Thymic Carcinoma: Unraveling Neuroendocrine Differentiation and Epithelial Cell Identity Loss
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Analysis of a Publicly Available Single-Cell RNA Sequencing Dataset of the Normal Human Thymus
2.2. Analysis of Publicly Available Bulk mRNA Expression and Methylation Datasets of Thymic Epithelial Tumors and Lung Cancers
2.3. Analysis of Our Cohort and Real-Time Quantitative Polymerase Chain Reaction
2.4. Statistical Analysis
3. Results
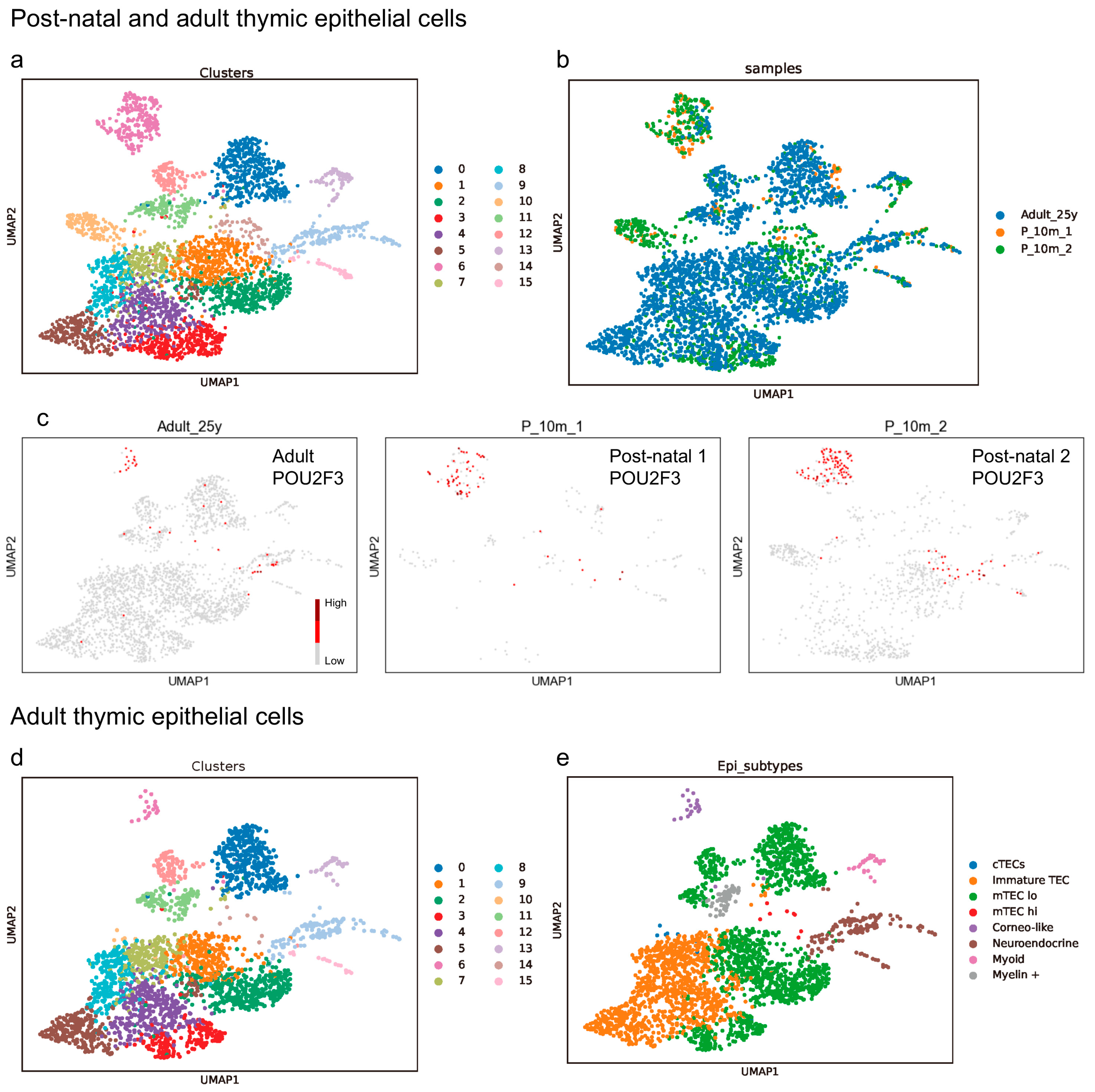
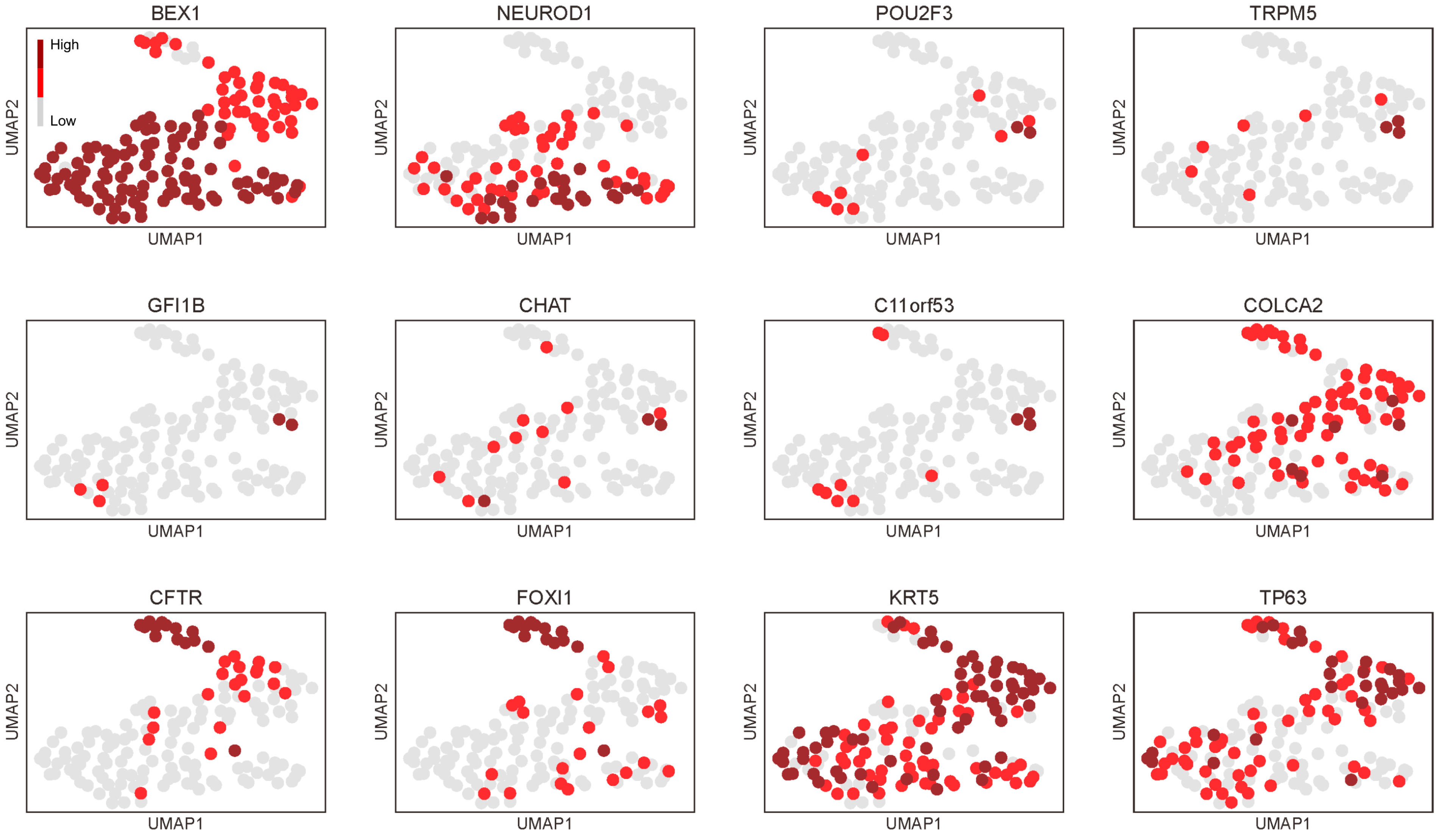
3.1. The Presence of Tuft Cells and Ionocytes within the Neuroendocrine Group in the Adult Thymus
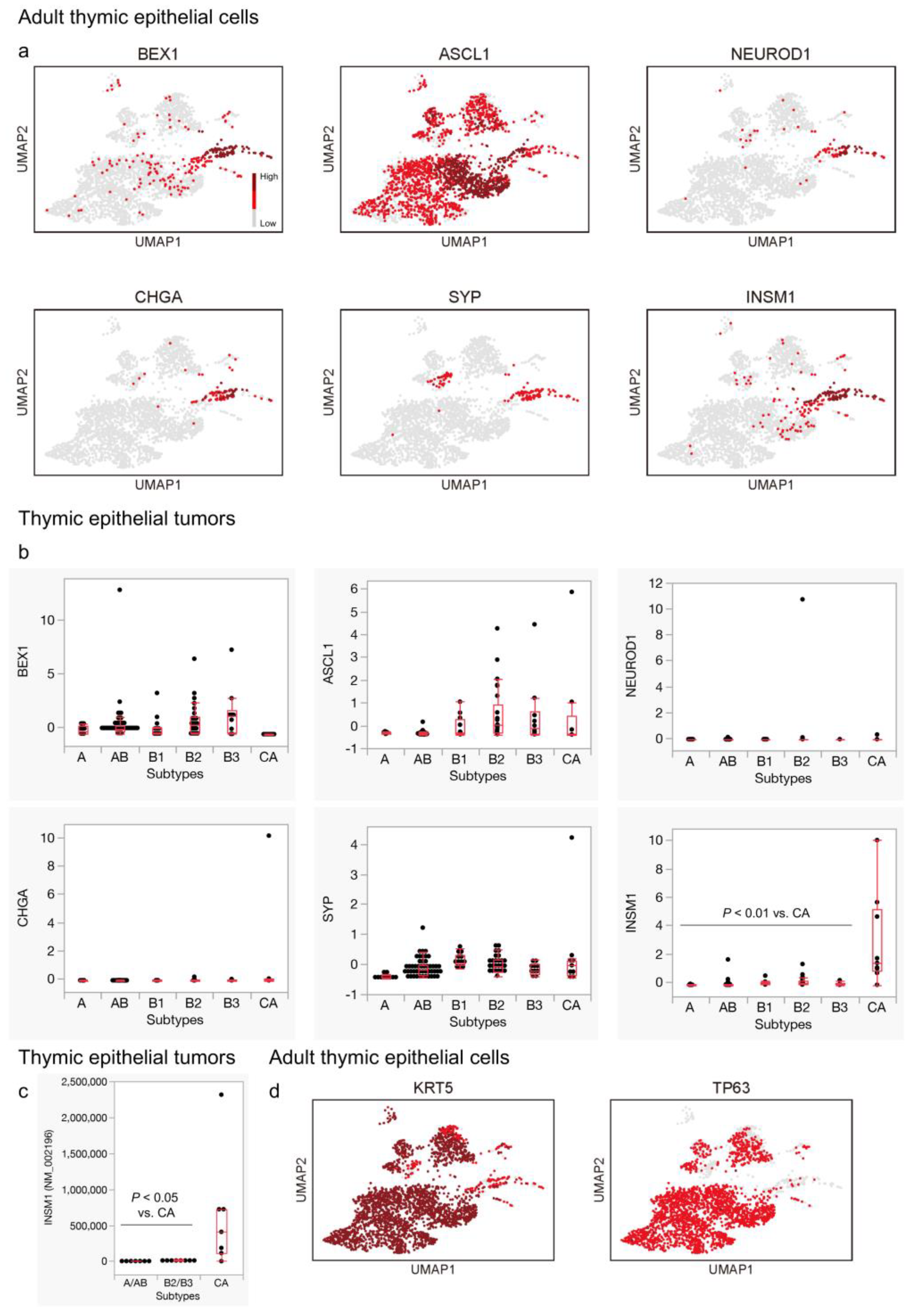
3.2. Expression of General Neuroendocrine Markers, except for INSM1, Is Not Remarkable in Thymic Carcinomas
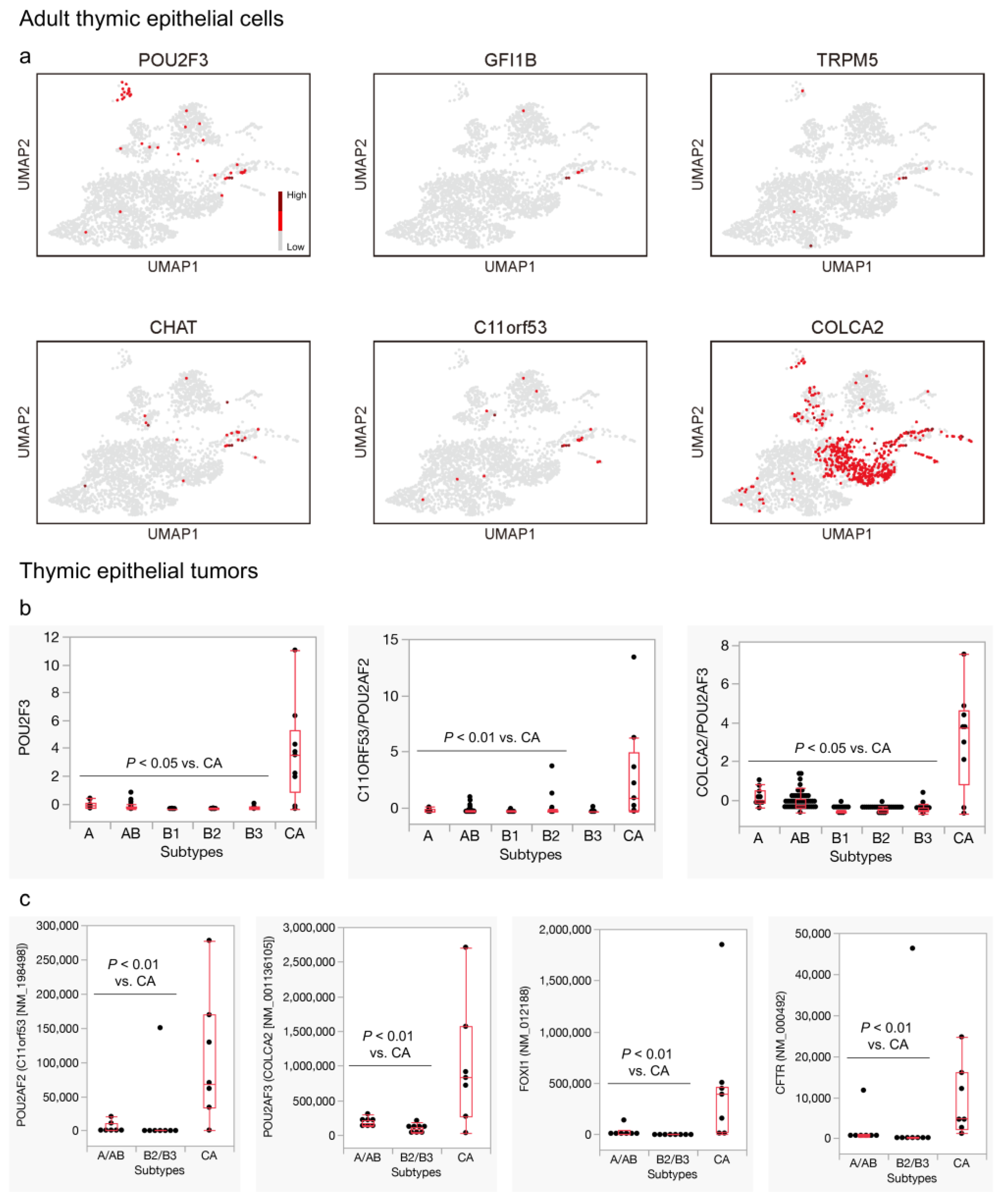
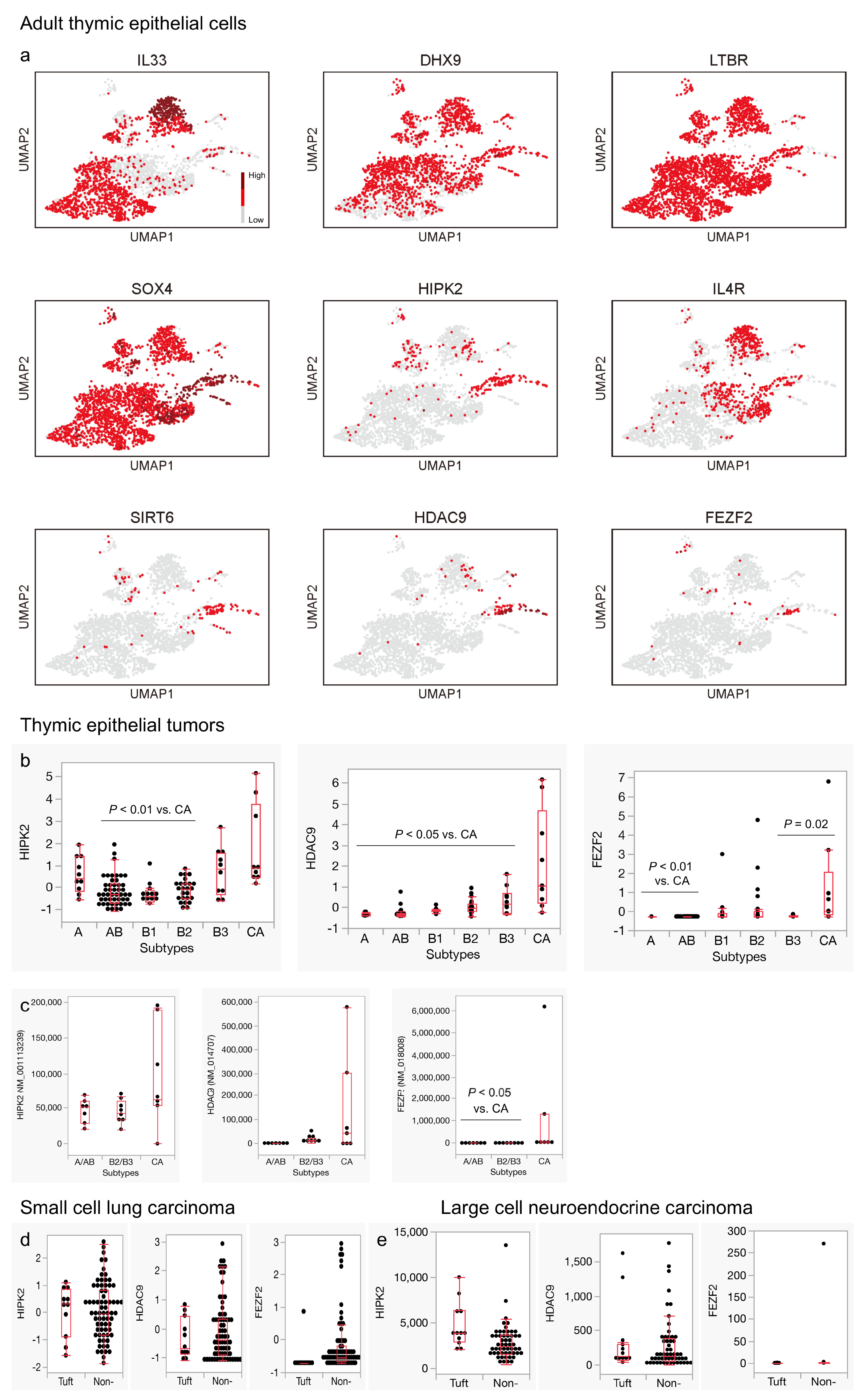
3.3. Genes Related to Thymic Tuft Cell Development Are Biasedly Expressed in the Neuroendocrine Group in the Adult Thymus and Significantly Expressed in Thymic Carcinomas
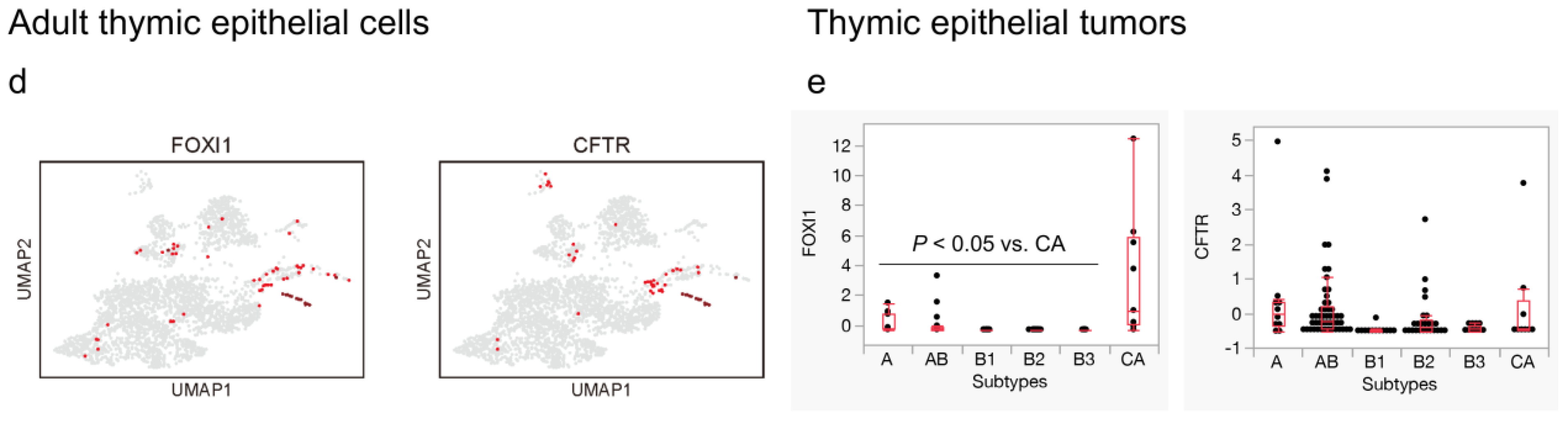
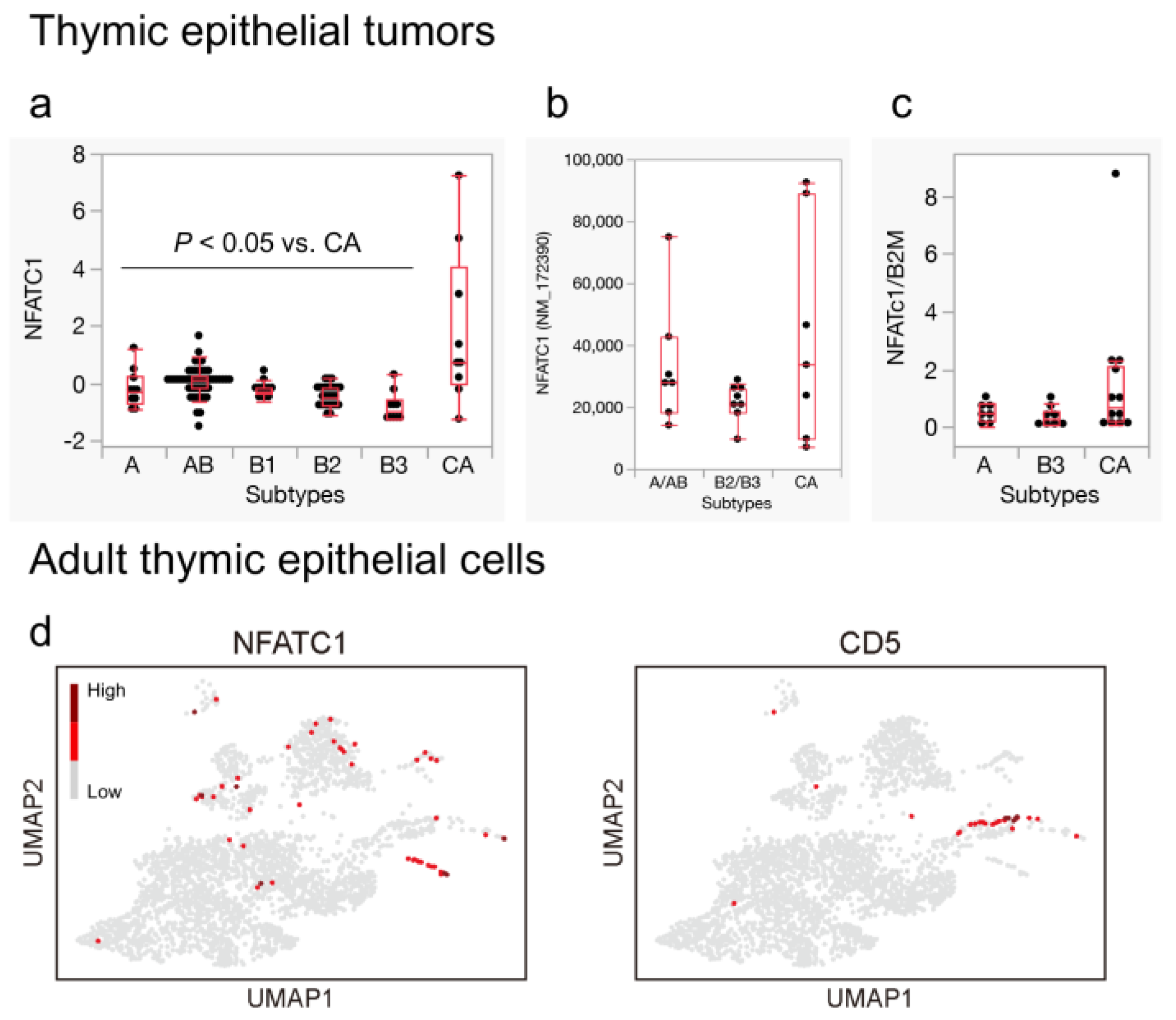
3.4. NFATC1 Expression in Thymic Carcinoma and Ionocytes in the Adult Thymus
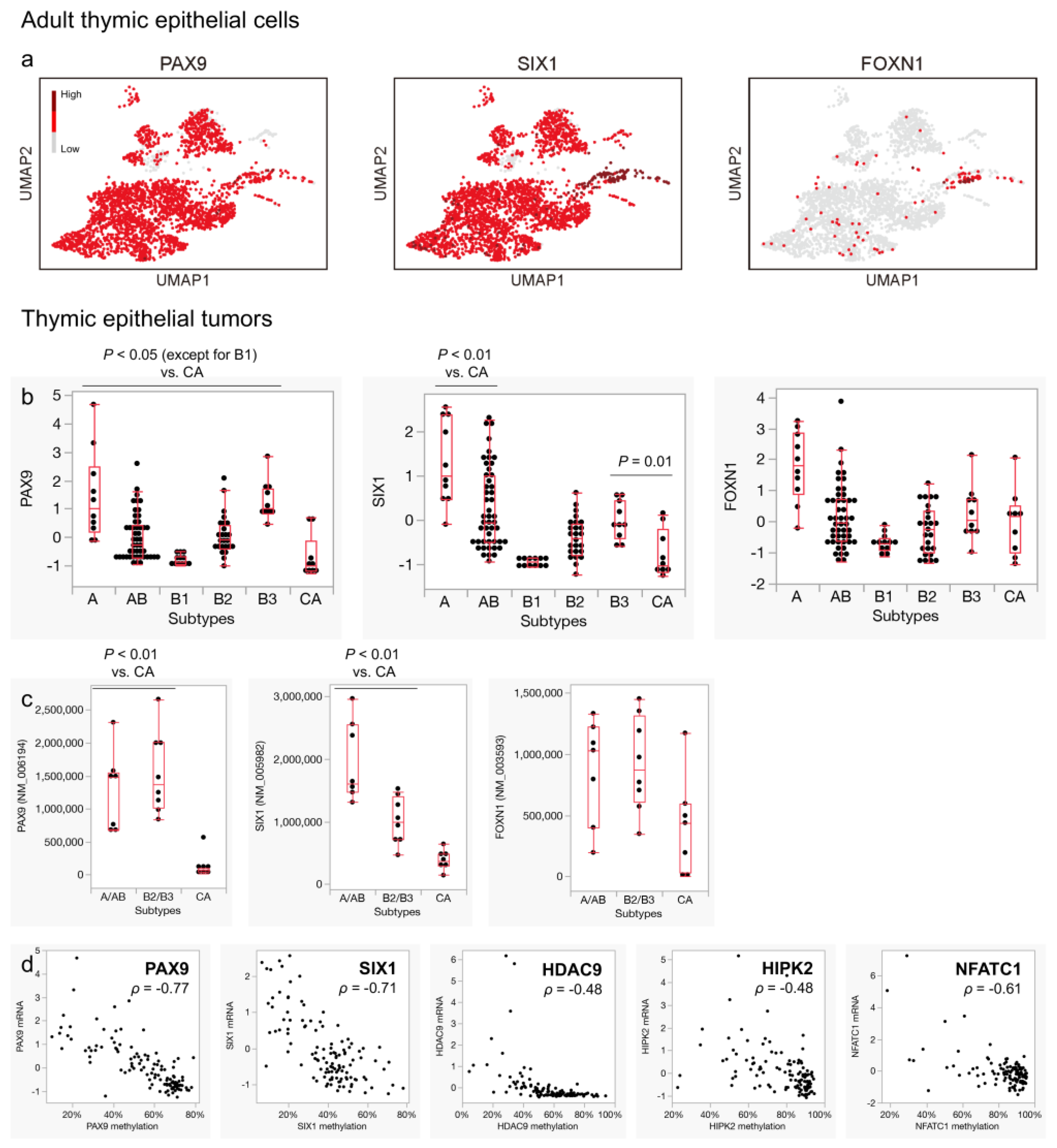
3.5. Expression of Pan-Thymic Epithelial Markers Is Suppressed in Thymic Carcinoma
3.6. Characteristic DNA Methylation Patterns May Be Linked to the Expression Profiles of Thymic Carcinoma
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marino, M.; Müller-Hermelink, H.K. Thymoma and thymic carcinoma. Relation of thymoma epithelial cells to the cortical and medullary differentiation of thymus. Virchows Arch. A Pathol. Anat. Histopathol. 1985, 407, 119–149. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Tomaru, U.; Ishizu, A.; Kiuchi, T.; Marukawa, K.; Matsuno, Y.; Kasahara, M. Expression of proteasome subunit β5t in thymic epithelial tumors. Am. J. Surg. Pathol. 2011, 35, 1296–1304. [Google Scholar] [CrossRef] [PubMed]
