Mass Spectrometry-Based Biomarkers to Detect Prostate Cancer: A Multicentric Study Based on Non-Invasive Urine Collection without Prior Digital Rectal Examination
Abstract
Simple Summary
Abstract
1. Introduction
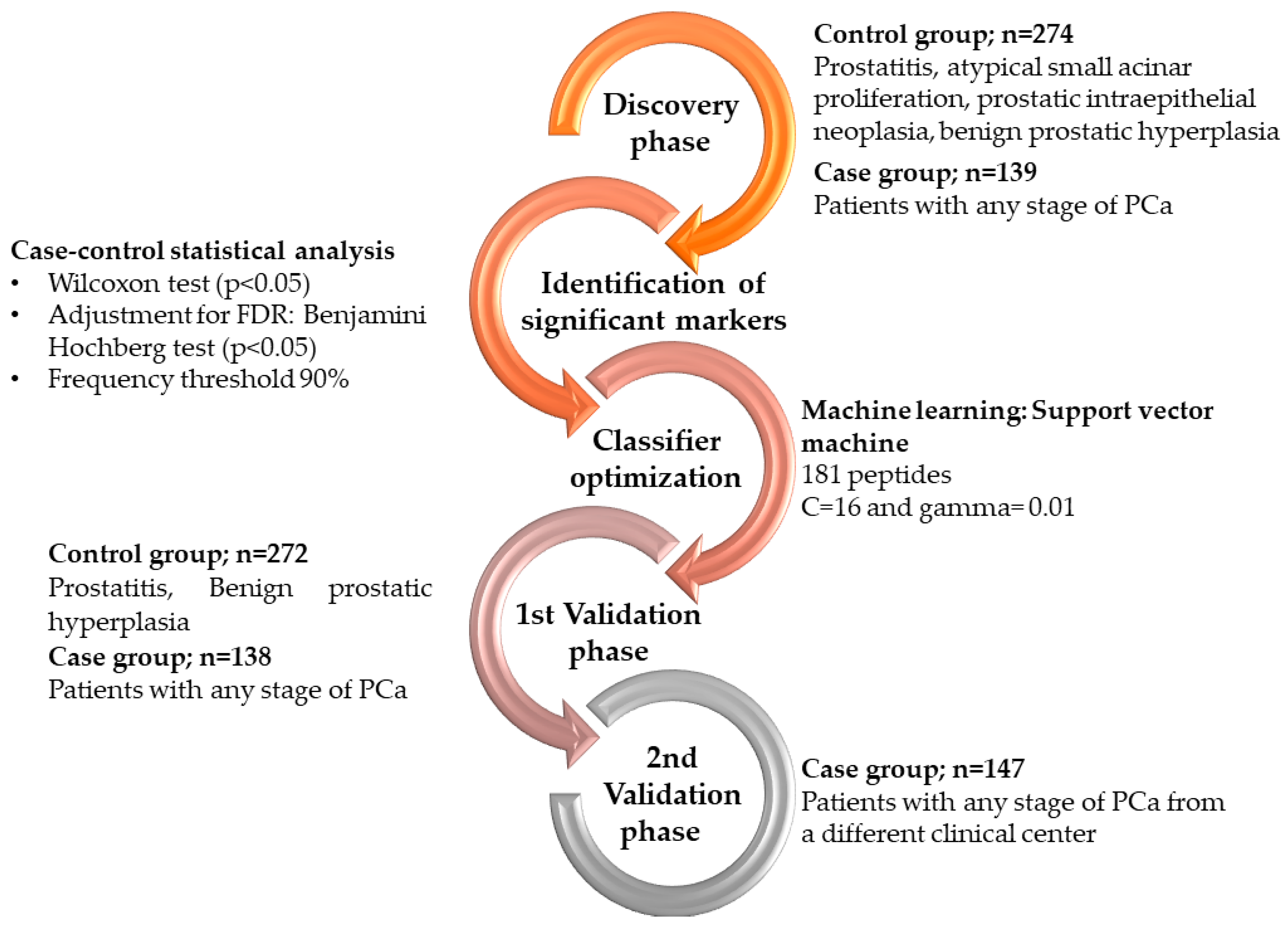
2. Materials and Methods
2.1. Patient Population and Characteristics
2.2. Sample Collection and Processing
2.3. Mass Spectrometry Analysis and Post-Acquisition Data Processing
2.4. Peptide Sequence Assignment
2.5. Statistical Analysis
2.6. Machine Learning Model Construction and Optimization
2.7. In Silico Protease Prediction and Bioinformatics Analysis
3. Results
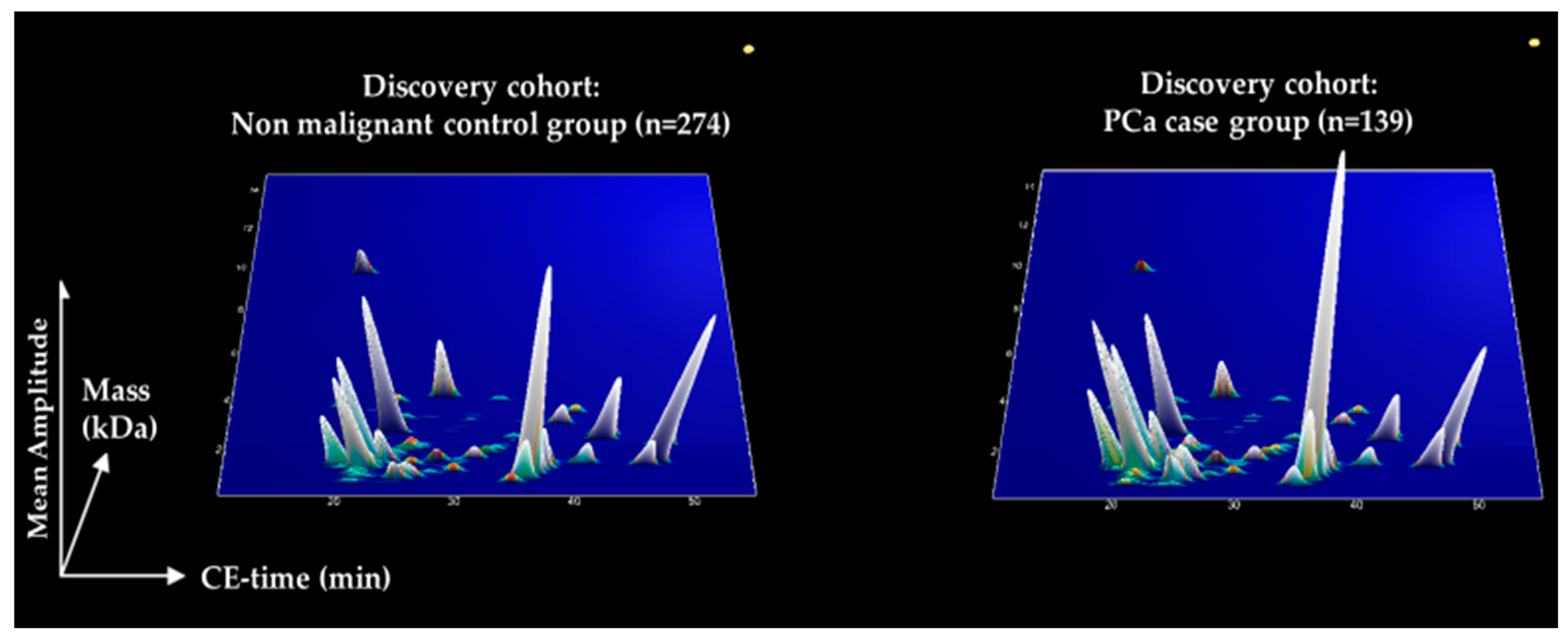
3.1. Discovery of Peptides with Significantly Altered Abundance in Urine for PCa
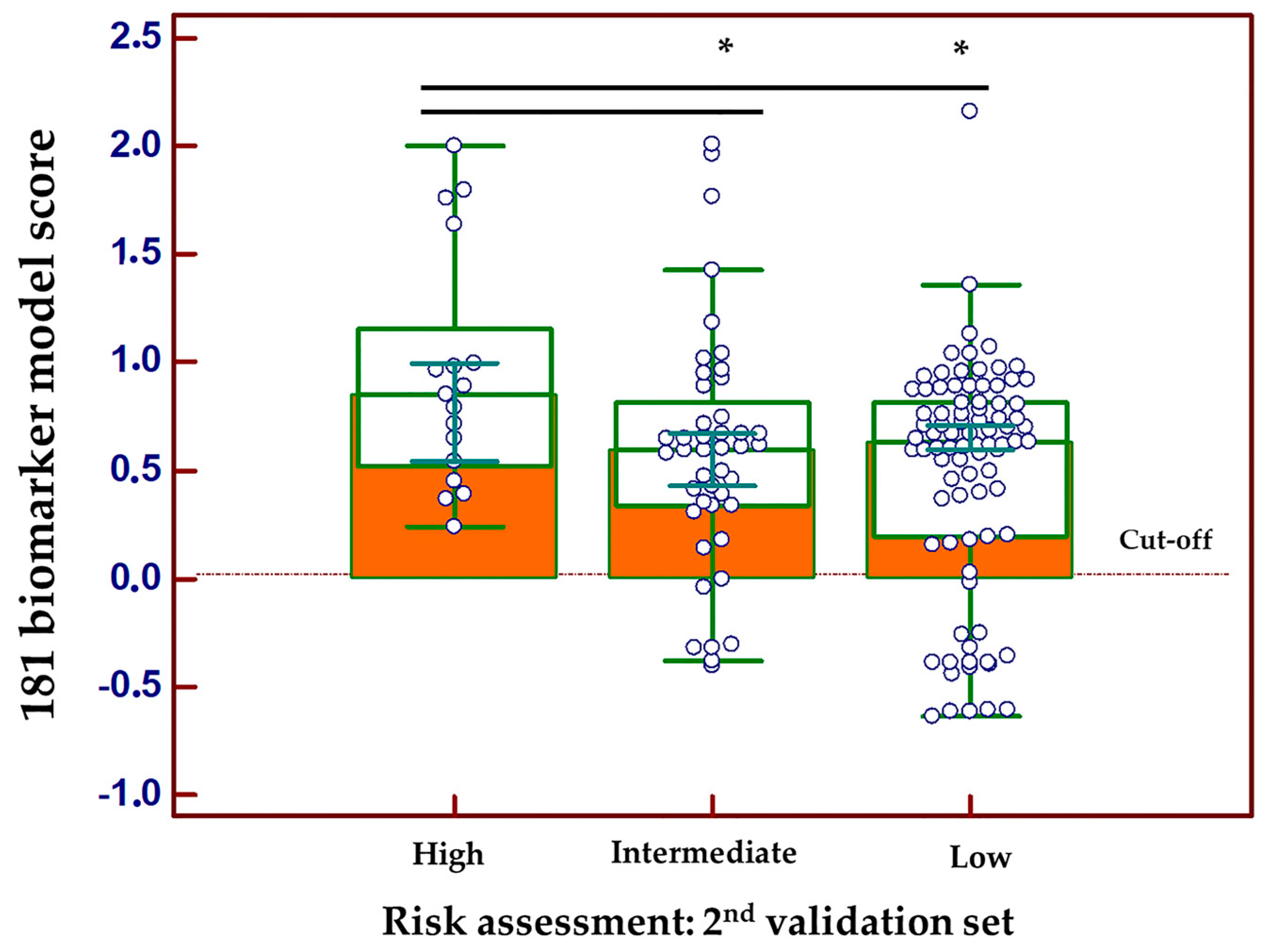
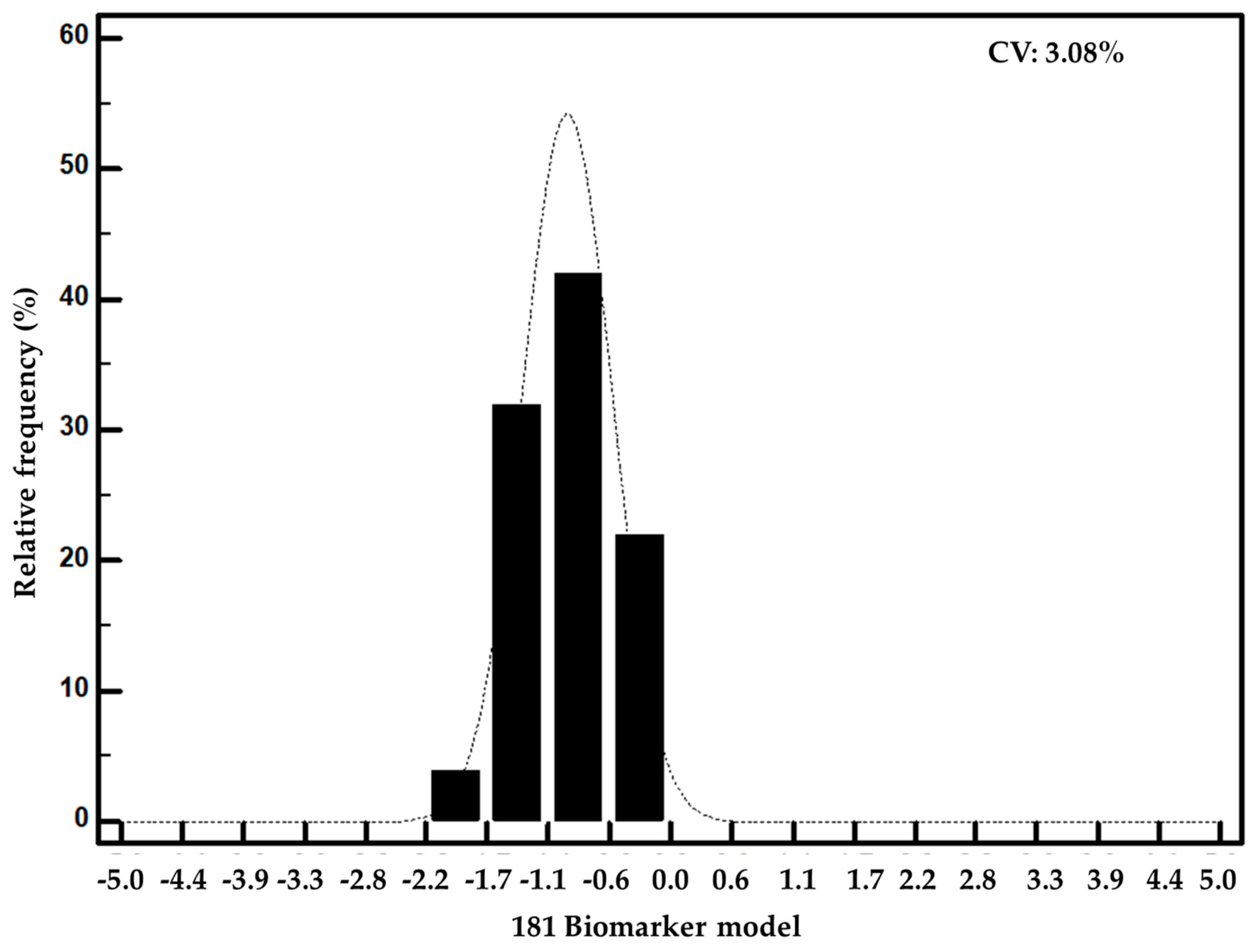
3.2. Development and Validation of a Biomarker Model Based on CE-MS Significant Peptides for PCa
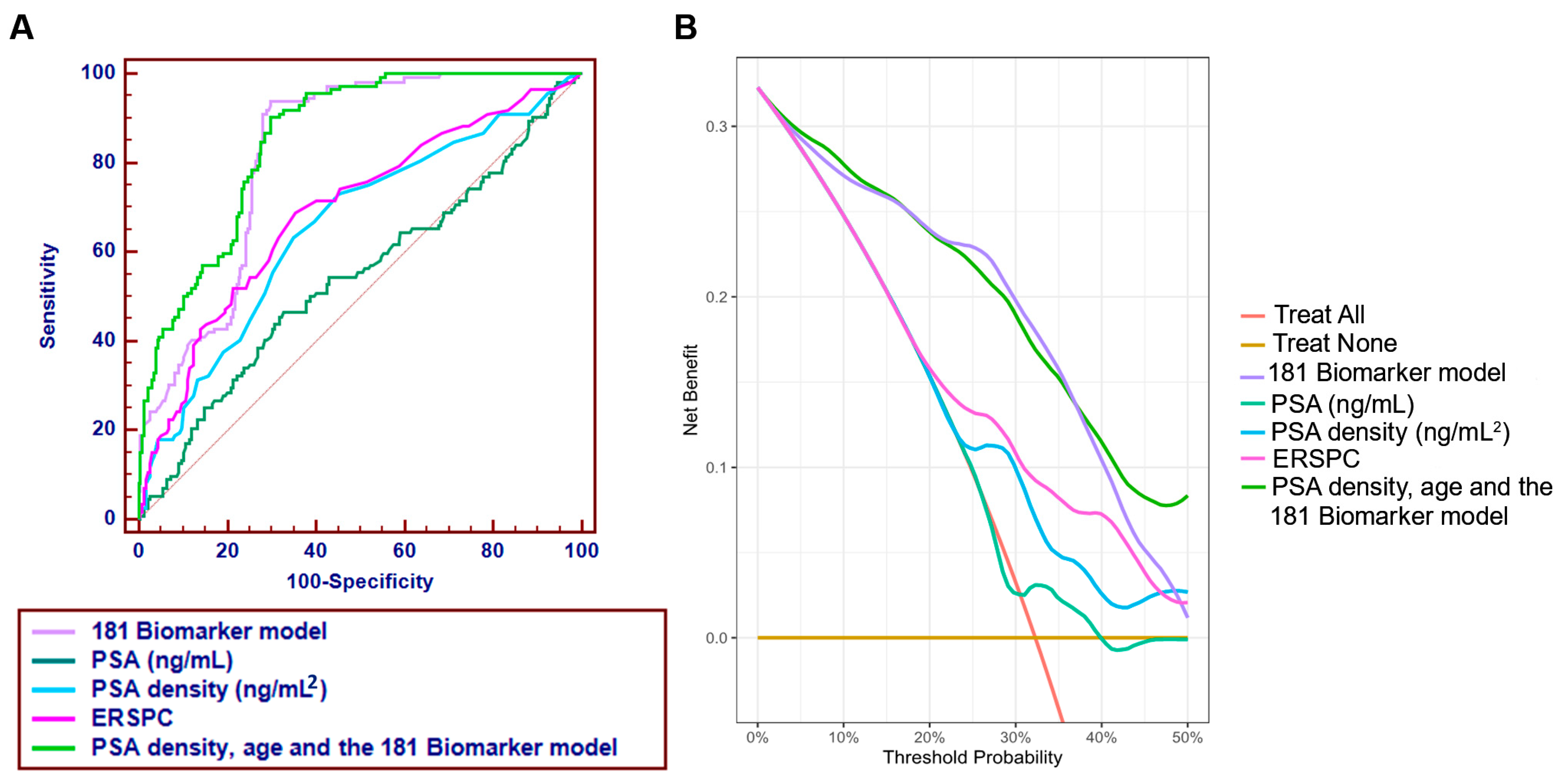
3.3. Comparator Models and Added Value over Clinical Standards
3.4. Correlation of the Peptide Profiling Data with PCa Progression
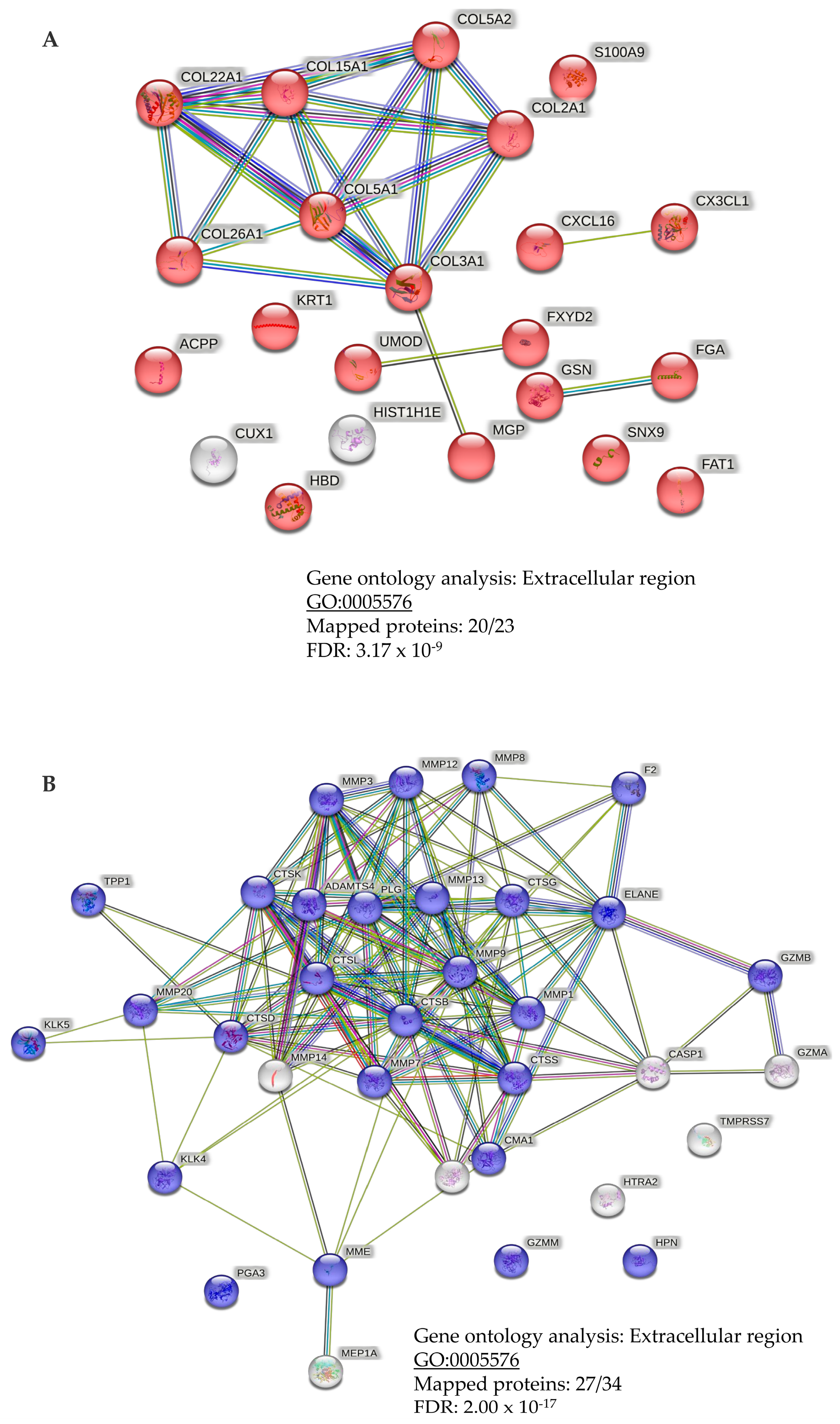
3.5. Link to Pathophysiology and Dysregulation of Proteases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Culp, M.B.; Soerjomataram, I.; Efstathiou, J.A.; Bray, F.; Jemal, A. Recent Global Patterns in Prostate Cancer Incidence and Mortality Rates. Eur. Urol. 2020, 77, 38–52. [Google Scholar] [CrossRef]
- Mottet, N.; van den Bergh, R.C.N.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer-2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2021, 79, 243–262. [Google Scholar] [CrossRef]
- Jemal, A.; Siegel, R.; Ward, E.; Murray, T.; Xu, J.; Smigal, C.; Thun, M.J. Cancer statistics. CA A Cancer J. Clin. 2006, 56, 106–130. [Google Scholar]
- Guichard, G.; Larre, S.; Gallina, A.; Lazar, A.; Faucon, H.; Chemama, S.; Allory, Y.; Patard, J.J.; Vordos, D.; Hoznek, A.; et al. Extended 21-sample needle biopsy protocol for diagnosis of prostate cancer in 1000 consecutive patients. Eur. Urol. 2007, 52, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Ankerst, D.P.; Miyamoto, R.; Nair, P.V.; Pollock, B.H.; Thompson, I.M.; Parekh, D.J. Yearly prostate specific antigen and digital rectal examination fluctuations in a screened population. J. Urol. 2009, 181, 2071–2075; discussion 2076. [Google Scholar] [CrossRef] [PubMed]
- Oerther, B.; Engel, H.; Bamberg, F.; Sigle, A.; Gratzke, C.; Benndorf, M. Cancer detection rates of the PI-RADSv2.1 assessment categories: Systematic review and meta-analysis on lesion level and patient level. Prostate Cancer Prostatic Dis. 2022, 25, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Pickersgill, N.A.; Vetter, J.M.; Andriole, G.L.; Shetty, A.S.; Fowler, K.J.; Mintz, A.J.; Siegel, C.L.; Kim, E.H. Accuracy and Variability of Prostate Multiparametric Magnetic Resonance Imaging Interpretation Using the Prostate Imaging Reporting and Data System: A Blinded Comparison of Radiologists. Eur. Urol. Focus 2020, 6, 267–272. [Google Scholar] [CrossRef]
- Gatti, M.; Faletti, R.; Calleris, G.; Giglio, J.; Berzovini, C.; Gentile, F.; Marra, G.; Misischi, F.; Molinaro, L.; Bergamasco, L.; et al. Prostate cancer detection with biparametric magnetic resonance imaging (bpMRI) by readers with different experience: Performance and comparison with multiparametric (mpMRI). Abdom. Radiol. 2019, 44, 1883–1893. [Google Scholar] [CrossRef]
