RSRC2 Expression Inhibits Malignant Progression of Triple-Negative Breast Cancer by Transcriptionally Regulating SCIN Expression
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatics Databases
2.2. Cell Culture and Lentiviral Transduction
2.3. Wound Healing Assay, Cell Migration and Invasion Assays and Plate Clonogenic Assay
2.4. Adhesion Assay
2.5. Luciferase Reporter Gene Experiment
2.6. RNA Extraction, Microarray Analysis and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.7. Chromatin Immunoprecipitation (ChIP) Assays
2.8. Western Blot Analysis
2.9. Animal Experiment
2.10. Statistical Analysis
3. Results
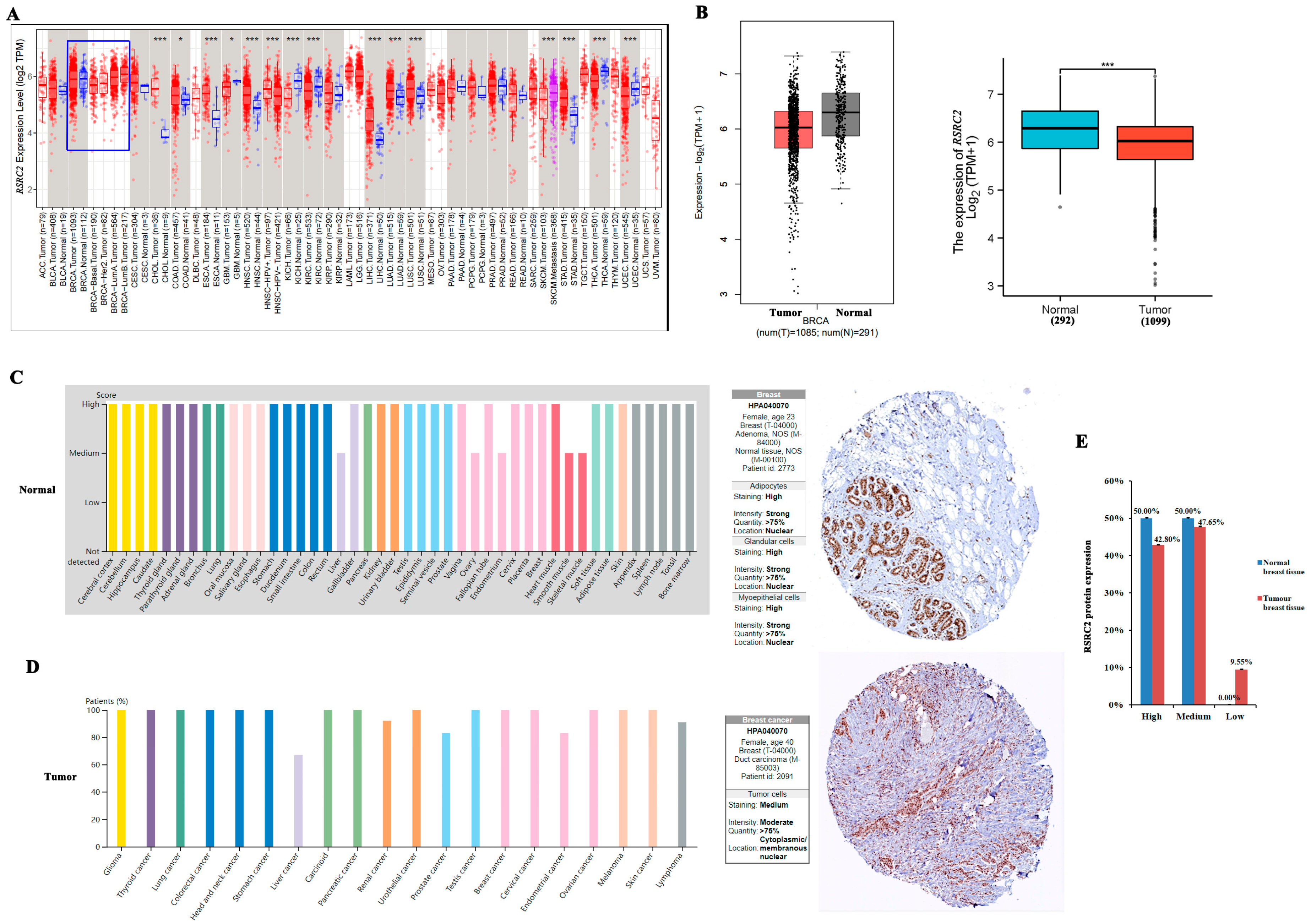
3.1. Expression of RSRC2 in Cancers by Bioinformatics Analysis
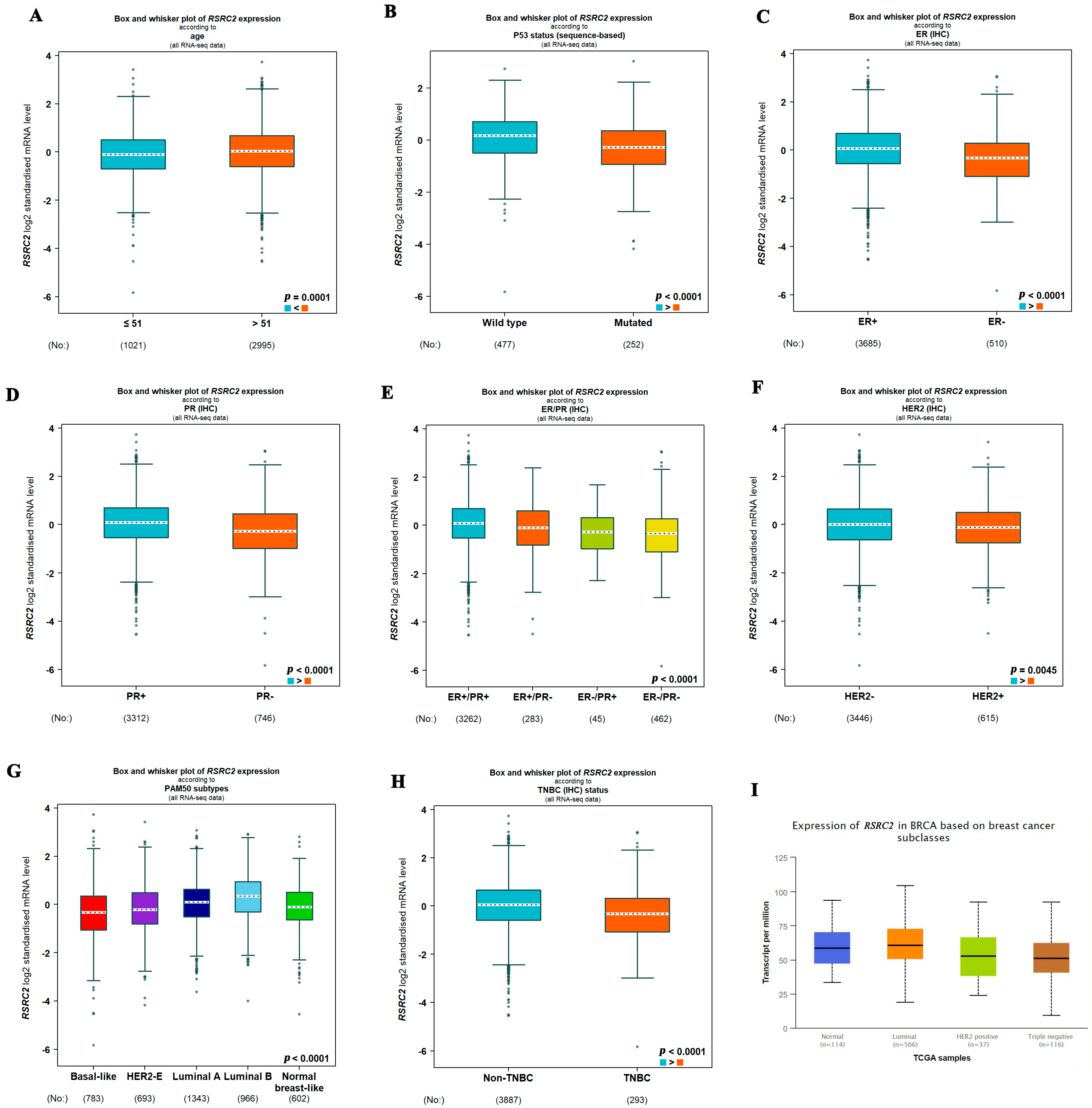
3.2. RSRC2 Expression Was Especially Lower in TNBC Than in Non-TNBC
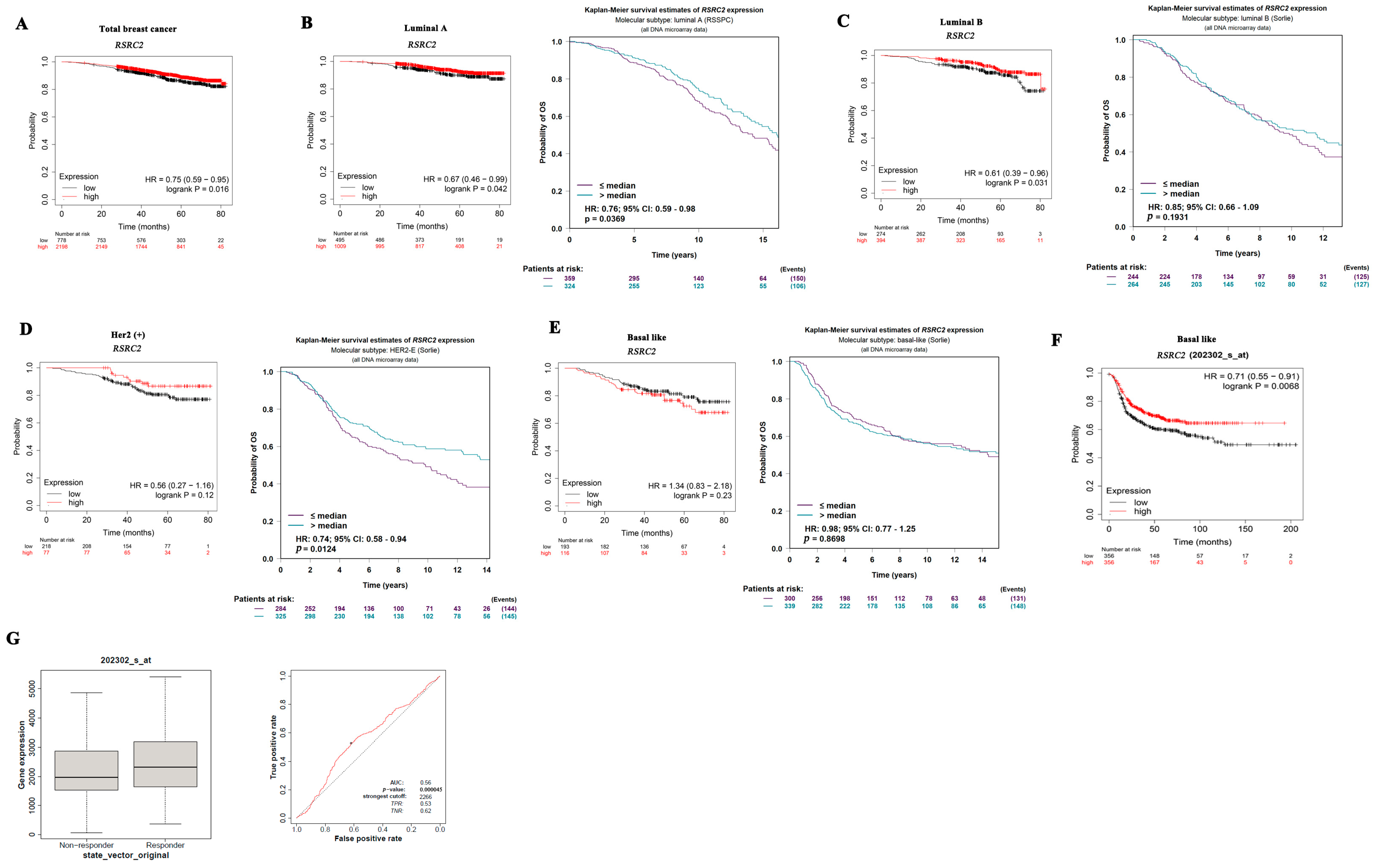
3.3. RSRC2 Expression Has Prognostic Value in Breast Cancer
3.4. Functional Enrichment Analysis of RSRC2 Coexpressed Genes in TNBC
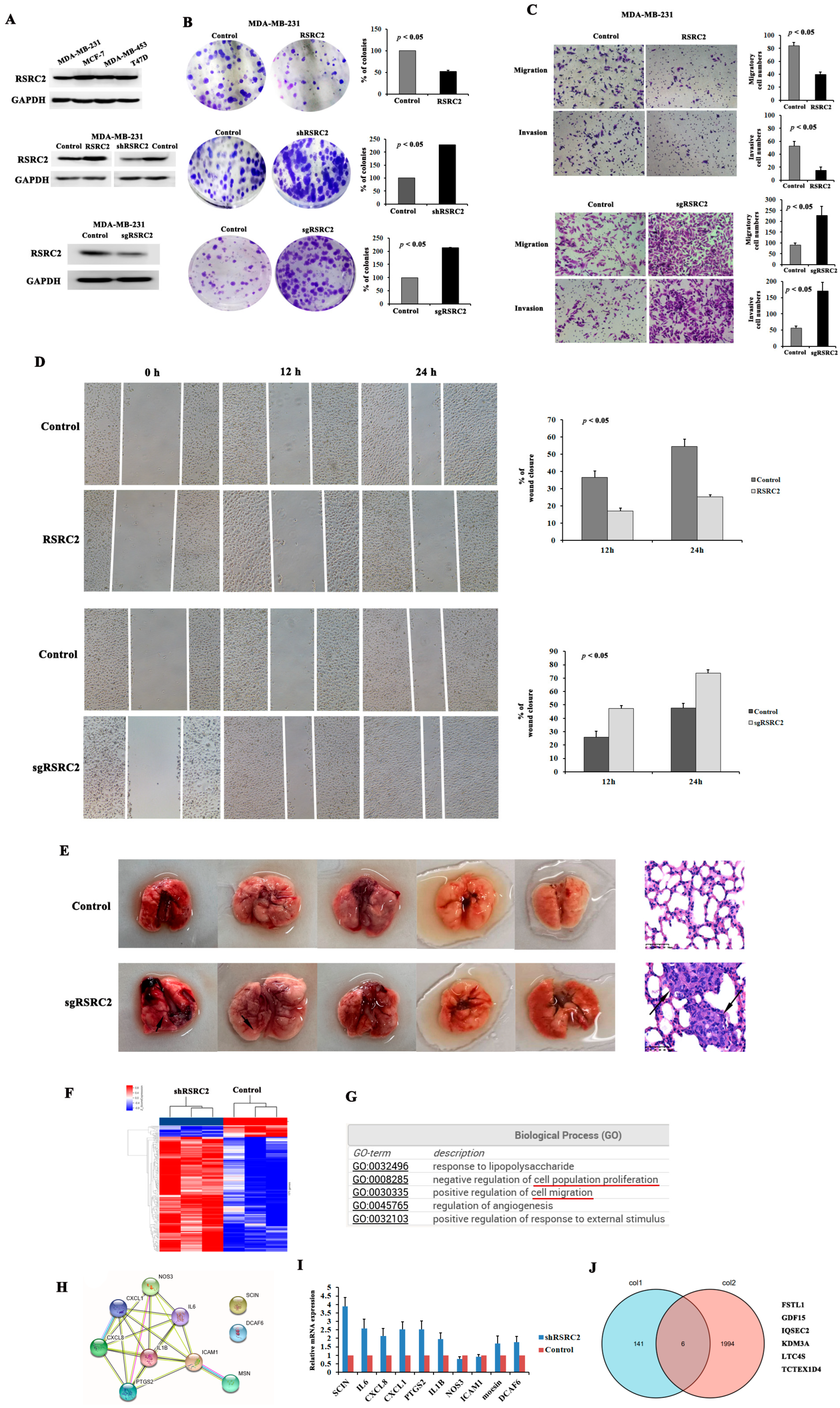
3.5. RSRC2-Downregulated Expression Promotes Migration, Invasion and Metastasis of MDA-MB-231 Cells
3.6. Global Regulation of the Transcriptome by RSRC2 in MDA-MB-231 Cells
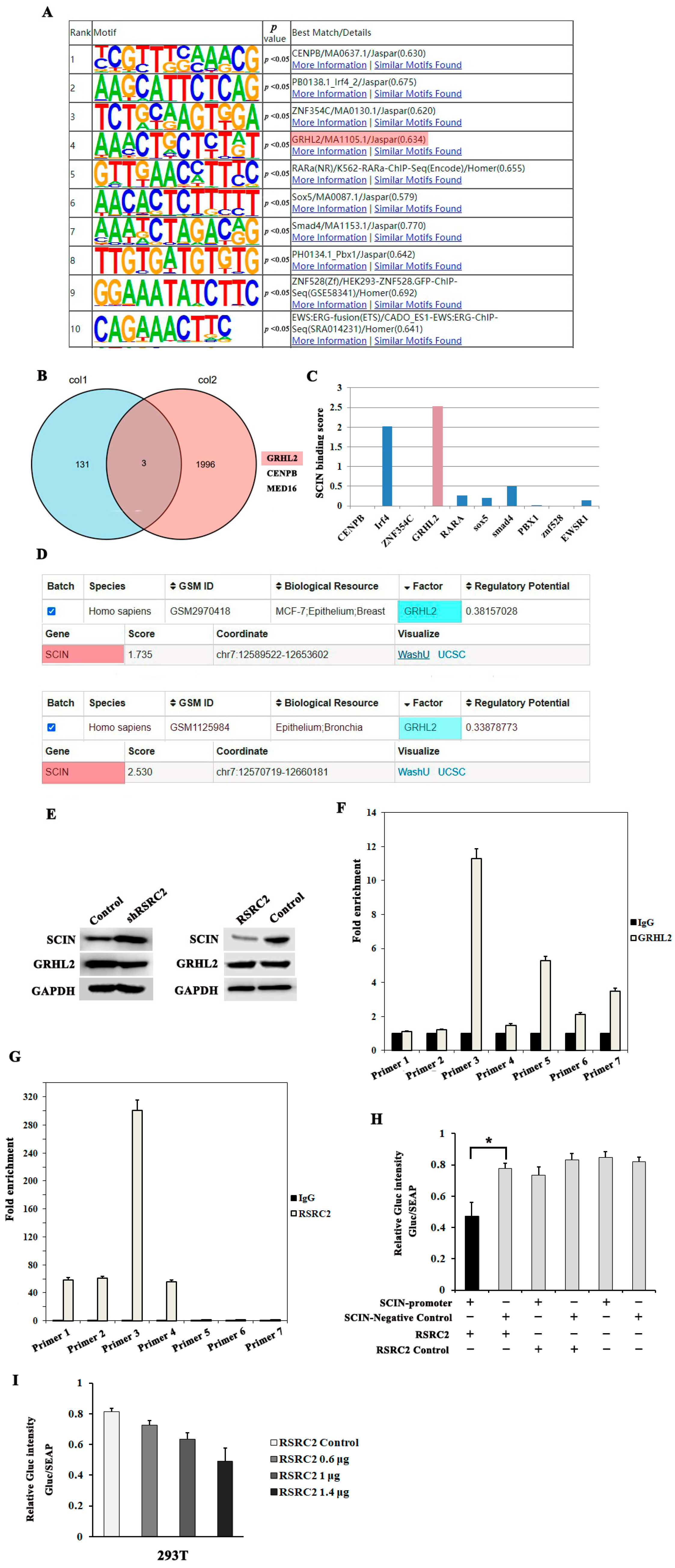
3.7. The Genomic Occupancy and Motif Combination by RSRC2
3.8. RSRC2 Might Directly Regulate SCIN Expression
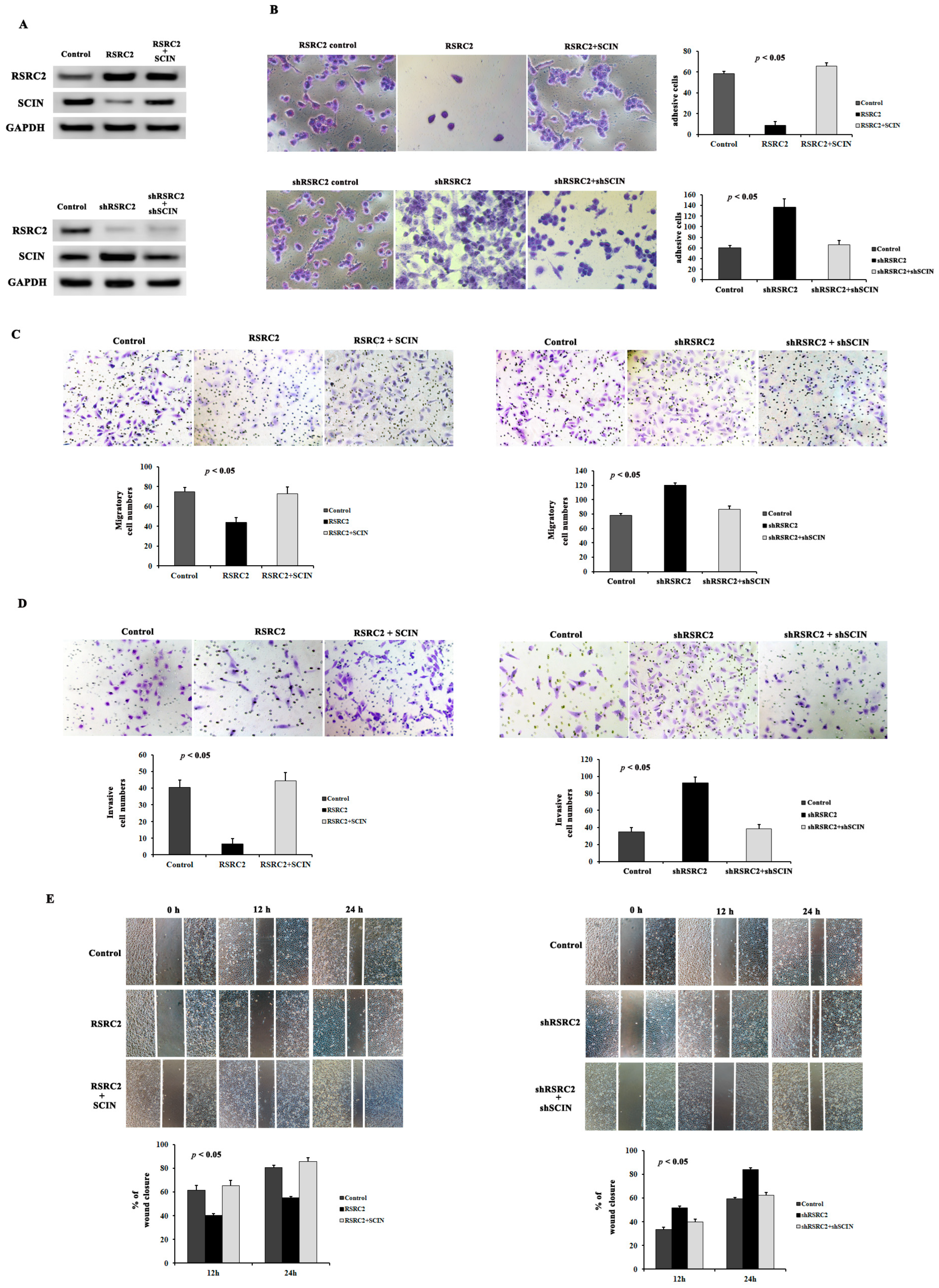
3.9. RSRC2 Inhibited Cell Adhesion, Clonality, Migration and Invasion Abilities by Suppressing SCIN Expression
