Exploring the Control of PARP1 Levels in High-Grade Serous Ovarian Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Established and Primary HGSOC Cells
2.2. Reagents and Treatments
2.3. Cytotoxicity Assay
2.4. Chromatin Immunoprecipitation (ChIP)-qPCR Assay
2.5. Luciferase Assay
2.6. Isolation of RNA
2.7. Real-time Quantitative PCR
2.8. Western Blot Analysis
2.9. Immunofluorescence Analysis
2.10. Kaplan–Meier Plotter Database Analysis
2.11. Statistical Analysis
3. Results
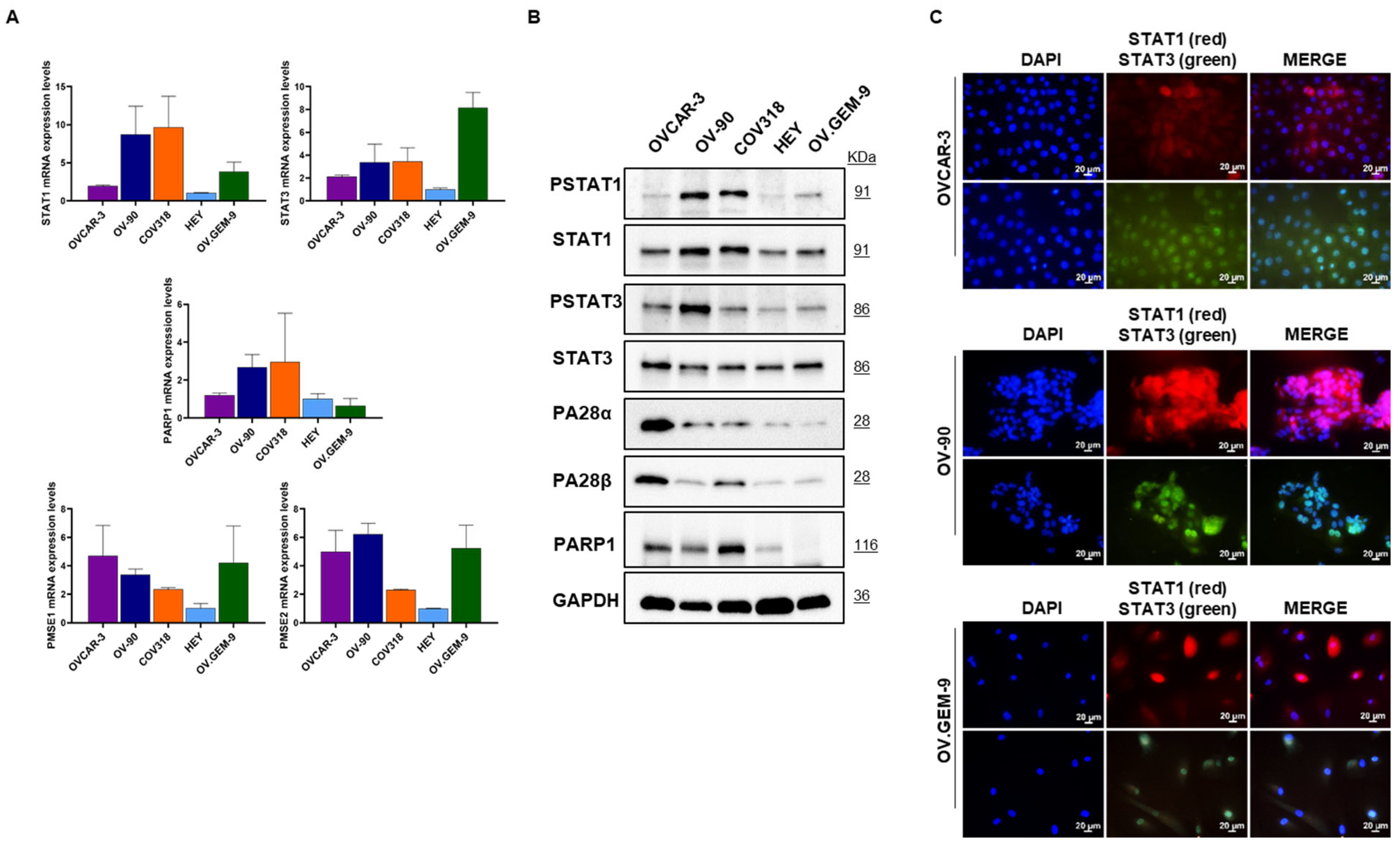