- Roobol, M.J.; Kranse, R.; Bangma, C.H.; van Leenders, A.G.; Blijenberg, B.G.; van Schaik, R.H.; Kirkels, W.J.; Otto, S.J.; van der Kwast, T.H.; de Koning, H.J.; et al. Screening for prostate cancer: Results of the Rotterdam section of the European randomized study of screening for prostate cancer. Eur. Urol. 2013, 64, 530–539. [Google Scholar] [CrossRef]
- Arnold, M.; Karim-Kos, H.E.; Coebergh, J.W.; Byrnes, G.; Antilla, A.; Ferlay, J.; Renehan, A.G.; Forman, D.; Soerjomataram, I. Recent trends in incidence of five common cancers in 26 European countries since 1988: Analysis of the European Cancer Observatory. Eur. J. Cancer 2015, 51, 1164–1187. [Google Scholar] [CrossRef] [PubMed]
- Center, M.M.; Jemal, A.; Lortet-Tieulent, J.; Ward, E.; Ferlay, J.; Brawley, O.; Bray, F. International variation in prostate cancer incidence and mortality rates. Eur. Urol. 2012, 61, 1079–1092. [Google Scholar] [CrossRef] [PubMed]
- Haas, G.P.; Delongchamps, N.; Brawley, O.W.; Wang, C.Y.; de la Roza, G. The worldwide epidemiology of prostate cancer: Perspectives from autopsy studies. Can. J. Urol. 2008, 15, 3866–3871. [Google Scholar] [PubMed]
- Aubin, S.M.; Reid, J.; Sarno, M.J.; Blase, A.; Aussie, J.; Rittenhouse, H.; Rittmaster, R.; Andriole, G.L.; Groskopf, J. PCA3 molecular urine test for predicting repeat prostate biopsy outcome in populations at risk: Validation in the placebo arm of the dutasteride REDUCE trial. J. Urol. 2010, 184, 1947–1952. [Google Scholar] [CrossRef] [PubMed]
- Van Neste, L.; Hendriks, R.J.; Dijkstra, S.; Trooskens, G.; Cornel, E.B.; Jannink, S.A.; de Jong, H.; Hessels, D.; Smit, F.P.; Melchers, W.J.; et al. Detection of High-grade Prostate Cancer Using a Urinary Molecular Biomarker-Based Risk Score. Eur. Urol. 2016, 70, 740–748. [Google Scholar] [CrossRef] [PubMed]
- Tomlins, S.A.; Day, J.R.; Lonigro, R.J.; Hovelson, D.H.; Siddiqui, J.; Kunju, L.P.; Dunn, R.L.; Meyer, S.; Hodge, P.; Groskopf, J.; et al. Urine TMPRSS2:ERG Plus PCA3 for Individualized Prostate Cancer Risk Assessment. Eur. Urol. 2016, 70, 45–53. [Google Scholar] [CrossRef]
- McKiernan, J.; Donovan, M.J.; Margolis, E.; Partin, A.; Carter, B.; Brown, G.; Torkler, P.; Noerholm, M.; Skog, J.; Shore, N.; et al. A Prospective Adaptive Utility Trial to Validate Performance of a Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer in Patients with Prostate-specific Antigen 2–10 ng/mL at Initial Biopsy. Eur. Urol. 2018, 74, 731–738. [Google Scholar] [CrossRef]
- Frantzi, M.; Gomez Gomez, E.; Blanca Pedregosa, A.; Valero Rosa, J.; Latosinska, A.; Culig, Z.; Merseburger, A.S.; Luque, R.M.; Requena Tapia, M.J.; Mischak, H.; et al. CE-MS-based urinary biomarkers to distinguish non-significant from significant prostate cancer. Br. J. Cancer 2019, 120, 1120–1128. [Google Scholar] [CrossRef]
- Latosinska, A.; Siwy, J.; Mischak, H.; Frantzi, M. Peptidomics and proteomics based on CE-MS as a robust tool in clinical application: The past, the present, and the future. Electrophoresis 2019, 40, 2294–2308. [Google Scholar] [CrossRef] [PubMed]
