The Vault Complex Is Significantly Involved in Therapeutic Responsiveness of Endocrine Tumors and Linked to Autophagy under Chemotherapeutic Conditions
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture, In Vivo Experiments and Gene Array
2.2. Immunofluorescence/Histochemistry
2.3. EV Isolation by Ultracentrifugation
2.4. EV Isolation by ExoQuick
2.5. Nanoparticle Tracking Analysis (NTA)
2.6. Si-RNA Experiments
2.7. RNA Isolation, Reverse Transcription and qRT-PCR
2.8. Western Blot
2.8.1. BON, NCI ASA Tumors
2.8.2. NCI EDPM Cells
2.8.3. BON, NCI TNF Cells
2.9. Statistical Analysis
3. Results
3.1. Investigation of Gene Transcripts Related to Therapeutic Responsiveness In Vivo
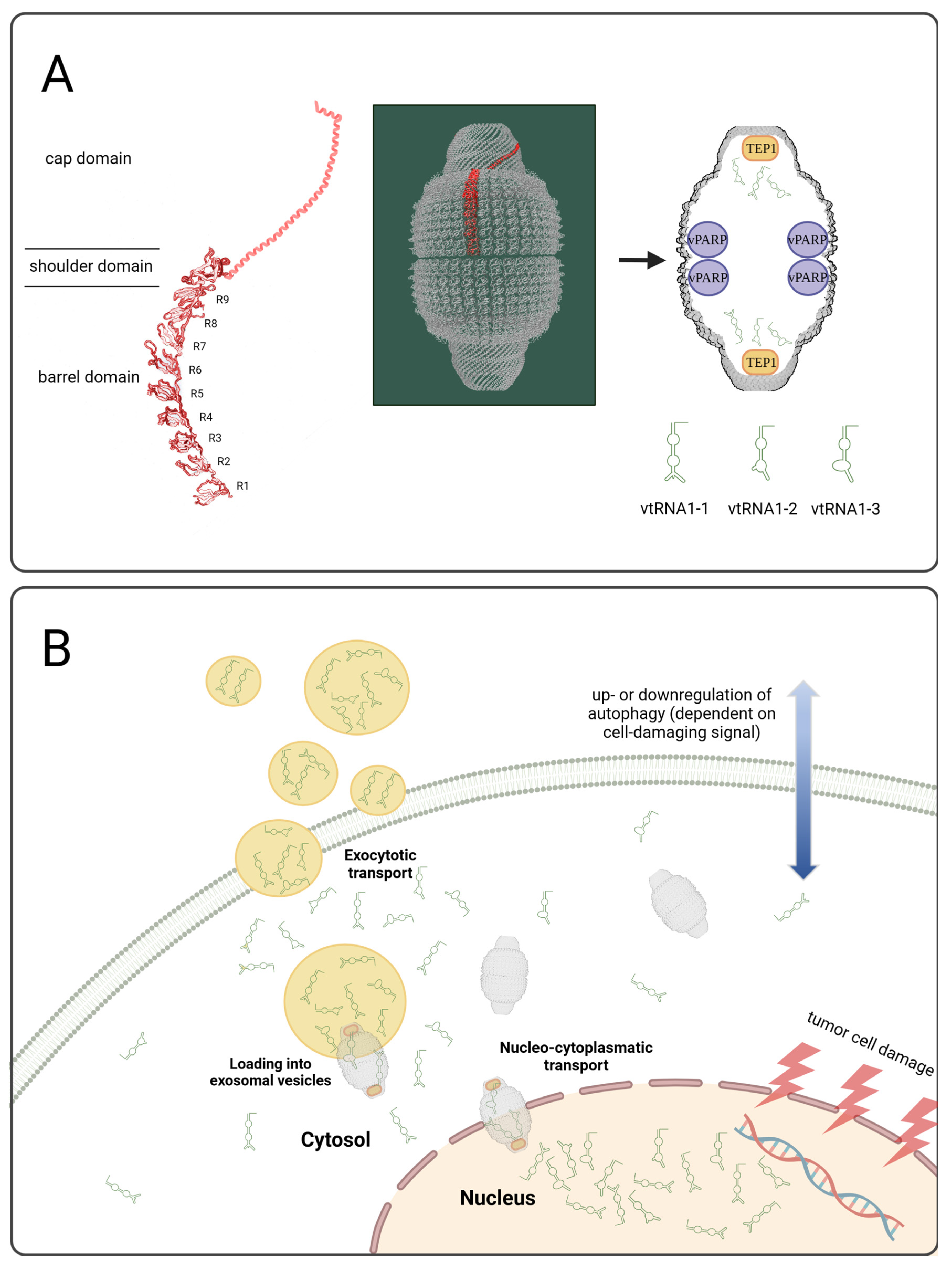
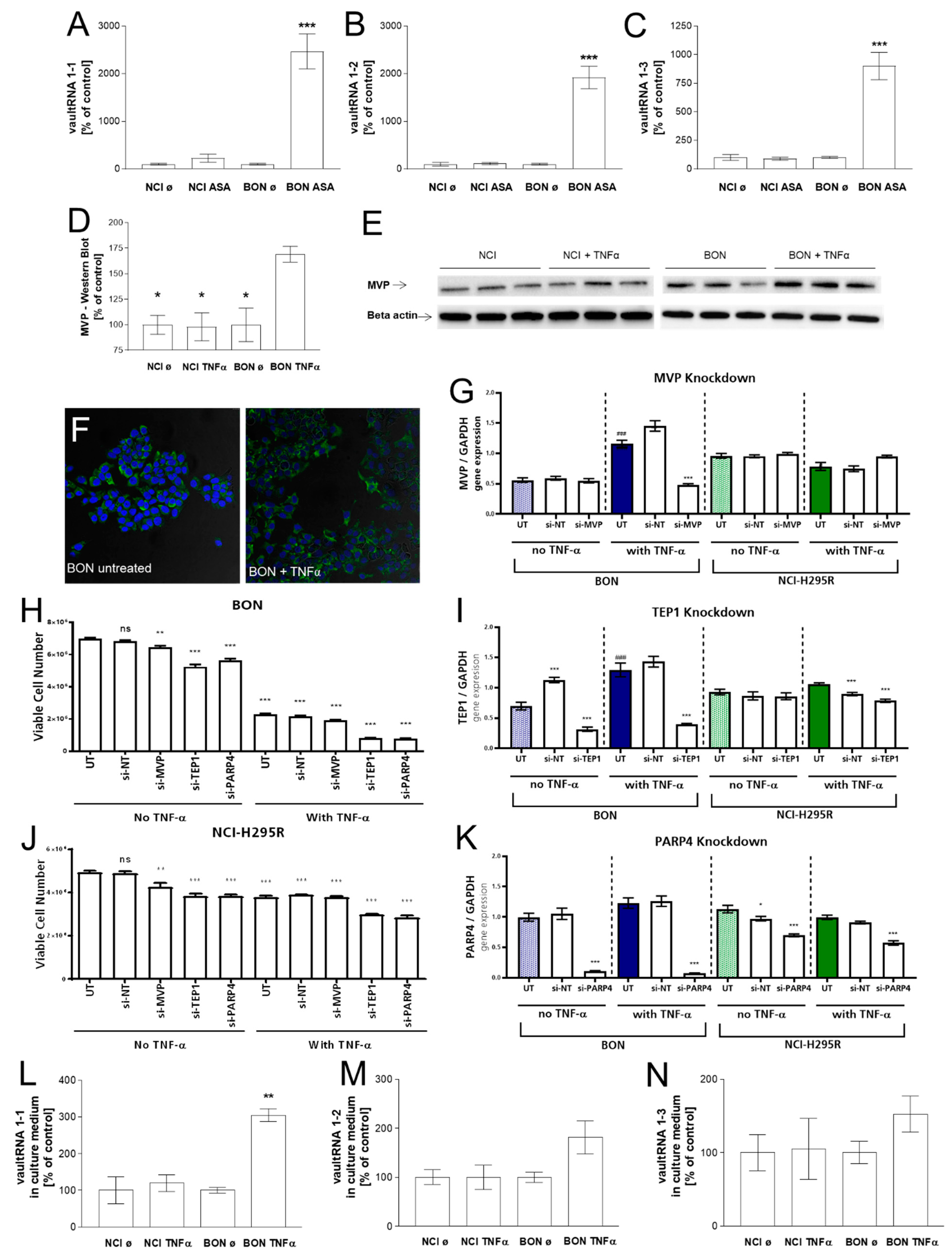
3.2. Investigation of the Vault Complex in Therapeutically Responding vs. Not-Responding Tumors
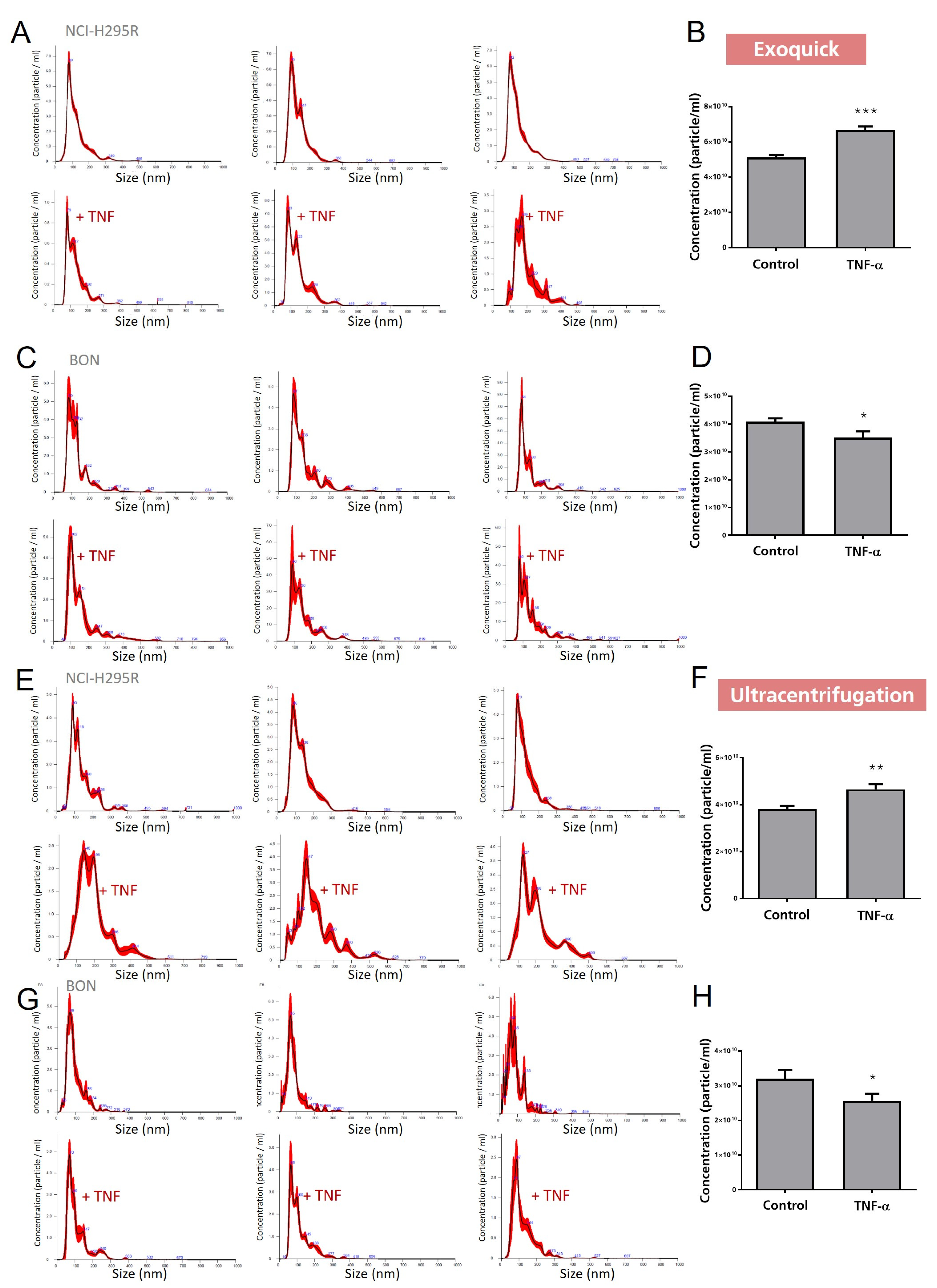
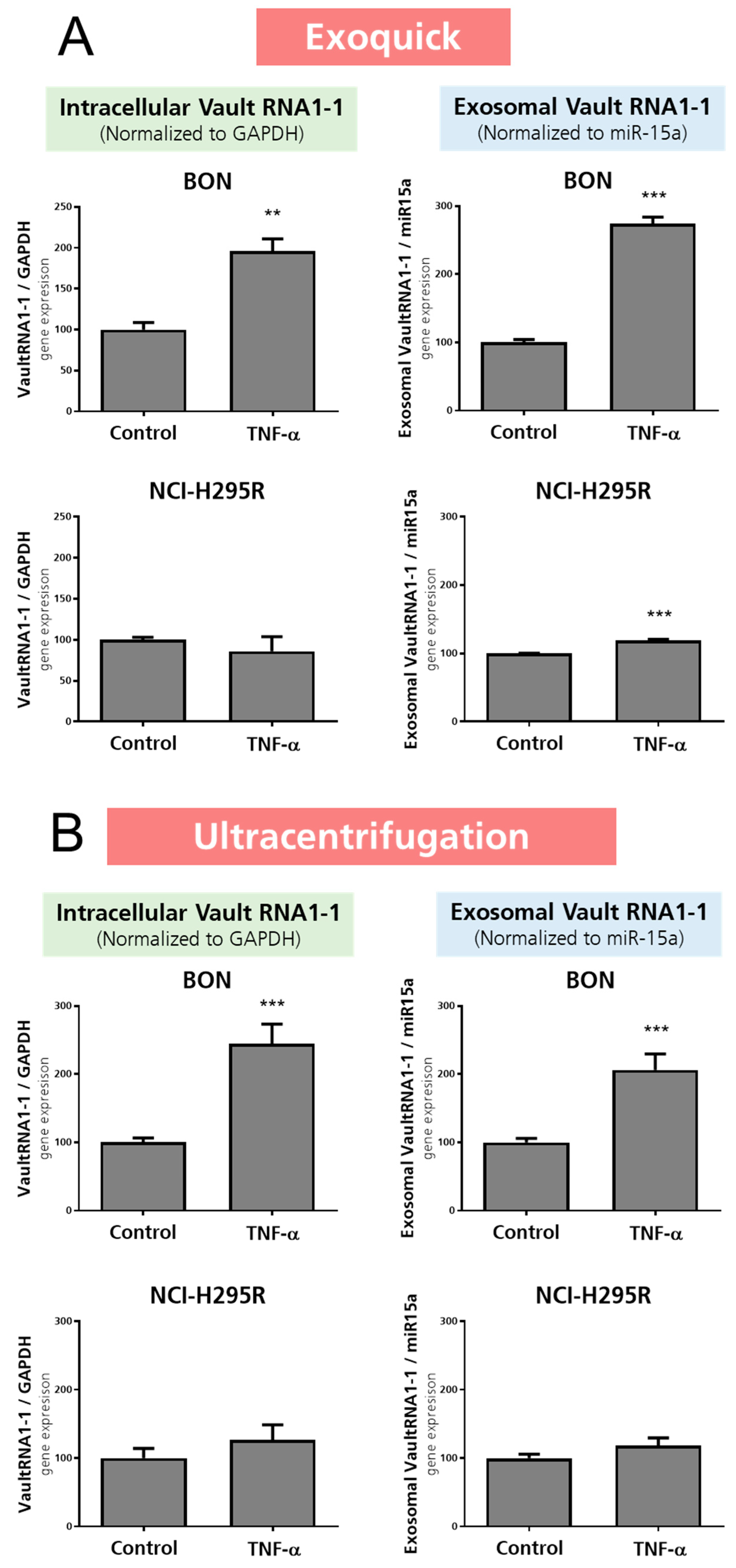
3.3. Investigation of Exosomal Release of TNFα-Treated BON and NCI-H295R Cells
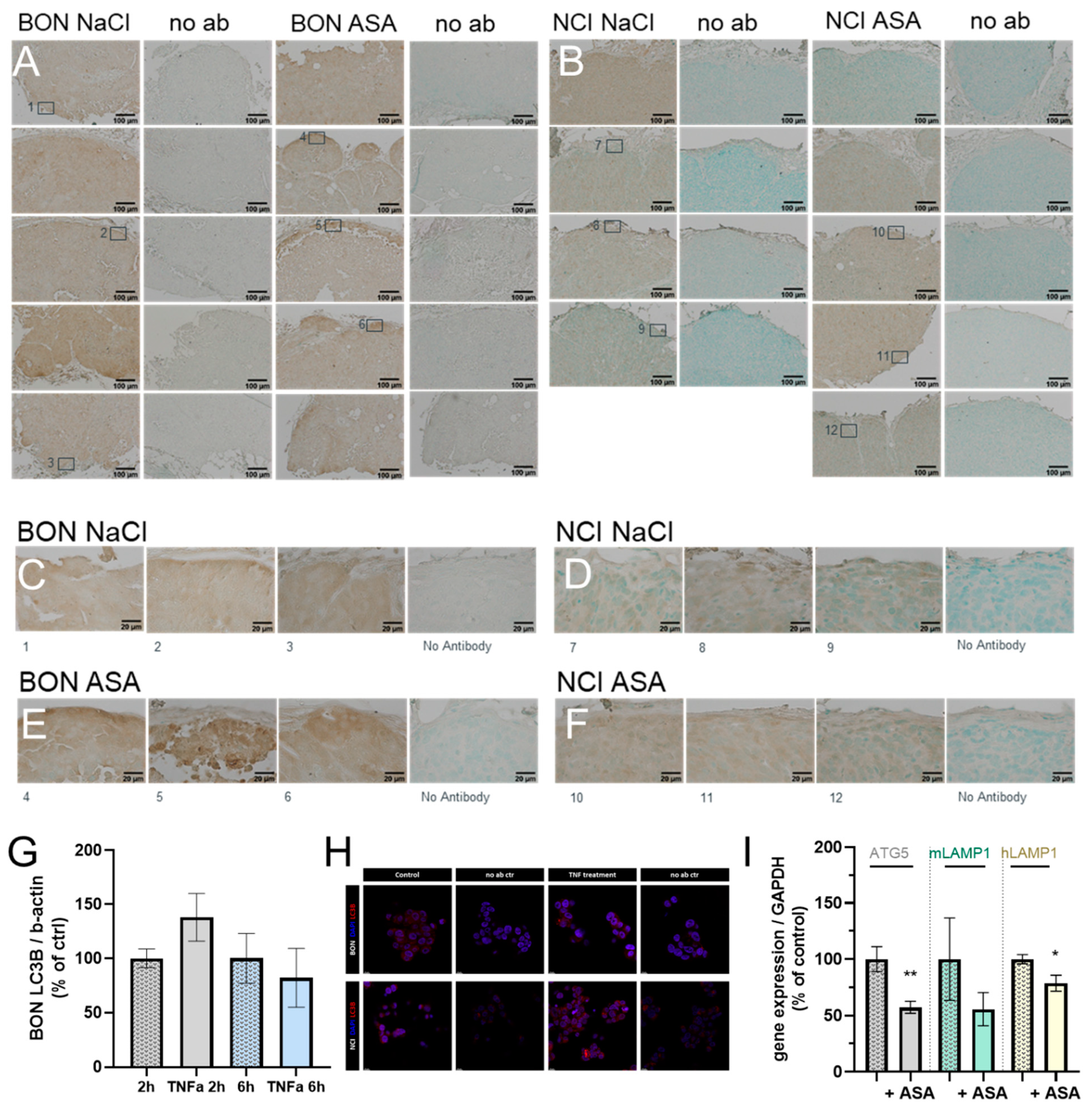
3.4. Correlation with Autophagic and Lysosomal Markers
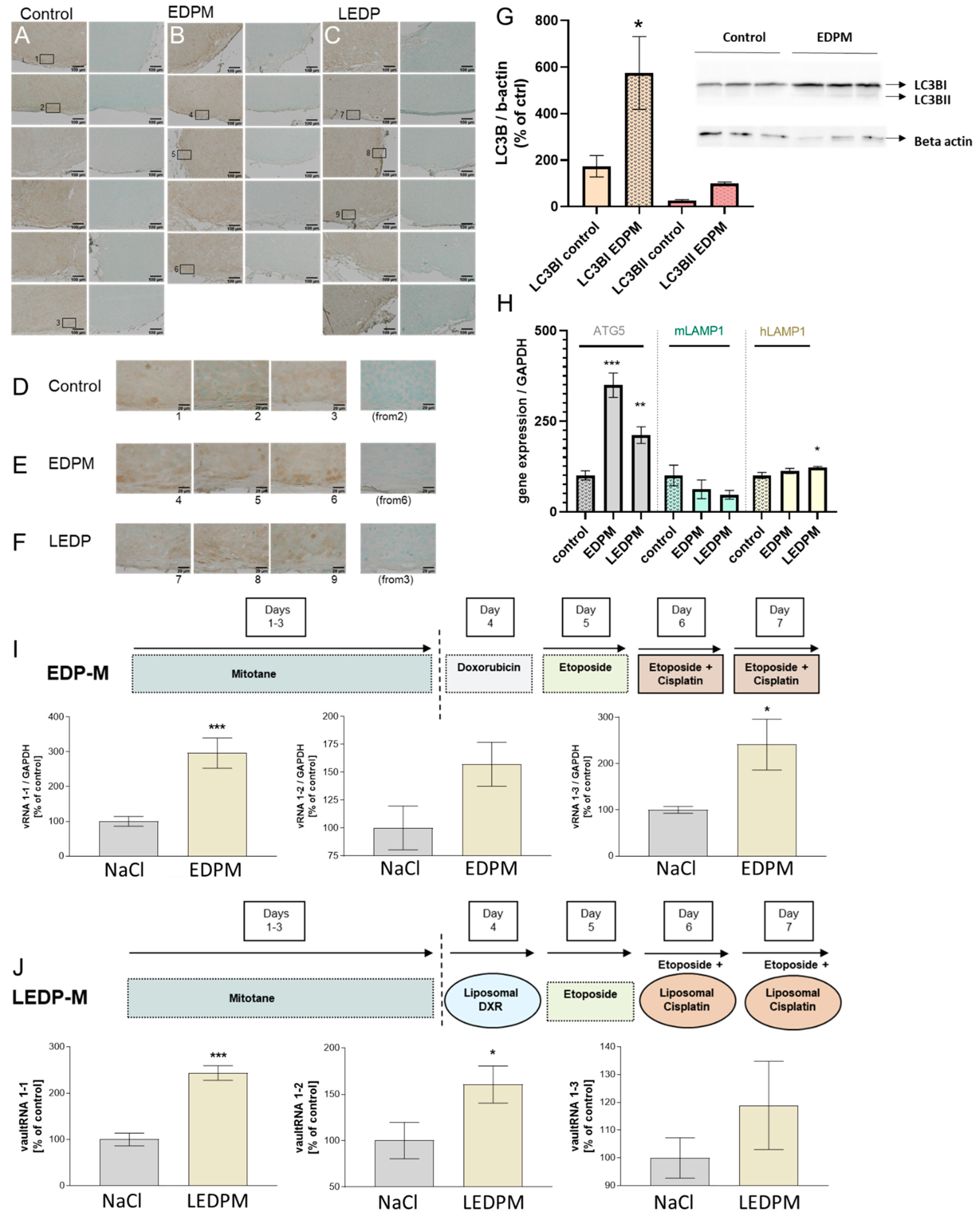
3.5. Proof of Principle in NCI-H295R Tumors and Correlation with Autophagic and Lysosomal Markers upon Chemotherapeutic Treatment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Silva, V.R.; Neves, S.P.; Santos, L.S.; Dias, R.B.; Bezerra, D.P. Challenges and Therapeutic Opportunities of Autophagy in Cancer Therapy. Cancers 2020, 12, 3461. [Google Scholar] [CrossRef]
- Tang, J.; Zhu, J.; Ye, Y.; Liu, Y.; He, Y.; Zhang, L.; Tang, D.; Qiao, C.; Feng, X.; Li, J.; et al. Inhibition LC3B can increase chemosensitivity of ovarian cancer cells. Cancer Cell Int. 2019, 19, 199. [Google Scholar] [CrossRef]
- Lei, Y.; Zhang, E.; Bai, L.; Li, Y. Autophagy in Cancer Immunotherapy. Cells 2022, 11, 2996. [Google Scholar] [CrossRef]