3.1. Characterization of Targets of Interest in Different HGSOC Cells
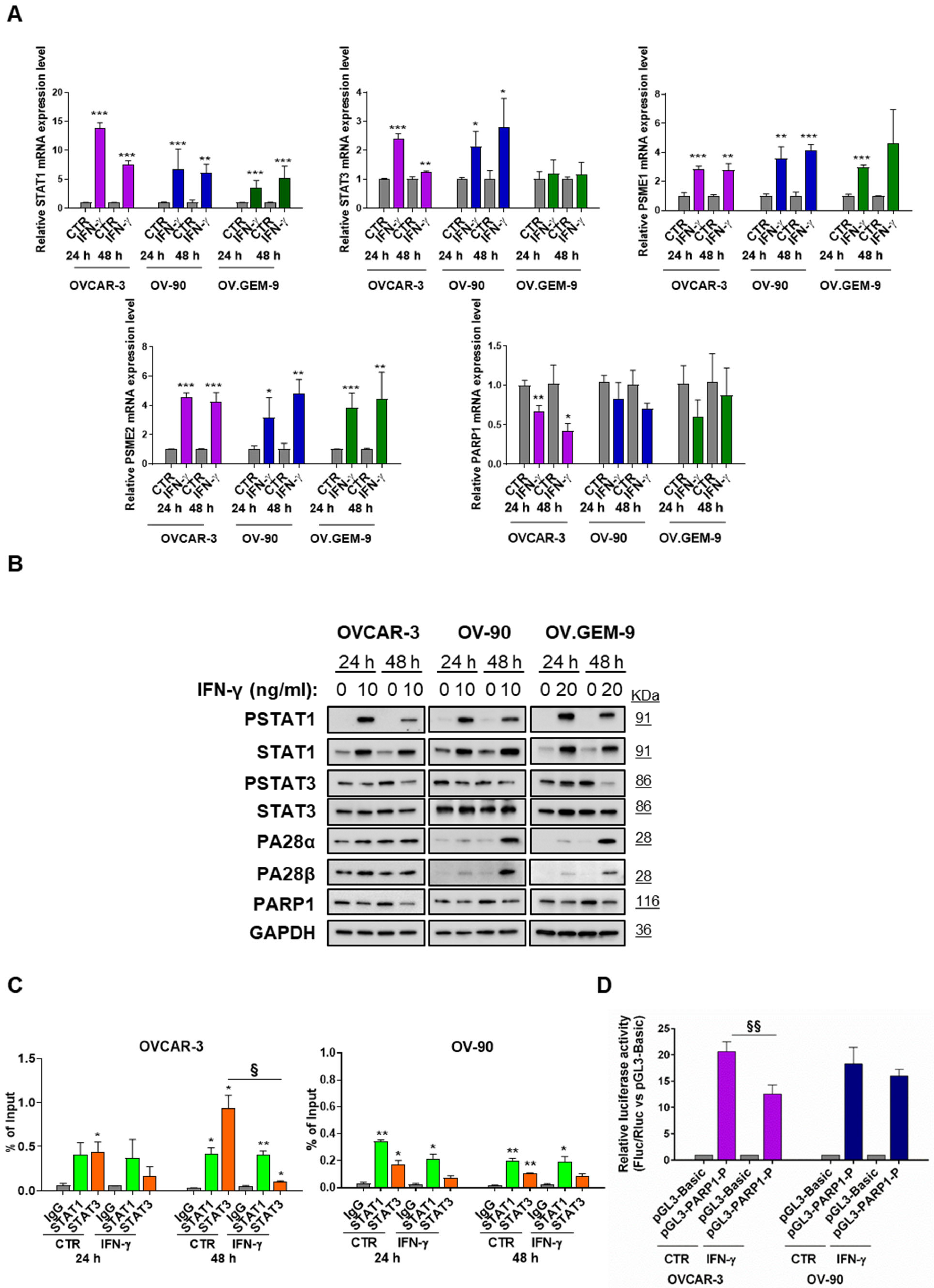
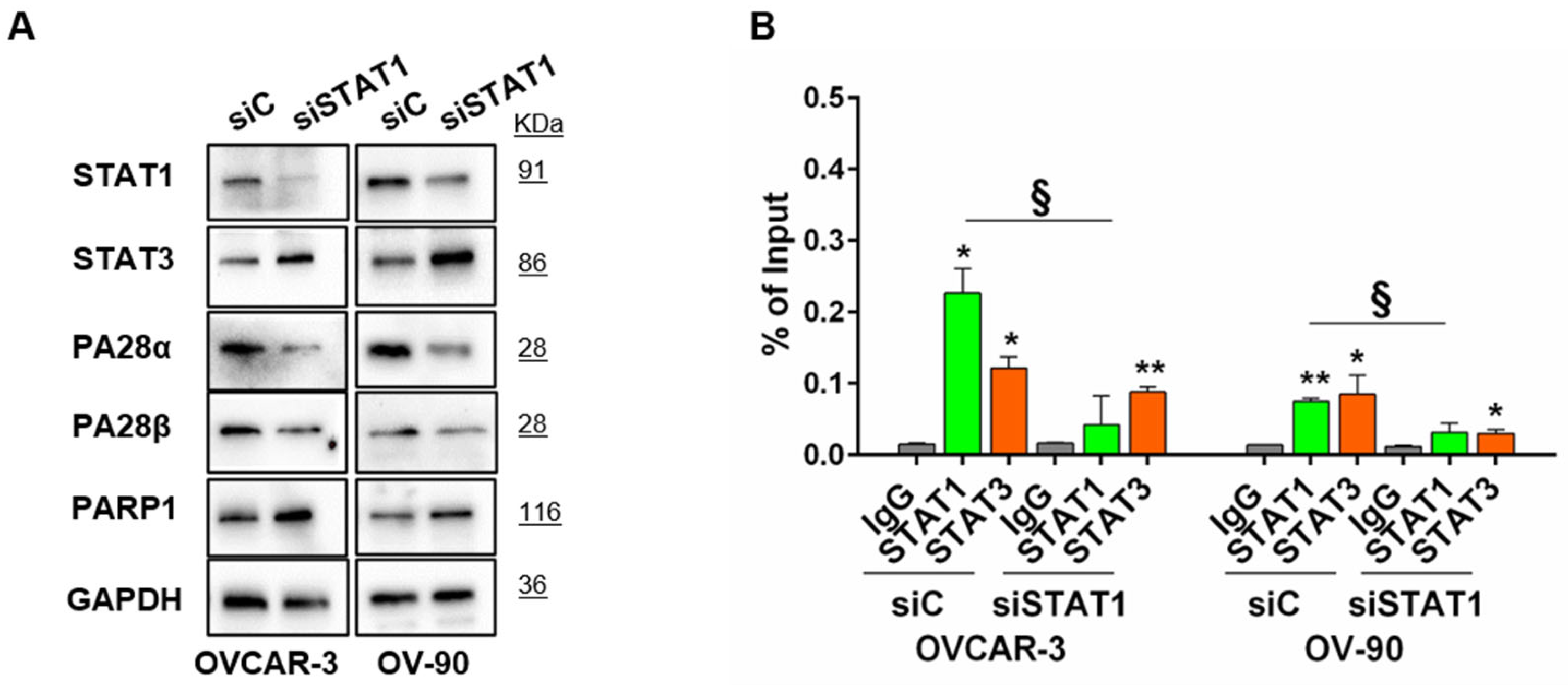
3.2. STAT1-Mediated Modulation of PARP1 Levels
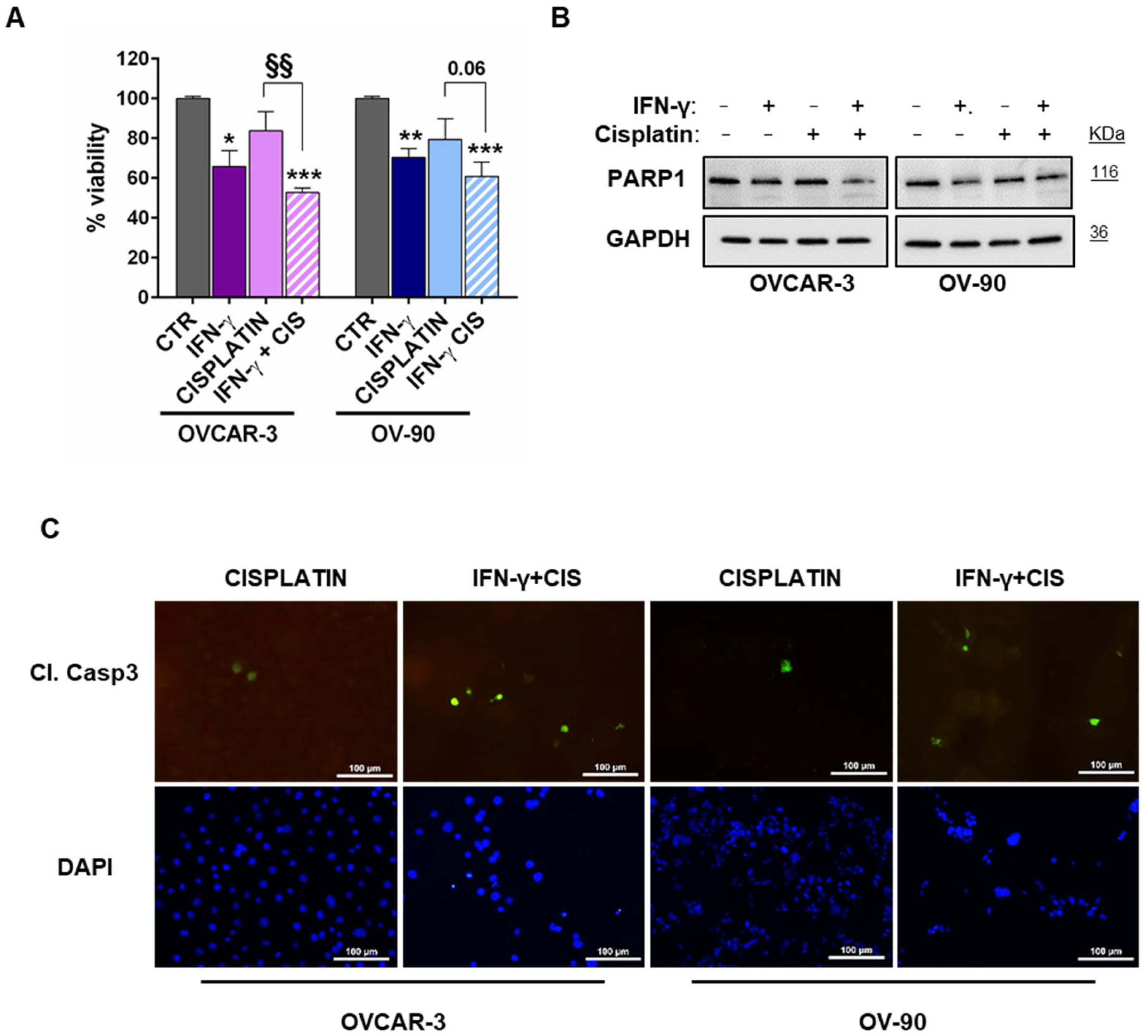
3.3. STAT1 Activation Sensitized HGSOC Cells to Cytotoxic Agent
3.4. STAT1 as a Prognostic/Predictive Factor in HGSOC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. 2020. Available online: https://gco.iarc.fr/today/data/factsheets/cancers/25-Ovary-fact-sheet.pdf (accessed on 4 April 2023).