- Frantzi, M.; Heidegger, I.; Roesch, M.C.; Gomez-Gomez, E.; Steiner, E.; Vlahou, A.; Mullen, W.; Guler, I.; Merseburger, A.S.; Mischak, H.; et al. Validation of diagnostic nomograms based on CE-MS urinary biomarkers to detect clinically significant prostate cancer. World J. Urol. 2022, 40, 2195–2203. [Google Scholar] [CrossRef]
- McShane, L.M.; Altman, D.G.; Sauerbrei, W.; Taube, S.E.; Gion, M.; Clark, G.M.; Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics. REporting recommendations for tumor MARKer prognostic studies (REMARK). Nat. Clin. Pract. Urol. 2005, 2, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Mischak, H.; Allmaier, G.; Apweiler, R.; Attwood, T.; Baumann, M.; Benigni, A.; Bennett, S.E.; Bischoff, R.; Bongcam-Rudloff, E.; Capasso, G.; et al. Recommendations for biomarker identification and qualification in clinical proteomics. Sci. Transl. Med. 2010, 2, 46ps42. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Gomez, E.; Carrasco-Valiente, J.; Blanca-Pedregosa, A.; Barco-Sanchez, B.; Fernandez-Rueda, J.L.; Molina-Abril, H.; Valero-Rosa, J.; Font-Ugalde, P.; Requena-Tapia, M.J. European Randomized Study of Screening for Prostate Cancer Risk Calculator: External Validation, Variability, and Clinical Significance. Urology 2017, 102, 85–91. [Google Scholar] [CrossRef]
- Heidegger, I.; Fritz, J.; Klocker, H.; Pichler, R.; Bektic, J.; Horninger, W. Age-Adjusted PSA Levels in Prostate Cancer Prediction: Updated Results of the Tyrol Prostate Cancer Early Detection Program. PLoS ONE 2015, 10, e0134134. [Google Scholar] [CrossRef]
- Epstein, J.I.; Allsbrook, W.C., Jr.; Amin, M.B.; Egevad, L.L.; Committee, I.G. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am. J. Surg. Pathol. 2005, 29, 1228–1242. [Google Scholar] [CrossRef]
- D’Amico, A.V.; Whittington, R.; Malkowicz, S.B.; Schultz, D.; Blank, K.; Broderick, G.A.; Tomaszewski, J.E.; Renshaw, A.A.; Kaplan, I.; Beard, C.J.; et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA 1998, 280, 969–974. [Google Scholar] [CrossRef]
- Metzger, J.; Negm, A.A.; Plentz, R.R.; Weismuller, T.J.; Wedemeyer, J.; Karlsen, T.H.; Dakna, M.; Mullen, W.; Mischak, H.; Manns, M.P.; et al. Urine proteomic analysis differentiates cholangiocarcinoma from primary sclerosing cholangitis and other benign biliary disorders. Gut 2013, 62, 122–130. [Google Scholar] [CrossRef]
- Mischak, H.; Kolch, W.; Aivaliotis, M.; Bouyssie, D.; Court, M.; Dihazi, H.; Dihazi, G.H.; Franke, J.; Garin, J.; Gonzalez de Peredo, A.; et al. Comprehensive human urine standards for comparability and standardization in clinical proteome analysis. Proteom. Clin. Appl. 2010, 4, 464–478. [Google Scholar] [CrossRef] [PubMed]
- Mischak, H.; Vlahou, A.; Ioannidis, J.P. Technical aspects and inter-laboratory variability in native peptide profiling: The CE-MS experience. Clin. Biochem. 2013, 46, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Zurbig, P.; Renfrow, M.B.; Schiffer, E.; Novak, J.; Walden, M.; Wittke, S.; Just, I.; Pelzing, M.; Neususs, C.; Theodorescu, D.; et al. Biomarker discovery by CE-MS enables sequence analysis via MS/MS with platform-independent separation. Electrophoresis 2006, 27, 2111–2125. [Google Scholar] [CrossRef]
- Frantzi, M.; Metzger, J.; Banks, R.E.; Husi, H.; Klein, J.; Dakna, M.; Mullen, W.; Cartledge, J.J.; Schanstra, J.P.; Brand, K.; et al. Discovery and validation of urinary biomarkers for detection of renal cell carcinoma. J. Proteom. 2014, 98, 44–58. [Google Scholar] [CrossRef] [PubMed]