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| RSRC2 | arginine/serine-rich coiled coil 2 |
| CHOL | cholangiocarcinoma |
| ChIP | chromatin immunoprecipitation |
| COAD | colon adenocarcinoma |
| ESCA | esophageal cancer |
| GBM | glioblastoma |
| HNSC | head and neck squamous cell carcinoma |
| Her2 | human epidermal growth factor receptor 2 |
| HTA | Human Transcriptome Array |
| KICH | kidney chromophobe cell carcinoma |
| KIRC | kidney renal clear-cell carcinoma |
| LIHC | liver hepatocellular carcinoma |
| LUAD | lung adenocarcinoma |
| LUSC | lung squamous cell carcinoma |
| SCIN | scinderin |
| STAD | stomach adenocarcinoma |
| THCA | thyroid carcinoma |
| TNBC | triple-negative breast cancer |
| UCEC | uterus endometrial carcinoma |
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef]
- Jiang, Y.-Z.; Ma, D.; Suo, C.; Shi, J.; Xue, M.; Hu, X.; Xiao, Y.; Yu, K.-D.; Liu, Y.-R.; Yu, Y.; et al. Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies. Cancer Cell 2019, 35, 428–440.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Q.; Yang, W.-K.; Wang, Y.-S.; Zhou, Q.; Lin, J.; Wei, X.-X.; Liang, T.; Liu, T.; Fan, W.-T.; et al. Development of an immune-related prognostic biomarker for triple-negative breast cancer. Ann. Med. 2022, 54, 1212–1220. [Google Scholar] [CrossRef]
- Kurehara, H.; Ishiguro, H.; Kimura, M.; Mitsui, A.; Ando, T.; Sugito, N.; Mori, R.; Takashima, N.; Ogawa, R.; Fujii, Y.; et al. A novel gene, RSRC2, inhibits cell proliferation and affects survival in esophageal cancer patients. Int. J. Oncol. 2007, 30, 421–428. [Google Scholar] [PubMed]