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, C.; De Felice, F.; Romito, A.; Iacobelli, V.; Sassu, C.M.; Corrado, G.; Ricci, C.; Scambia, G.; Fagotti, A. Chemotherapy resistance in epithelial ovarian cancer: Mechanisms and emerging treatments. Semin Cancer Biol. 2021, 77, 144–166. [Google Scholar] [CrossRef] [PubMed]
- DiSilvestro, P.; Banerjee, S.; Colombo, N.; Scambia, G.; Kim, B.G.; Oaknin, A.; Friedlander, M.; Lisyanskaya, A.; Floquet, A.; Leary, A.; et al. Overall Survival with Maintenance Olaparib at a 7-Year Follow-Up in Patients with Newly Diagnosed Advanced Ovarian Cancer and a BRCA Mutation: The SOLO1/GOG 3004 Trial. J. Clin. Oncol. 2022, 41, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Tattersall, A.; Ryan, N.; Wiggans, A.J.; Rogozińska, E.; Morrison, J. Poly(ADP-ribose) polymerase (PARP) inhibitors for the treatment of ovarian cancer. Cochrane Database Syst. Rev. 2022, 2, CD007929. [Google Scholar] [CrossRef]
- Rose, M.; Burgess, J.T.; O’Byrne, K.; Richard, D.J.; Bolderson, E. PARP Inhibitors: Clinical Relevance, Mechanisms of Action and Tumor Resistance. Front. Cell Dev. Biol. 2020, 8, 564601. [Google Scholar] [CrossRef]
- Vanacker, H.; Harter, P.; Labidi-Galy, S.I.; Banerjee, S.; Oaknin, A.; Lorusso, D.; Ray-Coquard, I. PARP-inhibitors in epithelial ovarian cancer: Actual positioning and future expectations. Cancer Treat. Rev. 2021, 99, 102255. [Google Scholar] [CrossRef]
- Noordermeer, S.M.; van Attikum, H. PARP Inhibitor Resistance: A Tug-of-War in BRCA-Mutated Cells. Trends Cell Biol. 2019, 29, 820–834. [Google Scholar] [CrossRef]
- Alvarez Secord, A.; O’Malley, D.M.; Sood, A.K.; Westin, S.N.; Liu, J.F. Rationale for combination PARP inhibitor and antiangiogenic treatment in advanced epithelial ovarian cancer: A review. Gynecol. Oncol. 2021, 162, 482–495. [Google Scholar] [CrossRef]
- Yu, H.; Jove, R. The STATs of cancer--new molecular targets come of age. Nat. Rev. Cancer 2004, 4, 97–105. [Google Scholar] [CrossRef]
- Erdogan, F.; Radu, T.B.; Orlova, A.; Qadree, A.K.; de Araujo, E.D.; Israelian, J.; Valent, P.; Mustjoki, S.M.; Herling, M.; Moriggl, R.; et al. JAK-STAT core cancer pathway: An integrative cancer interactome analysis. J. Cell Mol. Med. 2022, 26, 2049–2062. [Google Scholar] [CrossRef]
- Gao, S.; Zhang, W.; Yan, N.; Li, M.; Mu, X.; Yin, H.; Wang, J. The impact of STAT3 and phospho-STAT3 expression on the prognosis and clinicopathology of ovarian cancer: A systematic review and meta-analysis. J. Ovarian Res. 2021, 14, 164. [Google Scholar] [CrossRef]
- Li, X.; Wang, F.; Xu, X.; Zhang, J.; Xu, G. The Dual Role of STAT1 in Ovarian Cancer: Insight Into Molecular Mechanisms and Application Potentials. Front. Cell Dev. Biol. 2021, 9, 636595. [Google Scholar] [CrossRef]
- Koti, M.; Siu, A.; Clément, I.; Bidarimath, M.; Turashvili, G.; Edwards, A.; Rahimi, K.; Mes-Masson, A.M.; Squire, J.A. A distinct pre-existing inflammatory tumour microenvironment is associated with chemotherapy resistance in high-grade serous epithelial ovarian cancer. Br. J. Cancer 2015, 112, 1215–1222. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Z. STAT1 in cancer: Friend or foe? Discov. Med. 2017, 24, 19–29. [Google Scholar]