- Siwy, J.; Mullen, W.; Golovko, I.; Franke, J.; Zurbig, P. Human urinary peptide database for multiple disease biomarker discovery. Proteom. Clin. Appl. 2011, 5, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Dakna, M.; Harris, K.; Kalousis, A.; Carpentier, S.; Kolch, W.; Schanstra, J.P.; Haubitz, M.; Vlahou, A.; Mischak, H.; Girolami, M. Addressing the challenge of defining valid proteomic biomarkers and classifiers. BMC Bioinform. 2010, 11, 594. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.; Papadopoulos, T.; Mischak, H.; Mullen, W. Comparison of CE-MS/MS and LC-MS/MS sequencing demonstrates significant complementarity in natural peptide identification in human urine. Electrophoresis 2014, 35, 1060–1064. [Google Scholar] [CrossRef] [PubMed]
- Magalhaes, P.; Pontillo, C.; Pejchinovski, M.; Siwy, J.; Krochmal, M.; Makridakis, M.; Carrick, E.; Klein, J.; Mullen, W.; Jankowski, J.; et al. Comparison of Urine and Plasma Peptidome Indicates Selectivity in Renal Peptide Handling. Proteom. Clin. Appl. 2018, 12, e1700163. [Google Scholar] [CrossRef]
- Pejchinovski, M.; Klein, J.; Ramirez-Torres, A.; Bitsika, V.; Mermelekas, G.; Vlahou, A.; Mullen, W.; Mischak, H.; Jankowski, V. Comparison of higher energy collisional dissociation and collision-induced dissociation MS/MS sequencing methods for identification of naturally occurring peptides in human urine. Proteom. Clin. Appl. 2015, 9, 531–542. [Google Scholar] [CrossRef]
- Frantzi, M.; van Kessel, K.E.; Zwarthoff, E.C.; Marquez, M.; Rava, M.; Malats, N.; Merseburger, A.S.; Katafigiotis, I.; Stravodimos, K.; Mullen, W.; et al. Development and Validation of Urine-based Peptide Biomarker Panels for Detecting Bladder Cancer in a Multi-center Study. Clin. Cancer Res 2016, 22, 4077–4086. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B-Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Vickers, A.J.; Elkin, E.B. Decision curve analysis: A novel method for evaluating prediction models. Med. Decis. Mak. Int. J. Soc. Med. Decis. Mak. 2006, 26, 565–574. [Google Scholar] [CrossRef]
- Klein, J.; Eales, J.; Zurbig, P.; Vlahou, A.; Mischak, H.; Stevens, R. Proteasix: A tool for automated and large-scale prediction of proteases involved in naturally occurring peptide generation. Proteomics 2013, 13, 1077–1082. [Google Scholar] [CrossRef]
- Igarashi, Y.; Eroshkin, A.; Gramatikova, S.; Gramatikoff, K.; Zhang, Y.; Smith, J.W.; Osterman, A.L.; Godzik, A. CutDB: A proteolytic event database. Nucleic Acids Res. 2007, 35, D546–D549. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Gaudet, P.; Michel, P.A.; Zahn-Zabal, M.; Britan, A.; Cusin, I.; Domagalski, M.; Duek, P.D.; Gateau, A.; Gleizes, A.; Hinard, V.; et al. The neXtProt knowledgebase on human proteins: 2017 update. Nucleic Acids Res. 2017, 45, D177–D182. [Google Scholar] [CrossRef]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef]
- Mengual, L.; Frantzi, M.; Mokou, M.; Ingelmo-Torres, M.; Vlaming, M.; Merseburger, A.S.; Roesch, M.C.; Culig, Z.; Alcaraz, A.; Vlahou, A.; et al. Multicentric validation of diagnostic tests based on BC-116 and BC-106 urine peptide biomarkers for bladder cancer in two prospective cohorts of patients. Br. J. Cancer 2022, 127, 2043–2051. [Google Scholar] [CrossRef]