- Carrigan, P.E.; Bingham, J.L.; Srinvasan, S.; Brentnall, T.A.; Miller, L.J. Characterization of alternative spliceoforms and the RNA splicing machinery in pancreatic cancer. Pancreas 2011, 40, 281–288. [Google Scholar] [CrossRef]
- Park, J.S.; Young Yoon, S.; Kim, J.M.; Yeom, Y.I.; Kim, Y.S.; Kim, N.S. Identification of novel genes associated with the response to 5-FU treatment in gastric cancer cell lines using a cDNA microarray. Cancer Lett. 2004, 214, 19–33. [Google Scholar] [CrossRef]
- Dong, X.; Wang, J.; Tang, B.; Hao, Y.-X.; Li, P.-Y.; Li, S.-Y.; Yu, P.-W. The role and gene expression profile of SOCS3 in colorectal carcinoma. Oncotarget 2018, 9, 15984–15996. [Google Scholar] [CrossRef]
- Perilli, L.; Tessarollo, S.; Albertoni, L.; Curtarello, M.; Pastò, A.; Brunetti, E.; Fassan, M.; Rugge, M.; Indraccolo, S.; Amadori, A.; et al. Silencing of miR-182 is associated with modulation of tumorigenesis through apoptosis induction in an experimental model of colorectal cancer. BMC Cancer 2019, 19, 821. [Google Scholar] [CrossRef]
- Liu, T.; Sun, H.; Zhu, D.; Dong, X.; Liu, F.; Liang, X.; Chen, C.; Shao, B.; Wang, M.; Wang, Y.; et al. TRA2A Promoted Paclitaxel Resistance and Tumor Progression in Triple-Negative Breast Cancers via Regulating Alternative Splicing. Mol. Cancer Ther. 2017, 16, 1377–1388. [Google Scholar] [CrossRef]
- Huang, Y.; Du, X.; Chen, X.; Chen, C.; Wang, H.; Yang, Y.; Teng, L. MiR-301a-5p/SCIN promotes gastric cancer progression via regulating STAT3 and NF-kappaB signaling. J. Cancer 2021, 12, 5394–5403. [Google Scholar] [CrossRef]