- Ye, X.; Zhou, X.J.; Zhang, H. Exploring the Role of Autophagy-Related Gene 5 (ATG5) Yields Important Insights Into Autophagy in Autoimmune/Autoinflammatory Diseases. Front. Immunol. 2018, 9, 2334. [Google Scholar] [CrossRef] [PubMed]
- Eskelinen, E.L. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol. Aspects Med. 2006, 27, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Ballabio, A.; Bonifacino, J.S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 101–118. [Google Scholar] [CrossRef]
- Ferro, I.; Gavini, J.; Gallo, S.; Bracher, L.; Landolfo, M.; Candinas, D.; Stroka, D.M.; Polacek, N. The human vault RNA enhances tumorigenesis and chemoresistance through the lysosome in hepatocellular carcinoma. Autophagy 2022, 18, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Horos, R.; Buscher, M.; Kleinendorst, R.; Alleaume, A.M.; Tarafder, A.K.; Schwarzl, T.; Dziuba, D.; Tischer, C.; Zielonka, E.M.; Adak, A.; et al. The Small Non-coding Vault RNA1-1 Acts as a Riboregulator of Autophagy. Cell 2019, 176, 1054–1067.e1012. [Google Scholar] [CrossRef]
- Horos, R.; Buscher, M.; Sachse, C.; Hentze, M.W. Vault RNA emerges as a regulator of selective autophagy. Autophagy 2019, 15, 1463–1464. [Google Scholar] [CrossRef]
- Buscher, M.; Horos, R.; Hentze, M.W. ‘High vault-age’: Non-coding RNA control of autophagy. Open Biol. 2020, 10, 190307. [Google Scholar] [CrossRef]
- Kedersha, N.L.; Heuser, J.E.; Chugani, D.C.; Rome, L.H. Vaults. III. Vault ribonucleoprotein particles open into flower-like structures with octagonal symmetry. J. Cell Biol. 1991, 112, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Gallo, S.; Kong, E.; Ferro, I.; Polacek, N. Small but Powerful: The Human Vault RNAs as Multifaceted Modulators of Pro-Survival Characteristics and Tumorigenesis. Cancers 2022, 14, 2787. [Google Scholar] [CrossRef]