- Rébé, C.; Végran, F.; Berger, H.; Ghiringhelli, F. STAT3 activation: A key factor in tumor immunoescape. JAKSTAT 2013, 2, e23010. [Google Scholar] [CrossRef]
- Raspaglio, G.; Buttarelli, M.; Filippetti, F.; Battaglia, A.; Buzzonetti, A.; Scambia, G.; Gallo, D. Stat1 confers sensitivity to radiation in cervical cancer cells by controlling Parp1 levels: A new perspective for Parp1 inhibition. Cell Death Dis. 2021, 12, 933. [Google Scholar] [CrossRef]
- Barton, L.F.; Cruz, M.; Rangwala, R.; Deepe, G.S.; Monaco, J.J. Regulation of immunoproteasome subunit expression in vivo following pathogenic fungal infection. J. Immunol. 2002, 169, 3046–3052. [Google Scholar] [CrossRef]
- Van den Berg-Bakker, C.A.; Hagemeijer, A.; Franken-Postma, E.M.; Smit, V.T.; Kuppen, P.J.; van Ravenswaay Claasen, H.H.; Cornelisse, C.J.; Schrier, P.I. Establishment and characterization of 7 ovarian carcinoma cell lines and one granulosa tumor cell line: Growth features and cytogenetics. Int. J. Cancer 1993, 53, 613–620. [Google Scholar] [CrossRef]
- Hamilton, T.C.; Young, R.C.; McKoy, W.M.; Grotzinger, K.R.; Green, J.A.; Chu, E.W.; Whang-Peng, J.; Rogan, A.M.; Green, W.R.; Ozols, R.F. Characterization of a human ovarian carcinoma cell line (NIH:OVCAR-3) with androgen and estrogen receptors. Cancer Res. 1983, 43, 5379–5389. [Google Scholar]
- Provencher, D.M.; Lounis, H.; Champoux, L.; Tétrault, M.; Manderson, E.N.; Wang, J.C.; Eydoux, P.; Savoie, R.; Tonin, P.N.; Mes-Masson, A.M. Characterization of four novel epithelial ovarian cancer cell lines. Vitr. Cell. Dev. Biol. Anim. 2000, 36, 357–361. [Google Scholar] [CrossRef]
- Buick, R.N.; Pullano, R.; Trent, J.M. Comparative properties of five human ovarian adenocarcinoma cell lines. Cancer Res. 1985, 45, 3668–3676. [Google Scholar]
- Buttarelli, M.; Ciucci, A.; Palluzzi, F.; Raspaglio, G.; Marchetti, C.; Perrone, E.; Minucci, A.; Giacò, L.; Fagotti, A.; Scambia, G.; et al. Identification of a novel gene signature predicting response to first-line chemotherapy in BRCA wild-type high-grade serous ovarian cancer patients. J. Exp. Clin. Cancer Res. 2022, 41, 50. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Gyorffy, B.; Lánczky, A.; Szállási, Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr.-Relat. Cancer 2012, 19, 197–208. [Google Scholar] [CrossRef]
- Fekete, J.T.; Ősz, Á.; Pete, I.; Nagy, G.R.; Vereczkey, I.; Győrffy, B. Predictive biomarkers of platinum and taxane resistance using the transcriptomic data of 1816 ovarian cancer patients. Gynecol. Oncol. 2020, 156, 654–661. [Google Scholar] [CrossRef]
- Liu, Z.; Beach, J.A.; Agadjanian, H.; Jia, D.; Aspuria, P.J.; Karlan, B.Y.; Orsulic, S. Suboptimal cytoreduction in ovarian carcinoma is associated with molecular pathways characteristic of increased stromal activation. Gynecol. Oncol. 2015, 139, 394–400. [Google Scholar] [CrossRef]