- Qiao, X.; Zhou, Y.; Xie, W.; Wang, Y.; Zhang, Y.; Tian, T.; Dou, J.; Yang, X.; Shen, S.; Hu, J.; et al. Scinderin is a novel transcriptional target of BRMS1 involved in regulation of hepatocellular carcinoma cell apoptosis. Am. J. Cancer Res. 2018, 8, 1008–1018. [Google Scholar]
- Lai, X.; Su, W.; Zhao, H.; Yang, S.; Zeng, T.; Wu, W.; Wang, D. Loss of scinderin decreased expression of epidermal growth factor receptor and promoted apoptosis of castration-resistant prostate cancer cells. FEBS Open Bio 2018, 8, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.M.; Guo, J.M.; Chen, P.; Mao, L.G.; Feng, W.Y.; Le, D.H.; Li, K.Q. Suppression of scinderin modulates epithelialmesenchymal transition markers in highly metastatic gastric cancer cell line SGC7901. Mol. Med. Rep. 2014, 10, 2327–2333. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jian, W.; Zhang, X.; Wang, J.; Liu, Y.; Hu, C.; Wang, X.; Liu, R. Scinderin-knockdown inhibits proliferation and promotes apoptosis in human breast carcinoma cells. Oncol. Lett. 2018, 16, 3207–3214. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.X.; Liu, Y.L.; Kang, Y.F.; Lu, X.; Xu, K. MEX3A promotes nasopharyngeal carcinoma progression via the miR-3163/SCIN axis by regulating NF-kappaB signaling pathway. Cell Death Dis. 2022, 13, 420. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Li, J.; Zhu, D.; Niu, Z.; Pan, X.; Xu, P.; Ji, M.; Wei, Y.; Xu, J. Aberrant Scinderin Expression Correlates with Liver Metastasis and Poor Prognosis in Colorectal Cancer. Front. Pharmacol. 2019, 10, 1183. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-J.; Liu, J.-Y.; Chen, J.; Wu, Y.-X.; Yan, P.; Ji, C.-D.; Wang, Y.-X.; Xiang, D.-F.; Zhang, X.; Zhang, P.; et al. Scinderin promotes the invasion and metastasis of gastric cancer cells and predicts the outcome of patients. Cancer Lett. 2016, 376, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, B.; Zhao, X.; Zhao, N.; Sun, R.; Zhu, D.; Zhang, Y.; Li, Y.; Gu, Q.; Dong, X.; et al. Twist1-related miR-26b-5p suppresses epithelial-mesenchymal transition, migration and invasion by targeting SMAD1 in hepatocellular carcinoma. Oncotarget 2016, 7, 24383–24401. [Google Scholar] [CrossRef]
- Jiang, L.; You, C.; Xiao, Y.; Wang, H.; Su, G.-H.; Xia, B.-Q.; Zheng, R.-C.; Zhang, D.-D.; Jiang, Y.-Z.; Gu, Y.-J.; et al. Radiogenomic analysis reveals tumor heterogeneity of triple-negative breast cancer. Cell Rep. Med. 2022, 3, 100694. [Google Scholar] [CrossRef]
- Liu, T.; Sun, B.; Zhao, X.; Li, Y.; Zhao, X.; Liu, Y.; Yao, Z.; Gu, Q.; Dong, X.; Shao, B.; et al. USP44+ Cancer Stem Cell Subclones Contribute to Breast Cancer Aggressiveness by Promoting Vasculogenic Mimicry. Mol. Cancer Ther. 2015, 14, 2121–2131. [Google Scholar] [CrossRef]
- Youness, R.A.; Assal, R.A.; Abdel Motaal, A.; Gad, M.Z. A novel role of sONE/NOS3/NO signaling cascade in mediating hydrogen sulphide bilateral effects on triple negative breast cancer progression. Nitric Oxide Biol. Chem. 2018, 80, 12–23. [Google Scholar] [CrossRef]