- Kedersha, N.L.; Rome, L.H. Isolation and characterization of a novel ribonucleoprotein particle: Large structures contain a single species of small RNA. J. Cell Biol. 1986, 103, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Guerra, M.; Gonzalez-Alamos, M.; Llauro, A.; Casanas, A.; Querol-Audi, J.; de Pablo, P.J.; Verdaguer, N. Symmetry disruption commits vault particles to disassembly. Sci. Adv. 2022, 8, eabj7795. [Google Scholar] [CrossRef]
- Querol-Audi, J.; Casanas, A.; Uson, I.; Luque, D.; Caston, J.R.; Fita, I.; Verdaguer, N. The mechanism of vault opening from the high resolution structure of the N-terminal repeats of MVP. EMBO J. 2009, 28, 3450–3457. [Google Scholar] [CrossRef]
- Hantel, C.; Ozimek, A.; Lira, R.; Ragazzon, B.; Jackel, C.; Frantsev, R.; Reincke, M.; Bertherat, J.; Mussack, T.; Beuschlein, F. TNF alpha signaling is associated with therapeutic responsiveness to vascular disrupting agents in endocrine tumors. Mol. Cell Endocrinol. 2016, 423, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Hantel, C.; Jung, S.; Mussack, T.; Reincke, M.; Beuschlein, F. Liposomal polychemotherapy improves adrenocortical carcinoma treatment in a preclinical rodent model. Endocr. Relat. Cancer 2014, 21, 383–394. [Google Scholar] [CrossRef]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3.22.1–3.22.29. [Google Scholar] [CrossRef]
- Jung, S.; Nagy, Z.; Fassnacht, M.; Zambetti, G.; Weiss, M.; Reincke, M.; Igaz, P.; Beuschlein, F.; Hantel, C. Preclinical progress and first translational steps for a liposomal chemotherapy protocol against adrenocortical carcinoma. Endocr. Relat. Cancer 2016, 23, 825–837. [Google Scholar] [CrossRef]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. EMBO J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.A.; Outzen, H.; Overvatn, A.; Bjorkoy, G.; Johansen, T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar] [CrossRef]
- Amort, M.; Nachbauer, B.; Tuzlak, S.; Kieser, A.; Schepers, A.; Villunger, A.; Polacek, N. Expression of the vault RNA protects cells from undergoing apoptosis. Nat. Commun. 2015, 6, 7030. [Google Scholar] [CrossRef]