- Tofte, N.; Lindhardt, M.; Adamova, K.; Bakker, S.J.L.; Beige, J.; Beulens, J.W.J.; Birkenfeld, A.L.; Currie, G.; Delles, C.; Dimos, I.; et al. Early detection of diabetic kidney disease by urinary proteomics and subsequent intervention with spironolactone to delay progression (PRIORITY): A prospective observational study and embedded randomised placebo-controlled trial. Lancet. Diabetes Endocrinol. 2020, 8, 301–312. [Google Scholar] [CrossRef]
- Staessen, J.A.; Wendt, R.; Yu, Y.L.; Kalbitz, S.; Thijs, L.; Siwy, J.; Raad, J.; Metzger, J.; Neuhaus, B.; Papkalla, A.; et al. Predictive performance and clinical application of COV50, a urinary proteomic biomarker in early COVID-19 infection: A prospective multicentre cohort study. Lancet. Digit. Health 2022, 4, e727–e737. [Google Scholar] [CrossRef] [PubMed]
- Mavrogeorgis, E.; Mischak, H.; Latosinska, A.; Siwy, J.; Jankowski, V.; Jankowski, J. Reproducibility Evaluation of Urinary Peptide Detection Using CE-MS. Molecules 2021, 26, 7260. [Google Scholar] [CrossRef]
- Gronberg, H.; Adolfsson, J.; Aly, M.; Nordstrom, T.; Wiklund, P.; Brandberg, Y.; Thompson, J.; Wiklund, F.; Lindberg, J.; Clements, M.; et al. Prostate cancer screening in men aged 50-69 years (STHLM3): A prospective population-based diagnostic study. Lancet. Oncol. 2015, 16, 1667–1676. [Google Scholar] [CrossRef]
- Gaggar, A.; Jackson, P.L.; Noerager, B.D.; O’Reilly, P.J.; McQuaid, D.B.; Rowe, S.M.; Clancy, J.P.; Blalock, J.E. A novel proteolytic cascade generates an extracellular matrix-derived chemoattractant in chronic neutrophilic inflammation. J. Immunol. 2008, 180, 5662–5669. [Google Scholar] [CrossRef]
- O’Connell, S.P.; Frantzi, M.; Latosinska, A.; Webb, M.; Mullen, W.; Pejchinovski, M.; Salji, M.; Mischak, H.; Cooper, C.S.; Clark, J.; et al. A Model to Detect Significant Prostate Cancer Integrating Urinary Peptide and Extracellular Vesicle RNA Data. Cancers 2022, 14, 1995. [Google Scholar] [CrossRef]
- Chuang, T.D.; Chen, S.J.; Lin, F.F.; Veeramani, S.; Kumar, S.; Batra, S.K.; Tu, Y.; Lin, M.F. Human prostatic acid phosphatase, an authentic tyrosine phosphatase, dephosphorylates ErbB-2 and regulates prostate cancer cell growth. J. Biol. Chem. 2010, 285, 23598–23606. [Google Scholar] [CrossRef]
- Korbakis, D.; Gregorakis, A.K.; Scorilas, A. Quantitative analysis of human kallikrein 5 (KLK5) expression in prostate needle biopsies: An independent cancer biomarker. Clin. Chem. 2009, 55, 904–913. [Google Scholar] [CrossRef]
- Michael, I.P.; Pampalakis, G.; Mikolajczyk, S.D.; Malm, J.; Sotiropoulou, G.; Diamandis, E.P. Human tissue kallikrein 5 is a member of a proteolytic cascade pathway involved in seminal clot liquefaction and potentially in prostate cancer progression. J. Biol. Chem. 2006, 281, 12743–12750. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Dong, B.; Yan, Y.; Hu, G.; Xu, Y. Association between MMP-2 expression and prostate cancer: A meta-analysis. Biomed. Rep. 2016, 4, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Miao, C.; Liang, C.; Shao, P.; Wang, Z.; Li, J. Silencing CAPN2 Expression Inhibited Castration-Resistant Prostate Cancer Cells Proliferation and Invasion via AKT/mTOR Signal Pathway. Biomed. Res. Int. 2017, 2017, 2593674. [Google Scholar] [CrossRef]