- Kwon, K.-M.; Chung, T.-W.; Kwak, C.-H.; Choi, H.-J.; Kim, K.-W.; Ha, S.-H.; Cho, S.-H.; Lee, Y.-C.; Ha, K.-T.; Lee, M.-J.; et al. Disialyl GD2 ganglioside suppresses ICAM-1-mediated invasiveness in human breast cancer MDA-MB231 cells. Int. J. Biol. Sci. 2017, 13, 265–275. [Google Scholar] [CrossRef]
- Wu, Q.; Chen, D.; Luo, Q.; Yang, Q.; Zhao, C.; Zhang, D.; Zeng, Y.; Huang, L.; Zhang, Z.; Qi, Z. Extracellular matrix protein 1 recruits moesin to facilitate invadopodia formation and breast cancer metastasis. Cancer Lett. 2018, 437, 44–55. [Google Scholar] [CrossRef]
- Chen, B.; Song, L.; Nie, X.; Lin, F.; Yu, Z.; Kong, W.; Qi, X.; Wang, W. CXCL1 Regulated by miR-302e Is Involved in Cell Viability and Motility of Colorectal Cancer via Inhibiting JAK-STAT Signaling Pathway. Front. Oncol. 2020, 10, 577229. [Google Scholar] [CrossRef] [PubMed]
- Ren, Q.; Hou, Y.; Li, X.; Fan, X. Silence of TPPP3 suppresses cell proliferation, invasion and migration via inactivating NF-kappaB/COX2 signal pathway in breast cancer cell. Cell Biochem. Funct. 2020, 38, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Xie, M.; Meng, Z.; Lo, C.Y.; Chan, F.L.; Jiang, L.; Meng, X.; Yao, X. Endolysosomal ion channel MCOLN2 (Mucolipin-2) promotes prostate cancer progression via IL-1beta/NF-kappaB pathway. Br. J. Cancer 2021, 125, 1420–1431. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Bi, L.; Wang, Y.; Zhang, X.; Hou, Z.; Wang, Q.; Snijders, A.M.; Mao, J.-H. Integrative analysis of multi-omics data reveals distinct impacts of DDB1-CUL4 associated factors in human lung adenocarcinomas. Sci. Rep. 2017, 7, 333. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Ding, Y.; Wu, N.; Jiang, J.; Huang, Y.; Zhang, F.; Wang, H.; Zhou, Q.; Yang, Y.; Zhuo, W.; et al. FSTL1 promotes growth and metastasis in gastric cancer by activating AKT related pathway and predicts poor survival. Am. J. Cancer Res. 2021, 11, 712–728. [Google Scholar] [PubMed]
- Lv, C.; Li, S.; Zhao, J.; Yang, P.; Yang, C. M1 Macrophages Enhance Survival and Invasion of Oral Squamous Cell Carcinoma by Inducing GDF15-Mediated ErbB2 Phosphorylation. ACS Omega 2022, 7, 11405–11414. [Google Scholar] [CrossRef]
- Meng, C.; Xia, S.; He, Y.; Tang, X.; Zhang, G.; Zhou, T. Discovery of Prognostic Signature Genes for Overall Survival Prediction in Gastric Cancer. Comput. Math. Methods Med. 2020, 2020, 5479279. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, R.; Zhang, L.; Wang, C.; Dong, Z.; Feng, J.; Luo, M.; Zhang, Y.; Xu, Z.; Lv, S.; et al. Downregulation of miR-335 exhibited an oncogenic effect via promoting KDM3A/YAP1 networks in clear cell renal cell carcinoma. Cancer Gene Ther. 2022, 29, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.-L.; Luo, T.-Q.; Li, J.-N.; Xue, Z.-C.; Wang, Y.; Zhang, Y.; Chen, Y.-B.; Peng, C. Development and Validation of a Prognostic Classifier Based on Lipid Metabolism-Related Genes in Gastric Cancer. Front. Mol. Biosci. 2021, 8, 691143. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, M.; Shi, M.; Zhang, T.; Zhang, Z.; Cui, Q.; Yang, S.; Li, Z. A Survival-Related Competitive Endogenous RNA Network of Prognostic lncRNAs, miRNAs, and mRNAs in Wilms Tumor. Front. Oncol. 2021, 11, 608433. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Li, Y.; Li, Y.; Li, F.; Che, N.; Ni, C.; Zhao, N.; Zhao, X.; Liu, T. GRHL2 Expression Functions in Breast Cancer Aggressiveness and Could Serve as Prognostic and Diagnostic Biomarker for Breast Cancer. Clin. Med. Insights Oncol. 2022, 16, 11795549221109511. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, N.; Ni, C.; Fan, S.; Che, N.; Li, Y.; Wang, S.; Li, Y.; Dong, X.; Guo, Y.; Zhao, X.; et al. RSRC2 Expression Inhibits Malignant Progression of Triple-Negative Breast Cancer by Transcriptionally Regulating SCIN Expression. Cancers 2024, 16, 15. https://doi.org/10.3390/cancers16010015
Zhao N, Ni C, Fan S, Che N, Li Y, Wang S, Li Y, Dong X, Guo Y, Zhao X, et al. RSRC2 Expression Inhibits Malignant Progression of Triple-Negative Breast Cancer by Transcriptionally Regulating SCIN Expression. Cancers. 2024; 16(1):15. https://doi.org/10.3390/cancers16010015
Chicago/Turabian StyleZhao, Nan, Chunsheng Ni, Shuai Fan, Na Che, Yanlei Li, Song Wang, Yongli Li, Xueyi Dong, Yuhong Guo, Xiulan Zhao, and et al. 2024. "RSRC2 Expression Inhibits Malignant Progression of Triple-Negative Breast Cancer by Transcriptionally Regulating SCIN Expression" Cancers 16, no. 1: 15. https://doi.org/10.3390/cancers16010015
APA StyleZhao, N., Ni, C., Fan, S., Che, N., Li, Y., Wang, S., Li, Y., Dong, X., Guo, Y., Zhao, X., & Liu, T. (2024). RSRC2 Expression Inhibits Malignant Progression of Triple-Negative Breast Cancer by Transcriptionally Regulating SCIN Expression. Cancers, 16(1), 15. https://doi.org/10.3390/cancers16010015