- Li, F.; Chen, Y.; Zhang, Z.; Ouyang, J.; Wang, Y.; Yan, R.; Huang, S.; Gao, G.F.; Guo, G.; Chen, J.L. Robust expression of vault RNAs induced by influenza A virus plays a critical role in suppression of PKR-mediated innate immunity. Nucleic. Acids Res. 2015, 43, 10321–10337. [Google Scholar] [CrossRef] [PubMed]
- Lam, N.; Sugden, B. LMP1, a viral relative of the TNF receptor family, signals principally from intracellular compartments. EMBO J. 2003, 22, 3027–3038. [Google Scholar] [CrossRef] [PubMed]
- Lavorgna, A.; Harhaj, E.W. EBV LMP1: New and shared pathways to NF-kappaB activation. Proc. Natl. Acad. Sci. USA 2012, 109, 2188–2189. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Ren, Y.; Hu, X.; Mu, J.; Samykutty, A.; Zhuang, X.; Deng, Z.; Kumar, A.; Zhang, L.; Merchant, M.L.; et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat. Commun. 2017, 8, 14448. [Google Scholar] [CrossRef]
- Bishani, A.; Chernolovskaya, E.L. Activation of Innate Immunity by Therapeutic Nucleic Acids. Int. J. Mol. Sci. 2021, 22, 13360. [Google Scholar] [CrossRef]
- Petes, C.; Odoardi, N.; Gee, K. The Toll for Trafficking: Toll-Like Receptor 7 Delivery to the Endosome. Front. Immunol. 2017, 8, 1075. [Google Scholar] [CrossRef]
- Jiang, G.M.; Tan, Y.; Wang, H.; Peng, L.; Chen, H.T.; Meng, X.J.; Li, L.L.; Liu, Y.; Li, W.F.; Shan, H. The relationship between autophagy and the immune system and its applications for tumor immunotherapy. Mol. Cancer 2019, 18, 17. [Google Scholar] [CrossRef]
| Entrez Gene ID | Abbreviation | RNA Type | BON | NCI | |
|---|---|---|---|---|---|
| 56664 | vault RNA1-1 | VTRNA1-1 | non-coding | 16.6 | 1.8 |
| 677823 | small nucleolar RNA, H/ACA box 42 | SNORA42 | non-coding | 10.1 | |
| 677775 | small Cajal body-specific RNA 5 | SCARNA5 | non-coding | 8.2 | |
| 677808 | small nucleolar RNA, H/ACA box 23 | SNORA23 | non-coding | 8.0 | 7.6 |