- Tomaru, U.; Yamada, Y.; Ishizu, A.; Kuroda, T.; Matsuno, Y.; Kasahara, M. Proteasome subunit β5t expression in cervical ectopic thymoma. J. Clin. Pathol. 2012, 65, 858–859. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Tomaru, U.; Ishizu, A.; Kiuchi, T.; Kasahara, M.; Matsuno, Y. Expression of thymoproteasome subunit β5t in type AB thymoma. J. Clin. Pathol. 2014, 67, 276–278. [Google Scholar] [CrossRef] [PubMed]
- Ströbel, P.; Hartmann, E.; Rosenwald, A.; Kalla, J.; Ott, G.; Friedel, G.; Schalke, B.; Kasahara, M.; Tomaru, U.; Marx, A. Corticomedullary differentiation and maturational arrest in thymomas. Histopathology 2014, 64, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, D.; Henley, J.D.; Chiriboga, L.; Yee, H. Diagnostic utility of thymic epithelial markers CD205 (DEC205) and Foxn1 in thymic epithelial neoplasms. Am. J. Surg. Pathol. 2007, 31, 1038–1044. [Google Scholar] [CrossRef]
- Matsumoto, M.; Ohmura, T.; Hanibuchi, Y.; Ichimura-Shimizu, M.; Saijo, Y.; Ogawa, H.; Miyazawa, R.; Morimoto, J.; Tsuneyama, K.; Oya, T. AIRE illuminates the feature of medullary thymic epithelial cells in thymic carcinoma. Cancer Med. 2023, 12, 9843–9848. [Google Scholar] [CrossRef]
- Li, H.; Ren, B.; Yu, S.; Gao, H.; Sun, P.L. The clinicopathological significance of thymic epithelial markers expression in thymoma and thymic carcinoma. BMC Cancer 2023, 23, 161. [Google Scholar] [CrossRef]
- Kashima, J.; Hashimoto, T.; Yoshida, A.; Goto, Y.; Ushiku, T.; Ohe, Y.; Watanabe, S.I.; Yatabe, Y. Insulinoma-associated-1 (INSM1) expression in thymic squamous cell carcinoma. Virchows Arch. 2022, 481, 893–901. [Google Scholar] [CrossRef]
- WHO Classification of Tumours Editorial Board. Thoracic Tumours, 5th ed.; International Agency for Research on Cancer: Lyon, France, 2021.
- Miller, C.N.; Proekt, I.; von Moltke, J.; Wells, K.L.; Rajpurkar, A.R.; Wang, H.; Rattay, K.; Khan, I.S.; Metzger, T.C.; Pollack, J.L.; et al. Thymic tuft cells promote an IL-4-enriched medulla and shape thymocyte development. Nature 2018, 559, 627–631. [Google Scholar] [CrossRef]