- Winter, R.N.; Kramer, A.; Borkowski, A.; Kyprianou, N. Loss of caspase-1 and caspase-3 protein expression in human prostate cancer. Cancer Res. 2001, 61, 1227–1232. [Google Scholar]
- Roufas, C.; Chasiotis, D.; Makris, A.; Efstathiades, C.; Dimopoulos, C.; Zaravinos, A. The Expression and Prognostic Impact of Immune Cytolytic Activity-Related Markers in Human Malignancies: A Comprehensive Meta-analysis. Front Oncol. 2018, 8, 27. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, J.; Schiffer, E.; Mannello, F.; Horn, L.C.; Ganzer, R.; Stolzenburg, J.U. Protease Expression Levels in Prostate Cancer Tissue Can Explain Prostate Cancer-Associated Seminal Biomarkers-An Explorative Concept Study. Int. J. Mol. Sci. 2017, 18, 976. [Google Scholar] [CrossRef]
- Corcoran, N.M.; Hovens, C.M.; Hong, M.K.; Pedersen, J.; Casey, R.G.; Connolly, S.; Peters, J.; Harewood, L.; Gleave, M.E.; Goldenberg, S.L.; et al. Underestimation of Gleason score at prostate biopsy reflects sampling error in lower volume tumours. BJU Int. 2012, 109, 660–664. [Google Scholar] [CrossRef] [PubMed]
- Nafie, S.; Wanis, M.; Khan, M. The Efficacy of Transrectal Ultrasound Guided Biopsy Versus Transperineal Template Biopsy of the Prostate in Diagnosing Prostate Cancer in Men with Previous Negative Transrectal Ultrasound Guided Biopsy. Urol. J. 2017, 14, 3008–3012. [Google Scholar] [PubMed]

| Baseline Characteristics | Discovery Phase (n = 413) | 1st Validation Phase (n = 410) | p-Value (1st Validation vs. Discovery) | 2nd Validation Phase (n = 147) | p-Value (2nd Validation vs. Discovery) |
|---|---|---|---|---|---|
| Median age (95% CI; yr) | 64.0 (63.4–64.8) | 64.0 (63.0–65.0) | 0.6488 ¥ | 66.0 (64.2–67.0) | 0.3414 ¥ |
| PSA median (95% CI; ng/mL) | 5.4 (5.1–5.8) | 5.1 (4.8–5.4) | 0.6537 ¥ | 5.2 (4.5–5.9) | 0.6298 ¥ |
| Digital Rectal Examination (normal/suspicious/NA) | 339/74 | 340/70 | 0.7676 * | 90/20/37 | 0.3534 * |
| Previous biopsies (Y/N) | 109/304 | 99/311 | 0.5258 * | 0.8033 * | |
| Prostate volume (95% CI; mL) | 36.0 (34–39; n = 364) | 35.0 (34–37; n = 357) | 0.4416 ¥ | 40.0 (35–45; n = 135) | 0.0305 ¥ |
| PSA density (95% CI; ng/mL2) | 0.14 (0.13–0.15; n = 364) | 0.14 (0.13–0.15; n = 357) | 0.9379 ¥ | 0.14 (0.12–0.15; n = 135) | 0.5568 ¥ |
| Median urinary creatinine (95% CI; mmol/L) | 8.0 (7.3–8.3) | 7.8 (7.3–8.3) | 0.3696 ¥ | 8.8 (7.6–10.3) | 0.1163 ¥ |
| Disease pathology | |||||
| 65 (46.8%) | 66 (47.8%) | 0.9132 * | 99 (67.4%) | 0.1838 * |
GS 4 + 3 | 49 (35.3%)/ 14 (10.0%) | 46 (33.3%)/ 15 (10.9%) | 31 (21.1%) 4 (2.7%) | ||
| 6 (4.3%) | 8 (5.9%) | 8 (2.7%) | ||
| 5 (3.6%) | 3 (2.2%) | 5 (5.4%) | ||
| Non-PCa aetiologies | |||||
| 241 (88.0%) | 241 (88.6%) | 0.1515 * | - | |
| 18 (6.6%) | 16 (5.9%) | - | ||
| 15 (5.4%) | 15 (5.5%) | - | ||
| Baseline Characteristics | Group 1: Non-PCa | Group 2: PCa | p-Value Group 1 vs. Group 2 |
|---|---|---|---|
| 63.0 (57–69) | 66.0 (61–71) | <0.0001 ¥ |
| 5.1 (3.8–6.9) | 5.7 (4.0–8.0) | 0.0023 ¥ |
| 60/486 | 84/193 | 0.0249 * |
| 159/387 | 49/228 | 0.8646 * |
| 38.0 (29–52; n = 476) | 30.0 (22.9–43.1; n = 245) | <0.0001 ¥ |
| 0.13 (0.09–0.18; n = 476) | 0.18 (0.13–0.26; n = 245) | <0.0001 ¥ |
| 7.7 (5.5–10.3) | 8.0 (5.8–10.6) | 0.3636 ¥ |
| 181 Biomarker Model | PSAD | ERSPC | Diagnostic Nomogram | |||||
|---|---|---|---|---|---|---|---|---|
| Sensitivity Thresholds | Specificity | 95% CI | Specificity | 95% CI | Specificity | 95% CI | Specificity | 95% CI |