| 677774 | small Cajal body-specific RNA 1 | SCARNA1 | non-coding | 7.2 | 3.7 |
| 619565 | small nucleolar RNA, H/ACA box 52 | SNORA52 | non-coding | 6.6 | 3.1 |
| 677772 | small Cajal body-specific RNA 6 | SCARNA6 | non-coding | 6.4 | 2.6 |
| 677806 | small nucleolar RNA, H/ACA box 20 | SNORA20 | non-coding | 6.0 | 3.2 |
| 677819 | small nucleolar RNA, H/ACA box 37 | SNORA37 | non-coding | 5.7 | 2.6 |
| 677801 | small nucleolar RNA, H/ACA box 14A | SNORA14A | non-coding | 5.6 | 1.5 |
| 1349 | cytochrome c oxidase subunit VIIb | COX7B | coding | 5.4 | 1.8 |
| 56663 | vault RNA1-2 | VTRNA1-2 | non-coding | 5.0 | 2.3 |
| 26829 | RNA, U5E small nuclear 1 | RNU5E-1 | non-coding | 4.9 | 2.8 |
| 26784 | small nucleolar RNA, H/ACA box 64 | SNORA64 | non-coding | 4.4 | 2.8 |
| 677797 | small nucleolar RNA, H/ACA box 7B | SNORA7B | non-coding | 4.4 | 2.0 |
| 619505 | small nucleolar RNA, H/ACA box 21 | SNORA21 | non-coding | 4.2 | 1.9 |
| 6044 | small nucleolar RNA, H/ACA box 62 | SNORA62 | non-coding | 4.1 | |
| 677802 | small nucleolar RNA, H/ACA box 14B | SNORA14B | non-coding | 4.1 | |
| 692148 | small Cajal body-specific RNA 10 | SCARNA10 | non-coding | 4.1 | 2.7 |
| 677837 | small nucleolar RNA, H/ACA box 60 | SNORA60 | non-coding | 4.1 | 1.5 |
| 574040 | small nucleolar RNA, H/ACA box 6 | SNORA6 | non-coding | 4.1 | |
| 677811 | small nucleolar RNA, H/ACA box 28 | SNORA28 | non-coding | 4.0 | 2.2 |
| 6286 | S100 calcium binding protein P | S100P | coding | 3.9 | |
| 677825 | small nucleolar RNA, H/ACA box 44 | SNORA44 | non-coding | 3.9 | |
| 692158 | small nucleolar RNA, H/ACA box 57 | SNORA57 | non-coding | 3.8 | 2.0 |
| 677781 | small Cajal body-specific RNA 16 | SCARNA16 | non-coding | 3.8 | |
| 619568 | small nucleolar RNA, H/ACA box 4 | SNORA4 | non-coding | 3.6 | |
| 26824 | RNA, U11 small nuclear | RNU11 | non-coding | 3.6 | |
| 677773 | small Cajal body-specific RNA 23 | SCARNA23 | non-coding | 3.5 | 1.8 |