- Bornstein, C.; Nevo, S.; Giladi, A.; Kadouri, N.; Pouzolles, M.; Gerbe, F.; David, E.; Machado, A.; Chuprin, A.; Tóth, B.; et al. Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature 2018, 559, 622–626. [Google Scholar] [CrossRef] [PubMed]
- Gerbe, F.; Sidot, E.; Smyth, D.J.; Ohmoto, M.; Matsumoto, I.; Dardalhon, V.; Cesses, P.; Garnier, L.; Pouzolles, M.; Brulin, B.; et al. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 2016, 529, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Howitt, M.R.; Lavoie, S.; Michaud, M.; Blum, A.M.; Tran, S.V.; Weinstock, J.V.; Gallini, C.A.; Redding, K.; Margolskee, R.F.; Osborne, L.C.; et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 2016, 351, 1329–1333. [Google Scholar] [CrossRef] [PubMed]
- von Moltke, J.; Ji, M.; Liang, H.E.; Locksley, R.M. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 2016, 529, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Radovich, M.; Pickering, C.R.; Felau, I.; Ha, G.; Zhang, H.; Jo, H.; Hoadley, K.A.; Anur, P.; Zhang, J.; McLellan, M.; et al. The Integrated Genomic Landscape of Thymic Epithelial Tumors. Cancer Cell 2018, 33, 244–258.e210. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, l1. [Google Scholar] [CrossRef]
- Yamada, Y.; Simon-Keller, K.; Belharazem-Vitacolonnna, D.; Bohnenberger, H.; Kriegsmann, M.; Kriegsmann, K.; Hamilton, G.; Graeter, T.; Preissler, G.; Ott, G.; et al. A Tuft Cell-Like Signature Is Highly Prevalent in Thymic Squamous Cell Carcinoma and Delineates New Molecular Subsets Among the Major Lung Cancer Histotypes. J. Thorac. Oncol. 2021, 16, 1003–1016. [Google Scholar] [CrossRef]
- Yamada, Y.; Bohnenberger, H.; Kriegsmann, M.; Kriegsmann, K.; Sinn, H.-P.; Goto, N.; Nakanishi, Y.; Seno, H.; Chigusa, Y.; Fujimoto, M.; et al. Tuft cell-like carcinomas: Novel cancer subsets present in multiple organs sharing a unique gene expression signature. Br. J. Cancer 2022, 127, 1876–1885. [Google Scholar] [CrossRef]