| 80.0 | 71.4 | 65.1–76.6 | 37.1 | 23.4–56.0 | 40.7 | 28.2–57.8 | 72.8 | 65.4–78.2 |
| 90.0 | 70.4 | 58.4–77.0 | 19.2 | 8.33–34.7 | 23.0 | 11.9–37.2 | 67.5 | 56.4–74.9 |
| 95.0 | 55.5 | 32.4–74.1 | 7.9 | 3.6–15.6 | 12.9 | 2.8–22.5 | 56.0 | 40.5–66.7 |
| 97.5 | 31.5 | 1.2–54.6 | 4.9 | 1.6–10.1 | 4.1 | 0.7–15.9 | 45.3 | 14.0–58.6 |
| Mass [Da] | CE-Time [min] | Peptide Sequence | Protein Name | p-Value | Spearman’s Rho |
|---|---|---|---|---|---|
| 1353.66 | 25.88 | PVGpSGKDGANGIpG | Collagen alpha-1(II) | 0.0051 | −0.098 |
| 3718.72 | 32.42 | SGPPGRAGEPGLQGPAGPpGEKGEPGDDGpSGAEGPpGPQG | Collagen alpha- 1(II) | 0.0100 | −0.090 |
| 2280.97 | 26.16 | ADGQpGAKGEQGEAGQKGDAGApGP | Collagen alpha- 1(II) | 0.0116 | −0.088 |
| 2412.11 | 27.18 | RGGAGPPGpEGGKGAAGPpGpPGAAGTpG | Collagen alpha-1(III) | 0.0377 | 0.072 |
| 1873.83 | 31.95 | PPGpTGPGGDKGDTGPpGPQG | Collagen alpha-1(III) | 0.0341 | 0.074 |
| 1141.51 | 26.28 | EpGRDGVpGGpG | Collagen alpha-1(III) | 0.0341 | 0.074 |
| 2130.97 | 32.98 | GpTGpIGPpGpAGQPGDKGEGGAP | Collagen alpha-1(III) | 0.0335 | 0.074 |
| 2507.13 | 22.82 | ApGQNGEPGGkGERGAPGEkGEGGPpG | Collagen alpha-1(III) | 0.0322 | 0.075 |
| 2663.21 | 23.57 | NRGERGSEGSPGHpGQpGPPGpPGApGP | Collagen alpha-1(III) | 0.0302 | 0.076 |
| 1794.80 | 24.01 | GNDGApGKNGERGGpGGpGP | Collagen alpha-1(III) | 0.0285 | 0.076 |
| 1531.68 | 39.25 | GLpGPpGSNGNpGPpGP | Collagen alpha-1(III) | 0.0271 | 0.077 |
| 1796.84 | 21.01 | ApGPQGpRGDKGETGERG | Collagen alpha-1(III) | 0.0243 | 0.079 |
| 883.41 | 23.48 | PpGENGKpG | Collagen alpha-1(III) | 0.0144 | 0.085 |
| 2264.05 | 22.67 | KGDAGApGApGGKGDAGApGERGpPG | Collagen alpha-1(III) | 0.0085 | 0.092 |
| 2135.96 | 25.80 | GDAGApGApGGKGDAGApGERGPpG | Collagen alpha-1(III) | 0.0080 | 0.092 |
| 1594.73 | 23.13 | ApGGKGDAGApGERGpPG | Collagen alpha-1(III) | 0.0029 | 0.104 |
| 2679.19 | 23.56 | PGMPGADGpPGHPGKEGppGEKGGQGpPG | Collagen alpha-1(V) | 0.0244 | 0.078 |
| 1522.73 | 22.99 | KGDpGpAGLpGKDGpP | Collagen alpha-1(V) | 0.0158 | 0.084 |
| 1176.56 | 26.86 | KPGTDVFmGpP | Collagen alpha-1(XV) | 0.0053 | 0.097 |
| 2226.96 | 33.46 | GNSGEKGDQGFQGQPGFPGPpGP | Collagen alpha-1(XVI) | 0.0046 | 0.099 |
| 3023.39 | 24.65 | ppGAKGQEGAHGAPGAAGNPGAPGHVGAPGPSGpP | Collagen alpha-1(XXII) | 0.0382 | 0.072 |
| 1540.74 | 39.81 | GPpGVPGpPGpGGSPGLP | Collagen alpha-1(XXII) | 0.0344 | 0.074 |
| 1536.72 | 19.91 | KDGPnGPpGpPGTKGE | Collagen alpha-1(XXII) | 0.0328 | 0.074 |
| 935.45 | 23.82 | GRpGPpGPpG | Collagen alpha-1(XXVI) | 0.0326 | −0.075 |
| 1240.54 | 27.23 | ApGEDGRpGPpGS | Collagen alpha-2(V) | 0.0425 | 0.071 |
| 2480.21 | 23.24 | EAGENQKQPEKNAGPTARTSATVP | C-X-C chemokine 16 | 0.0361 | 0.073 |
| 1728.76 | 36.62 | ESVVLEPEAT | Fractalkine | 0.0011 | 0.114 |
| 2272.24 | 23.91 | SETAPAAPAAPAPAEKTPVKKKA | Histone H1.4 | 0.0070 | 0.094 |
| 937.46 | 34.16 | PVQGQQQGP | Homeobox protein cut-like 1 | 0.0080 | 0.092 |
| 1276.71 | 19.96 | KVVAGVANALAHK | Hemoglobin delta | 0.0086 | 0.091 |
| 879.50 | 19.95 | KLGHPDTL | Protein S100-A9 | 0.0088 | 0.091 |
| 2567.13 | 34.83 | ATPLYINI | Protocadherin Fat 1 | 0.0131 | 0.086 |
| 2501.11 | 34.31 | ASTAQASSSAASNNHQVGSGNDPWSA | Sorting nexin-9 | 0.0378 | 0.072 |
| 1294.62 | 19.43 | ADHEGTHSTKRG | Fibrinogen alpha chain | 0.0378 | 0.072 |