| 85495 | ribonuclease P RNA component H1 | RPPH1 | non-coding | 3.4 | 1.7 |
| 26776 | small nucleolar RNA, H/ACA box 71B | SNORA71B | non-coding | 3.3 | |
| 541471 | uncharacterized LOC541471 | LOC541471 | coding | 3.3 | |
| 100033436 | small nucleolar RNA, C/D box 116-25 | SNORD116-25 | non-coding | 3.3 | 1.8 |
| 114599 | small nucleolar RNA, C/D box 15B | SNORD15B | non-coding | 3.2 | 1.9 |
| 677809 | small nucleolar RNA, H/ACA box 24 | SNORA24 | non-coding | 3.2 | 2.4 |
| 401466 | chromosome 8 open reading frame 59 | C8orf59 | coding | 3.0 | |
| 100151683 | RNA, U4atac small nuclear (U12-dependent splicing) | RNU4ATAC | non-coding | 3.0 | 1.6 |
| 692225 | small nucleolar RNA, C/D box 94 | SNORD94 | non-coding | 3.0 | |
| 100033438 | small nucleolar RNA, C/D box 116-26 | SNORD116-26 | non-coding | 3.0 | 2.4 |
| 4477 | microseminoprotein, beta- | MSMB | coding | 2.9 | |
| 677829 | small nucleolar RNA, H/ACA box 49 | SNORA49 | non-coding | 2.9 | 1.8 |
| 9446 | glutathione S-transferase omega 1 | GSTO1 | coding | 2.9 | |
| 93081 | testis expressed 30 | TEX30 | coding | 2.9 | |
| 51642 | mitochondrial ribosomal protein L48 | MRPL48 | coding | 2.8 | 1.9 |
| 5203 | prefoldin subunit 4 | PFDN4 | coding | 2.8 | |
| 677793 | small nucleolar RNA, H/ACA box 2A | SNORA2A | non-coding | 2.8 | |
| 521 | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E | ATP5I | coding | 2.8 | |
| 3957 | lectin, galactoside-binding, soluble, 2 | LGALS2 | coding | 2.8 | |
| 4709 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa | NDUFB3 | coding | 2.8 | 1.6 |
| 51503 | CWC15 spliceosome-associated protein homolog (S. cerevisiae) | CWC15 | coding | 2.7 | 1.7 |
| 6750 | somatostatin | SST | coding | 2.7 | |
| 594839 | small nucleolar RNA, H/ACA box 33 | SNORA33 | non-coding | 2.7 | |
| 25826 | small nucleolar RNA, C/D box 82 | SNORD82 | non-coding | 2.7 | 1.6 |
| 6201 | ribosomal protein S7 | RPS7 | coding | 2.7 | 1.9 |