- Yamada, Y.; Sugimoto, A.; Hoki, M.; Yoshizawa, A.; Hamaji, M.; Date, H.; Haga, H.; Marx, A. POU2F3 beyond thymic carcinomas: Expression across the spectrum of thymomas hints to medullary differentiation in type A thymoma. Virchows Arch. 2022, 480, 843–851. [Google Scholar] [CrossRef]
- Bautista, J.L.; Cramer, N.T.; Miller, C.N.; Chavez, J.; Berrios, D.I.; Byrnes, L.E.; Germino, J.; Ntranos, V.; Sneddon, J.B.; Burt, T.D.; et al. Single-cell transcriptional profiling of human thymic stroma uncovers novel cellular heterogeneity in the thymic medulla. Nat. Commun. 2021, 12, 1096. [Google Scholar] [CrossRef] [PubMed]
- Plasschaert, L.W.; Žilionis, R.; Choo-Wing, R.; Savova, V.; Knehr, J.; Roma, G.; Klein, A.M.; Jaffe, A.B. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 2018, 560, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Montoro, D.T.; Haber, A.L.; Biton, M.; Vinarsky, V.; Lin, B.; Birket, S.E.; Yuan, F.; Chen, S.; Leung, H.M.; Villoria, J.; et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 2018, 560, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, K.; Matsuno, Y.; Kunitoh, H.; Maeshima, A.; Asamura, H.; Tsuchiya, R. Immunohistochemical KIT (CD117) expression in thymic epithelial tumors. Chest 2005, 128, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.C.; Chen, P.C.; Chiang, H. KIT (CD117) is frequently overexpressed in thymic carcinomas but is absent in thymomas. J. Pathol. 2004, 202, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Hishima, T.; Fukayama, M.; Fujisawa, M.; Hayashi, Y.; Arai, K.; Funata, N.; Koike, M. CD5 expression in thymic carcinoma. Am. J. Pathol. 1994, 145, 268–275. [Google Scholar] [PubMed]
- Kaira, K.; Murakami, H.; Serizawa, M.; Koh, Y.; Abe, M.; Ohde, Y.; Takahashi, T.; Kondo, H.; Nakajima, T.; Yamamoto, N. MUC1 expression in thymic epithelial tumors: MUC1 may be useful marker as differential diagnosis between type B3 thymoma and thymic carcinoma. Virchows Arch. 2011, 458, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Klingbeil, O.; He, X.Y.; Wu, X.S.; Arun, G.; Lu, B.; Somerville, T.D.D.; Milazzo, J.P.; Wilkinson, J.E.; Demerdash, O.E.; et al. POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Genes. Dev. 2018, 32, 915–928. [Google Scholar] [CrossRef]