| 1013.37 | 25.06 | cDDYRLcE | Matrix Gla Protein | 0.0433 | 0.070 |
| 1159.61 | 26.41 | SGSVIDQSRVL | Uromodulin | 0.0440 | −0.070 |
| 1099.49 | 28.06 | DGGGSPKGDVDP | Sodium/potassium-transporting ATPase subunit gamma | 0.0406 | −0.071 |
| 1934.79 | 19.91 | GSGGSSYGSGGGSYGSGGGGGGGRG | Keratin; type II cytoskeletal 1 | 0.0335 | −0.074 |
| 1732.78 | 28.30 | WVGTGASEAEKTGAQEL | Gelsolin | 0.0126 | −0.087 |
| 976.58 | 20.52 | KELKFVTL | Prostatic acid phosphatase | 0.0022 | −0.107 |
| Protease | Uniprot ID | Symbol | # CS | Xcorr Score |
|---|---|---|---|---|
| Matrix metalloproteinase-20 | O60882 | MMP20 | 10 | 618.96 |
| Matrix metalloproteinase-25 | Q9NPA2 | MMP25 | 73 | 283.10 |
| Stromelysin-1 | P08254 | MMP3 | 93 | 260.70 |
| Kallikrein-5 | Q9Y337 | KLK5 | 6 | 250.59 |
| 72 kDa type IV collagenase | P08253 | MMP2 | 53 | 233.08 |
| Calpain-2 catalytic subunit | P17655 | CAPN2 | 146 | 231.49 |
| Transmembrane protease serine 7 | Q7RTY8 | TMPRSS7 | 78 | 220.89 |
| Caspase-1 | P29466 | CASP1 | 8 | 178.55 |
| Macrophage metalloelastase | P39900 | MMP12 | 134 | 162.13 |
| Calpain-1 catalytic subunit | P07384 | CAPN1 | 147 | 156.59 |
| Cathepsin K | P43235 | CTSK | 77 | −66.68 |
| Meprin A subunit alpha | Q16819 | MEP1A | 109 | −92.86 |
| Kallikrein-4 | Q9Y5K2 | KLK4 | 18 | −114.74 |
| Prothrombin | P00734 | F2 | 5 | −192.73 |
| Granzyme A | P12544 | GZMA | 13 | −220.95 |
| Plasminogen | P00747 | PLG | 13 | −220.95 |
| Cathepsin G | P08311 | CTSG | 36 | −340.28 |
| Serine protease hepsin | P05981 | HPN | 15 | −363.28 |
| Chymase | P23946 | CMA1 | 5 | −500.00 |
| Tripeptidyl-peptidase 1 | O14773 | TPP1 | 5 | −500.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frantzi, M.; Culig, Z.; Heidegger, I.; Mokou, M.; Latosinska, A.; Roesch, M.C.; Merseburger, A.S.; Makridakis, M.; Vlahou, A.; Blanca-Pedregosa, A.; et al. Mass Spectrometry-Based Biomarkers to Detect Prostate Cancer: A Multicentric Study Based on Non-Invasive Urine Collection without Prior Digital Rectal Examination. Cancers 2023, 15, 1166. https://doi.org/10.3390/cancers15041166
Frantzi M, Culig Z, Heidegger I, Mokou M, Latosinska A, Roesch MC, Merseburger AS, Makridakis M, Vlahou A, Blanca-Pedregosa A, et al. Mass Spectrometry-Based Biomarkers to Detect Prostate Cancer: A Multicentric Study Based on Non-Invasive Urine Collection without Prior Digital Rectal Examination. Cancers. 2023; 15(4):1166. https://doi.org/10.3390/cancers15041166
Chicago/Turabian StyleFrantzi, Maria, Zoran Culig, Isabel Heidegger, Marika Mokou, Agnieszka Latosinska, Marie C. Roesch, Axel S. Merseburger, Manousos Makridakis, Antonia Vlahou, Ana Blanca-Pedregosa, and et al. 2023. "Mass Spectrometry-Based Biomarkers to Detect Prostate Cancer: A Multicentric Study Based on Non-Invasive Urine Collection without Prior Digital Rectal Examination" Cancers 15, no. 4: 1166. https://doi.org/10.3390/cancers15041166
APA StyleFrantzi, M., Culig, Z., Heidegger, I., Mokou, M., Latosinska, A., Roesch, M. C., Merseburger, A. S., Makridakis, M., Vlahou, A., Blanca-Pedregosa, A., Carrasco-Valiente, J., Mischak, H., & Gomez-Gomez, E. (2023). Mass Spectrometry-Based Biomarkers to Detect Prostate Cancer: A Multicentric Study Based on Non-Invasive Urine Collection without Prior Digital Rectal Examination. Cancers, 15(4), 1166. https://doi.org/10.3390/cancers15041166