- Ding, L.; Chen, X.; Xu, X.; Qian, Y.; Liang, G.; Yao, F.; Yao, Z.; Wu, H.; Zhang, J.; He, Q.; et al. PARP1 Suppresses the Transcription of PD-L1 by Poly(ADP-Ribosyl)ating STAT3. Cancer Immunol. Res. 2019, 7, 136–149. [Google Scholar] [CrossRef]
- Gupte, R.; Nandu, T.; Kraus, W.L. Nuclear ADP-ribosylation drives IFNγ-dependent STAT1α enhancer formation in macrophages. Nat. Commun. 2021, 12, 3931. [Google Scholar] [CrossRef]
- Martincuks, A.; Song, J.; Kohut, A.; Zhang, C.; Li, Y.J.; Zhao, Q.; Mak, E.; Rodriguez-Rodriguez, L.; Yu, H.; Cristea, M. PARP Inhibition Activates STAT3 in Both Tumor and Immune Cells Underlying Therapy Resistance and Immunosuppression In Ovarian Cancer. Front. Oncol. 2021, 11, 724104. [Google Scholar] [CrossRef]
- Long, L.L.; Ma, S.C.; Guo, Z.Q.; Zhang, Y.P.; Fan, Z.; Liu, L.J.; Liu, L.; Han, D.D.; Leng, M.X.; Wang, J.; et al. PARP inhibition induces synthetic lethality and adaptive immunity in LKB1-mutant lung cancer. Cancer Res. 2022, 83, 568–581. [Google Scholar] [CrossRef]
- Shah, J.B.; Pueschl, D.; Wubbenhorst, B.; Fan, M.; Pluta, J.; D’Andrea, K.; Hubert, A.P.; Shilan, J.S.; Zhou, W.; Kraya, A.A.; et al. Analysis of matched primary and recurrent BRCA1/2 mutation-associated tumors identifies recurrence-specific drivers. Nat. Commun. 2022, 13, 6728. [Google Scholar] [CrossRef] [PubMed]
- Au, K.K.; Le Page, C.; Ren, R.; Meunier, L.; Clément, I.; Tyrishkin, K.; Peterson, N.; Kendall-Dupont, J.; Childs, T.; Francis, J.A.; et al. STAT1-associated intratumoural TH1 immunity predicts chemotherapy resistance in high-grade serous ovarian cancer. J. Pathol. Clin. Res. 2016, 2, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Josahkian, J.A.; Saggioro, F.P.; Vidotto, T.; Ventura, H.T.; Candido Dos Reis, F.J.; de Sousa, C.B.; Tiezzi, D.G.; de Andrade, J.M.; Koti, M.; Squire, J.A. Increased STAT1 Expression in High Grade Serous Ovarian Cancer Is Associated with a Better Outcome. Int. J. Gynecol. Cancer 2018, 28, 459–465. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raspaglio, G.; Buttarelli, M.; Cappoli, N.; Ciucci, A.; Fagotti, A.; Scambia, G.; Gallo, D. Exploring the Control of PARP1 Levels in High-Grade Serous Ovarian Cancer. Cancers 2023, 15, 2361. https://doi.org/10.3390/cancers15082361
Raspaglio G, Buttarelli M, Cappoli N, Ciucci A, Fagotti A, Scambia G, Gallo D. Exploring the Control of PARP1 Levels in High-Grade Serous Ovarian Cancer. Cancers. 2023; 15(8):2361. https://doi.org/10.3390/cancers15082361
Chicago/Turabian StyleRaspaglio, Giuseppina, Marianna Buttarelli, Natalia Cappoli, Alessandra Ciucci, Anna Fagotti, Giovanni Scambia, and Daniela Gallo. 2023. "Exploring the Control of PARP1 Levels in High-Grade Serous Ovarian Cancer" Cancers 15, no. 8: 2361. https://doi.org/10.3390/cancers15082361
APA StyleRaspaglio, G., Buttarelli, M., Cappoli, N., Ciucci, A., Fagotti, A., Scambia, G., & Gallo, D. (2023). Exploring the Control of PARP1 Levels in High-Grade Serous Ovarian Cancer. Cancers, 15(8), 2361. https://doi.org/10.3390/cancers15082361