- Hishima, T.; Fukayama, M.; Hayashi, Y.; Fujii, T.; Arai, K.; Shiozawa, Y.; Funata, N.; Koike, M. Neuroendocrine differentiation in thymic epithelial tumors with special reference to thymic carcinoma and atypical thymoma. Hum. Pathol. 1998, 29, 330–338. [Google Scholar] [CrossRef]
- Polański, K.; Young, M.D.; Miao, Z.; Meyer, K.B.; Teichmann, S.A.; Park, J.E. BBKNN: Fast batch alignment of single cell transcriptomes. Bioinformatics 2020, 36, 964–965. [Google Scholar] [CrossRef]
- Imoto, Y.; Nakamura, T.; Escolar, E.G.; Yoshiwaki, M.; Kojima, Y.; Yabuta, Y.; Katou, Y.; Yamamoto, T.; Hiraoka, Y.; Saitou, M. Resolution of the curse of dimensionality in single-cell RNA sequencing data analysis. Life Sci. Alliance 2022, 5, e202201591. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, J.; Ohmoto, M.; Yamaguchi, T.; Matsumoto, I.; Hirota, J. Skn-1a/Pou2f3 functions as a master regulator to generate Trpm5-expressing chemosensory cells in mice. PLoS ONE 2017, 12, e0189340. [Google Scholar] [CrossRef] [PubMed]
- Petrini, I.; Meltzer, P.S.; Kim, I.K.; Lucchi, M.; Park, K.S.; Fontanini, G.; Gao, J.; Zucali, P.A.; Calabrese, F.; Favaretto, A.; et al. A specific missense mutation in GTF2I occurs at high frequency in thymic epithelial tumors. Nat. Genet. 2014, 46, 844–849. [Google Scholar] [CrossRef] [PubMed]
- George, J.; Lim, J.S.; Jang, S.J.; Cun, Y.; Ozretić, L.; Kong, G.; Leenders, F.; Lu, X.; Fernández-Cuesta, L.; Bosco, G.; et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015, 524, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Belharazem-Vitacolonnna, D.; Bohnenberger, H.; Weiß, C.; Matsui, N.; Kriegsmann, M.; Kriegsmann, K.; Sinn, P.; Simon-Keller, K.; Hamilton, G.; et al. Pulmonary cancers across different histotypes share hybrid tuft cell/ionocyte-like molecular features and potentially druggable vulnerabilities. Cell Death Dis. 2022, 13, 979. [Google Scholar] [CrossRef] [PubMed]