| 677770 | small Cajal body-specific RNA 22 | SCARNA22 | non-coding | 2.7 | 2.2 |
| 677798 | small nucleolar RNA, H/ACA box 9 | SNORA9 | non-coding | 2.7 | |
| 26777 | small nucleolar RNA, H/ACA box 71A | SNORA71A | non-coding | 2.7 | 1.5 |
| 7012 | telomerase RNA component | TERC | non-coding | 2.7 | |
| 677777 | small Cajal body-specific RNA 12 | SCARNA12 | non-coding | 2.7 | 1.6 |
| 10247 | heat-responsive protein 12 | HRSP12 | coding | 2.6 | |
| 100033431 | small nucleolar RNA, C/D box 116-20 | SNORD116-20 | non-coding | 2.6 | 1.6 |
| 84300 | mitochondrial nucleoid factor 1 | MNF1 | coding | 2.6 | 1.6 |
| 3434 | interferon-induced protein with tetratricopeptide repeats 1 | IFIT1 | coding | 2.6 | 2.8 |
| 319103 | small nucleolar RNA, C/D box 8 | SNORD8 | non-coding | 2.6 | |
| 677814 | small nucleolar RNA, H/ACA box 31 | SNORA31 | non-coding | 2.6 | |
| 100033821 | small nucleolar RNA, C/D box 116-29 | SNORD116-29 | non-coding | 2.6 | 1.6 |
| 4338 | molybdenum cofactor synthesis 2 | MOCS2 | coding | 2.6 | 2.4 |
| 29950 | SERTA domain containing 1 | SERTAD1 | coding | 2.6 | 2.1 |
| 25906 | anaphase promoting complex subunit 15 | ANAPC15 | coding | 2.6 | 2.0 |
| 26828 | RNA, U5F small nuclear 1 | RNU5F-1 | non-coding | 2.6 | |
| 116937 | small nucleolar RNA, C/D box 83A | SNORD83A | non-coding | 2.6 | |
| 6206 | ribosomal protein S12 | RPS12 | coding | 2.6 | |
| 51053 | geminin, DNA replication inhibitor | GMNN | coding | 2.6 | |
| 100033420 | small nucleolar RNA, C/D box 116-8 | SNORD116-8 | non-coding | 2.6 | 2.2 |
| 57819 | LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) | LSM2 | non-coding | 2.6 | 1.8 |
| 677833 | small nucleolar RNA, H/ACA box 54 | SNORA54 | non-coding | 2.6 | |
| 56662 | vault RNA1-3 | VTRNA1-3 | non-coding | 2.5 | |
| 119392 | SWI5-dependent recombination repair 1 | SFR1 | coding | 2.5 | 1.6 |
| 341 | apolipoprotein C-I | APOC1 | coding | 2.5 | |
