ± 9 | 61 ± 6 |
| BMI (kg/m2) † | 25.67 ± 3.81 | 27.87 ± 4.16 |
| Histopathological analysis †† | ||
| NP | 20 (100.0%) | - |
| Hyperplastic polyps | - | 9 (26.5%) |
| Conventional adenomas | - | 25 (73.5%) |
| LGD | - | 20 |
| HGD | - | 5 |
| Histopathological Groups | |||
|---|---|---|---|
| Fecal SCFAs (mM) | Control (n = 20) | Hyperplastic Polyps (n = 9) | Conventional Adenomas (n = 25) |
| Acetic acid | 47.16 ± 22.35 | 37.98 ± 23.88 | 49.86 ± 23.46 |
| Propionic acid | 13.65 ± 8.08 | 14.10 ± 7.94 | 15.92 ± 8.42 |
| Butyric acid | 13.16 ± 9.73 | 9.38 ± 6.04 | 11.48 ± 6.67 |
| Isobutyric acid | 0.99 ± 1.11 | 1.08 ± 1.29 | 0.96 ± 1.01 |
| Isovaleric acid | 2.15 ± 1.85 | 2.06 ± 1.21 | 2.18 ± 1.76 |
| Valeric acid | 2.38 ± 1.77 | 1.33 ± 0.37 | 1.67 ± 1.01 |
| Caproic acid | 0.80 ± 1.26 | 0.17 ± 0.35 * | 0.23 ± 0.45 * |
| Diagnosis Group | Dependent Variable | Independent Variable | R2 | β | p |
|---|---|---|---|---|---|
| Control | Acetic acid | Lachnospiraceae | 0.301 | 0.581 | 0.007 |
| Propionic acid | Oscillospiraceae | 0.434 | −0.623 | 0.002 | |
| Coprobacillaceae | 0.434 | 0.376 | 0.044 | ||
| Butyric acid | Oscillospiraceae | 0.275 | −0.560 | 0.010 | |
| Polyps | Acetic acid | Oscillospiraceae | 0.240 | −0.512 | 0.002 |
| Propionic acid | Rikenellaceae | 0.232 | −0.505 | 0.002 | |
| Butyric acid | Rikenellaceae | 0.145 | −0.413 | 0.015 | |
| Isobutyric acid | Enterobacteriaceae | 0.267 | 0.537 | 0.001 | |
| Isovaleric acid | Lachnospiraceae | 0.115 | −0.376 | 0.028 | |
| Hyperplastic polyps | Propionic acid | Rikenellaceae | 0.388 | −0.681 | 0.043 |
| Butyric acid | Oscillospiraceae | 0.373 | −0.672 | 0.047 | |
| Isobutyric acid | Enterobacteriaceae | 0.943 | 0.975 | 0.000 | |
| Isovaleric acid | Enterobacteriaceae | 0.835 | 0.925 | 0.000 | |
| Conventional adenomas | Acetic acid | Rikenellaceae | 0.238 | −0.519 | 0.008 |
| Propionic acid | Christensenellaceae | 0.312 | −0.423 | 0.022 | |
| Coprobacillaceae | 0.312 | 0.367 | 0.044 | ||
| Butyric acid | Coprobacillaceae | 0.138 | 0.417 | 0.038 | |
| Low-grade dysplasia | Acetic acid | Rikenellaceae | 0.221 | −0.512 | 0.021 |
| Propionic acid | Eubacterium coprostanoligenes group | 0.494 | −0.544 | 0.04 | |
| Christensenellaceae | 0.494 | −0.457 | 0.013 | ||
| Isobutyric acid | Ruminococcaceae | 0.232 | −0.522 | 0.018 | |
| High-grade dysplasia | Isobutyric acid | Enterobacteriaceae | 0.706 | 0.883 | 0.047 |
| Caproic acid | Oscillospiraceae | 0.739 | 0.897 | 0.039 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-Saavedra, S.; Arboleya, S.; Nogacka, A.M.; González del Rey, C.; Suárez, A.; Diaz, Y.; Gueimonde, M.; Salazar, N.; González, S.; de los Reyes-Gavilán, C.G. Commensal Fecal Microbiota Profiles Associated with Initial Stages of Intestinal Mucosa Damage: A Pilot Study. Cancers 2024, 16, 104. https://doi.org/10.3390/cancers16010104
Ruiz-Saavedra S, Arboleya S, Nogacka AM, González del Rey C, Suárez A, Diaz Y, Gueimonde M, Salazar N, González S, de los Reyes-Gavilán CG. Commensal Fecal Microbiota Profiles Associated with Initial Stages of Intestinal Mucosa Damage: A Pilot Study. Cancers. 2024; 16(1):104. https://doi.org/10.3390/cancers16010104
Chicago/Turabian StyleRuiz-Saavedra, Sergio, Silvia Arboleya, Alicja M. Nogacka, Carmen González del Rey, Adolfo Suárez, Ylenia Diaz, Miguel Gueimonde, Nuria Salazar, Sonia González, and Clara G. de los Reyes-Gavilán. 2024. "Commensal Fecal Microbiota Profiles Associated with Initial Stages of Intestinal Mucosa Damage: A Pilot Study" Cancers 16, no. 1: 104. https://doi.org/10.3390/cancers16010104
APA StyleRuiz-Saavedra, S., Arboleya, S., Nogacka, A. M., González del Rey, C., Suárez, A., Diaz, Y., Gueimonde, M., Salazar, N., González, S., & de los Reyes-Gavilán, C. G. (2024). Commensal Fecal Microbiota Profiles Associated with Initial Stages of Intestinal Mucosa Damage: A Pilot Study. Cancers, 16(1), 104. https://doi.org/10.3390/cancers16010104