- George, J.; Walter, V.; Peifer, M.; Alexandrov, L.B.; Seidel, D.; Leenders, F.; Maas, L.; Müller, C.; Dahmen, I.; Delhomme, T.M.; et al. Integrative genomic profiling of large-cell neuroendocrine carcinomas reveals distinct subtypes of high-grade neuroendocrine lung tumors. Nat. Commun. 2018, 9, 1048. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.S.; He, X.Y.; Ipsaro, J.J.; Huang, Y.H.; Preall, J.B.; Ng, D.; Shue, Y.T.; Sage, J.; Egeblad, M.; Joshua-Tor, L.; et al. OCA-T1 and OCA-T2 are coactivators of POU2F3 in the tuft cell lineage. Nature 2022, 607, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Huang, H.; Wang, Y.; Sendinc, E.; Shi, Y. Selective regulation of tuft cell-like small cell lung cancer by novel transcriptional co-activators C11orf53 and COLCA2. Cell Discov. 2022, 8, 112. [Google Scholar] [CrossRef]
- Szczepanski, A.P.; Tsuboyama, N.; Watanabe, J.; Hashizume, R.; Zhao, Z.; Wang, L. POU2AF2/C11orf53 functions as a coactivator of POU2F3 by maintaining chromatin accessibility and enhancer activity. Sci. Adv. 2022, 8, eabq2403. [Google Scholar] [CrossRef]
- Xu, L.; Wei, C.; Chen, Y.; Wu, Y.; Shou, X.; Chen, W.; Lu, D.; Sun, H.; Li, W.; Yu, B.; et al. IL-33 induces thymic involution-associated naive T cell aging and impairs host control of severe infection. Nat. Commun. 2022, 13, 6881. [Google Scholar] [CrossRef]
- Dong, X.; Zhang, J.; Zhang, Q.; Liang, Z.; Xu, Y.; Zhao, Y.; Zhang, B. Cytosolic Nuclear Sensor Dhx9 Controls Medullary Thymic Epithelial Cell Differentiation by p53-Mediated Pathways. Front. Immunol. 2022, 13, 896472. [Google Scholar] [CrossRef] [PubMed]
- Mino, N.; Muro, R.; Ota, A.; Nitta, S.; Lefebvre, V.; Nitta, T.; Fujio, K.; Takayanagi, H. The transcription factor Sox4 is required for thymic tuft cell development. Int. Immunol. 2022, 34, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, J.; Lei, T.; Liang, Z.; Dong, X.; Sun, L.; Zhao, Y. Sirt6-mediated epigenetic modification of DNA accessibility is essential for Pou2f3-induced thymic tuft cell development. Commun. Biol. 2022, 5, 544. [Google Scholar] [CrossRef]
