Deep Learning with Transformer or Convolutional Neural Network in the Assessment of Tumor-Infiltrating Lymphocytes (TILs) in Breast Cancer Based on US Images: A Dual-Center Retrospective Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Image Acquisition
2.3. Clinical and Pathological Analysis
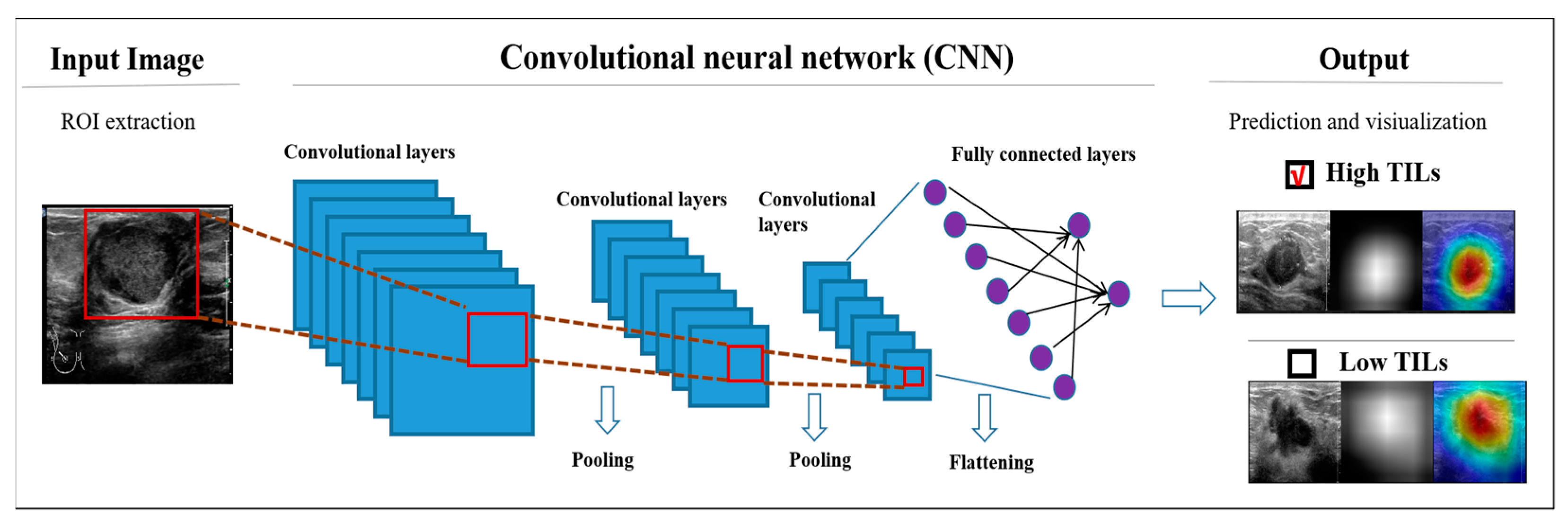
2.4. DL Models
2.5. Stratified Analysis to Assess the Diagnostic Value
2.6. Statistical Analysis
3. Results
3.1. Baseline Characters
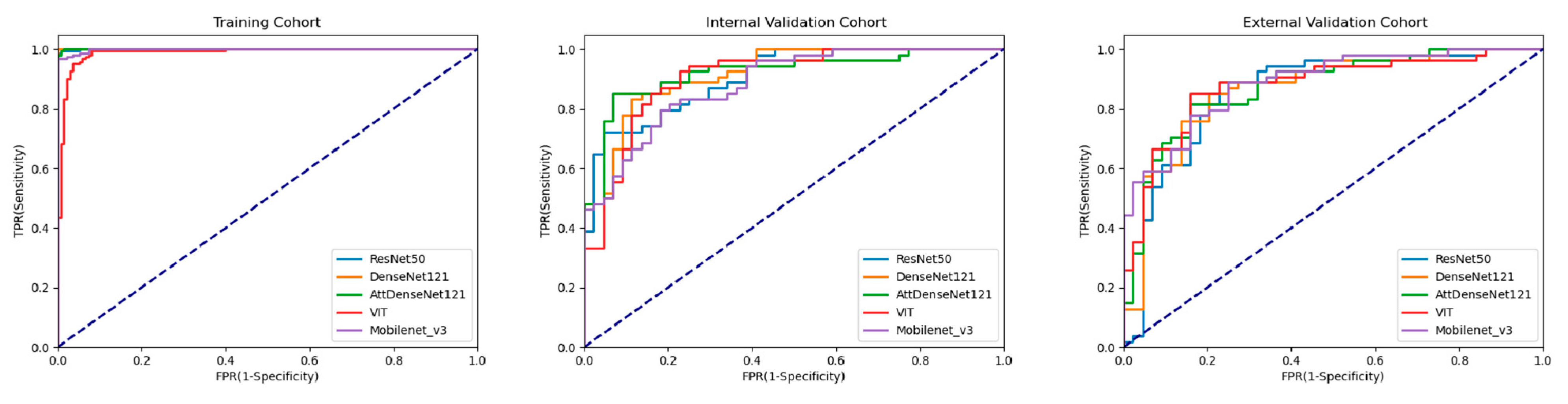
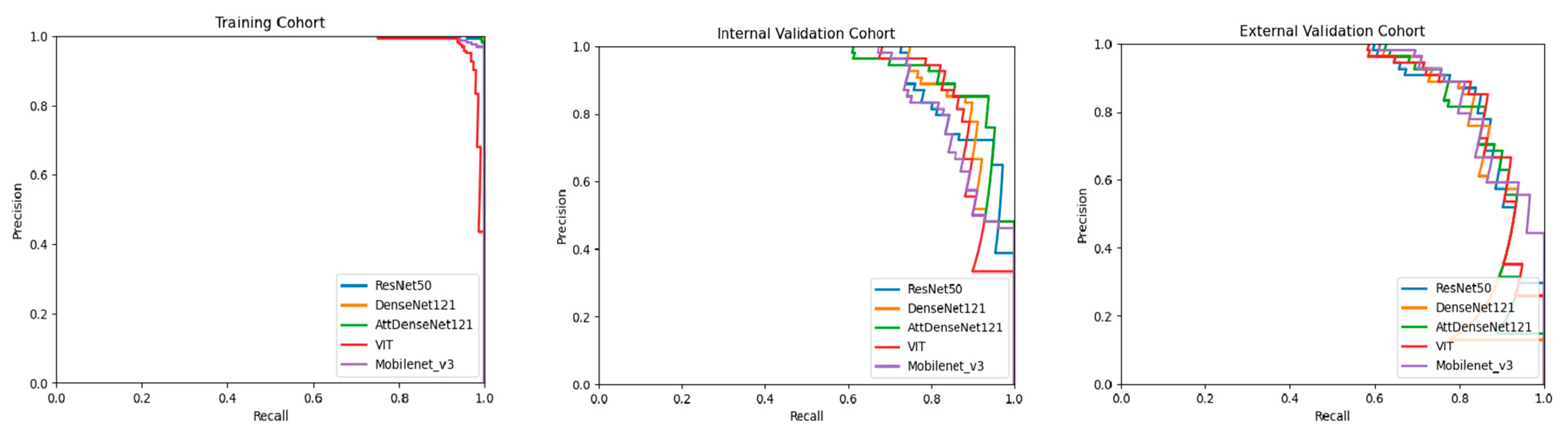
3.2. Performance of DL Models
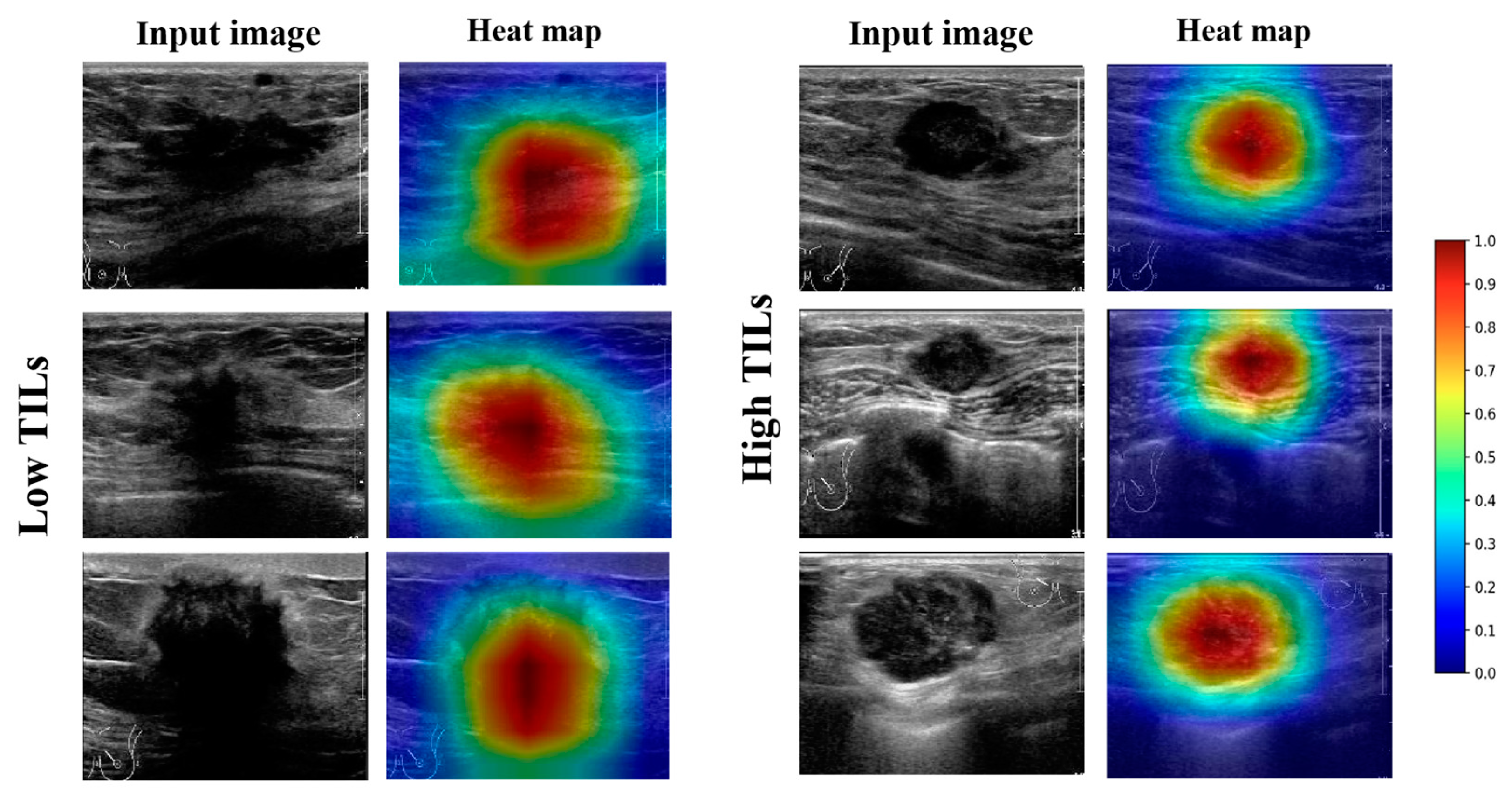
3.3. Visual Interpretation of the Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luen, S.J.; Savas, P.; Fox, S.B.; Salgado, R.; Loi, S. Tumour-infiltrating lymphocytes and the emerging role of immunotherapy in breast cancer. Pathology 2017, 49, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Perez, E.A.; Ballman, K.V.; Tenner, K.S.; Thompson, E.A.; Badve, S.S.; Bailey, H.; Baehner, F.L. Association of Stromal Tumor-Infiltrating Lymphocytes With Recurrence-Free Survival in the N9831 Adjuvant Trial in Patients With Early-Stage HER2-Positive Breast Cancer. JAMA Oncol. 