| 30836 | deoxynucleotidyltransferase, terminal, interacting protein 2 | DNTTIP2 | coding | 2.5 | 1.8 |
| 8365 | histone cluster 1, H4h | HIST1H4H | coding | 2.5 | |
| 6643 | sorting nexin 2 | SNX2 | coding | 2.5 | 2.7 |
| 200916 | ribosomal protein L22-like 1 | RPL22L1 | coding | 2.5 | |
| 26804 | small nucleolar RNA, C/D box 45B | SNORD45B | non-coding | 2.5 | −2.7 |
| 1347 | cytochrome c oxidase subunit VIIa polypeptide 2 (liver) | COX7A2 | coding | 2.5 | 1.5 |
| 174 | alpha-fetoprotein | AFP | coding | 2.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bornstein, S.; Shapiro, I.; Mazumdar, A.; Zitzmann, K.; Nölting, S.; Luca, E.; Beuschlein, F.; Sharma, A.; Hantel, C. The Vault Complex Is Significantly Involved in Therapeutic Responsiveness of Endocrine Tumors and Linked to Autophagy under Chemotherapeutic Conditions. Cancers 2023, 15, 1783. https://doi.org/10.3390/cancers15061783
Bornstein S, Shapiro I, Mazumdar A, Zitzmann K, Nölting S, Luca E, Beuschlein F, Sharma A, Hantel C. The Vault Complex Is Significantly Involved in Therapeutic Responsiveness of Endocrine Tumors and Linked to Autophagy under Chemotherapeutic Conditions. Cancers. 2023; 15(6):1783. https://doi.org/10.3390/cancers15061783
Chicago/Turabian StyleBornstein, Stefan, Igor Shapiro, Alekhya Mazumdar, Kathrin Zitzmann, Svenja Nölting, Edlira Luca, Felix Beuschlein, Ashish Sharma, and Constanze Hantel. 2023. "The Vault Complex Is Significantly Involved in Therapeutic Responsiveness of Endocrine Tumors and Linked to Autophagy under Chemotherapeutic Conditions" Cancers 15, no. 6: 1783. https://doi.org/10.3390/cancers15061783
APA StyleBornstein, S., Shapiro, I., Mazumdar, A., Zitzmann, K., Nölting, S., Luca, E., Beuschlein, F., Sharma, A., & Hantel, C. (2023). The Vault Complex Is Significantly Involved in Therapeutic Responsiveness of Endocrine Tumors and Linked to Autophagy under Chemotherapeutic Conditions. Cancers, 15(6), 1783. https://doi.org/10.3390/cancers15061783