- Naso, J.R.; Vrana, J.A.; Koepplin, J.W.; Molina, J.R.; Roden, A.C. EZH2 and POU2F3 Can Aid in the Distinction of Thymic Carcinoma from Thymoma. Cancers 2023, 15, 2274. [Google Scholar] [CrossRef] [PubMed]
- Arman, M.; Calvo, J.; Trojanowska, M.E.; Cockerill, P.N.; Santana, M.; López-Cabrera, M.; Vives, J.; Lozano, F. Transcriptional regulation of human CD5: Important role of Ets transcription factors in CD5 expression in T cells. J. Immunol. 2004, 172, 7519–7529. [Google Scholar] [CrossRef]
- Berland, R.; Wortis, H.H. Normal B-1a cell development requires B cell-intrinsic NFATc1 activity. Proc. Natl. Acad. Sci. USA 2003, 100, 13459–13464. [Google Scholar] [CrossRef]
- Garaud, S.; Morva, A.; Lemoine, S.; Hillion, S.; Bordron, A.; Pers, J.O.; Berthou, C.; Mageed, R.A.; Renaudineau, Y.; Youinou, P. CD5 promotes IL-10 production in chronic lymphocytic leukemia B cells through STAT3 and NFAT2 activation. J. Immunol. 2011, 186, 4835–4844. [Google Scholar] [CrossRef]
- Zhang, C.; Xin, H.; Zhang, W.; Yazaki, P.J.; Zhang, Z.; Le, K.; Li, W.; Lee, H.; Kwak, L.; Forman, S.; et al. CD5 Binds to Interleukin-6 and Induces a Feed-Forward Loop with the Transcription Factor STAT3 in B Cells to Promote Cancer. Immunity 2016, 44, 913–923. [Google Scholar] [CrossRef]
- Laclef, C.; Souil, E.; Demignon, J.; Maire, P. Thymus, kidney and craniofacial abnormalities in Six 1 deficient mice. Mech. Dev. 2003, 120, 669–679. [Google Scholar] [CrossRef]
- Romano, R.; Palamaro, L.; Fusco, A.; Giardino, G.; Gallo, V.; Del Vecchio, L.; Pignata, C. FOXN1: A Master Regulator Gene of Thymic Epithelial Development Program. Front. Immunol. 2013, 4, 187. [Google Scholar] [CrossRef]
- Peters, H.; Neubüser, A.; Kratochwil, K.; Balling, R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes. Dev. 1998, 12, 2735–2747. [Google Scholar] [CrossRef] [PubMed]
- Bleul, C.C.; Corbeaux, T.; Reuter, A.; Fisch, P.; Mönting, J.S.; Boehm, T. Formation of a functional thymus initiated by a postnatal epithelial progenitor cell. Nature 2006, 441, 992–996. [Google Scholar] [CrossRef] [PubMed]
- Bolden, J.E.; Peart, M.J.; Johnstone, R.W. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006, 5, 769–784. [Google Scholar] [CrossRef] [PubMed]
- Eckschlager, T.; Plch, J.; Stiborova, M.; Hrabeta, J. Histone Deacetylase Inhibitors as Anticancer Drugs. Int. J. Mol. Sci. 2017, 18, 1414. [Google Scholar] [CrossRef] [PubMed]
- Shou, J.; Jing, J.; Xie, J.; You, L.; Jing, Z.; Yao, J.; Han, W.; Pan, H. Nuclear factor of activated T cells in cancer development and treatment. Cancer Lett. 2015, 361, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Mancini, M.; Toker, A. NFAT proteins: Emerging roles in cancer progression. Nat. Rev. Cancer 2009, 9, 810–820. [Google Scholar] [CrossRef]
- Legrand, M.; Tallet, A.; Guyétant, S.; Samimi, M.; Ortonne, N.; Kervarrec, T. CD5 expression in Merkel cell carcinoma and extracutaneous neuroendocrine carcinomas. Pathology 2023, 55, 141–143. [Google Scholar] [CrossRef]
- Letourneau, A.; Santoni, F.A.; Bonilla, X.; Sailani, M.R.; Gonzalez, D.; Kind, J.; Chevalier, C.; Thurman, R.; Sandstrom, R.S.; Hibaoui, Y.; et al. Domains of genome-wide gene expression dysregulation in Down’s syndrome. Nature 2014, 508, 345–350. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamada, Y.; Iwane, K.; Nakanishi, Y.; Haga, H. Thymic Carcinoma: Unraveling Neuroendocrine Differentiation and Epithelial Cell Identity Loss. Cancers 2024, 16, 115. https://doi.org/10.3390/cancers16010115
Yamada Y, Iwane K, Nakanishi Y, Haga H. Thymic Carcinoma: Unraveling Neuroendocrine Differentiation and Epithelial Cell Identity Loss. Cancers. 2024; 16(1):115. https://doi.org/10.3390/cancers16010115
Chicago/Turabian StyleYamada, Yosuke, Kosuke Iwane, Yuki Nakanishi, and Hironori Haga. 2024. "Thymic Carcinoma: Unraveling Neuroendocrine Differentiation and Epithelial Cell Identity Loss" Cancers 16, no. 1: 115. https://doi.org/10.3390/cancers16010115
APA StyleYamada, Y., Iwane, K., Nakanishi, Y., & Haga, H. (2024). Thymic Carcinoma: Unraveling Neuroendocrine Differentiation and Epithelial Cell Identity Loss. Cancers, 16(1), 115. https://doi.org/10.3390/cancers16010115