2016, 2, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Dieci, M.V.; Miglietta, F.; Guarneri, V. Immune Infiltrates in Breast Cancer: Recent Updates and Clinical Implications. Cells 2021, 10, 223. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; von Minckwitz, G.; Brase, J.C.; Sinn, B.V.; Gade, S.; Kronenwett, R.; Pfitzner, B.M.; Salat, C.; Loi, S.; Schmitt, W.D.; et al. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2015, 33, 983–991. [Google Scholar] [CrossRef]
- Denkert, C.; von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Heppner, B.I.; Weber, K.E.; Budczies, J.; Huober, J.; Klauschen, F.; Furlanetto, J.; et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: A pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018, 19, 40–50. [Google Scholar] [CrossRef]
- Salgado, R.; Denkert, C.; Demaria, S.; Sirtaine, N.; Klauschen, F.; Pruneri, G.; Wienert, S.; Van den Eynden, G.; Baehner, F.L.; Penault-Llorca, F.; et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2015, 26, 259–271. [Google Scholar] [CrossRef]
- Çelebi, F.; Agacayak, F.; Ozturk, A.; Ilgun, S.; Ucuncu, M.; Iyigun, Z.E.; Ordu, Ç.; Pilanci, K.N.; Alco, G.; Gultekin, S.; et al. Usefulness of imaging findings in predicting tumor-infiltrating lymphocytes in patients with breast cancer. Eur. Radiol. 2020, 30, 2049–2057. [Google Scholar] [CrossRef]
- Candelaria, R.P.; Spak, D.A.; Rauch, G.M.; Huo, L.; Bassett, R.L.; Santiago, L.; Scoggins, M.E.; Guirguis, M.S.; Patel, M.M.; Whitman, G.J.; et al. BI-RADS Ultrasound Lexicon Descriptors and Stromal Tumor-Infiltrating Lymphocytes in Triple-Negative Breast Cancer. Acad Radiol. 2022, 29 (Suppl. 1), S35–S41. [Google Scholar] [CrossRef]
- Lee, H.J.; Lee, J.E.; Jeong, W.G.; Ki, S.Y.; Park, M.H.; Lee, J.S.; Nah, Y.K.; Lim, H.S. HER2-Positive Breast Cancer: Association of MRI and Clinicopathologic Features With Tumor-Infiltrating Lymphocytes. AJR Am. J. Roentgenol. 2022, 218, 258–269. [Google Scholar] [CrossRef]
- Ku, Y.J.; Kim, H.H.; Cha, J.H.; Shin, H.J.; Chae, E.Y.; Choi, W.J.; Lee, H.J.; Gong, G. Predicting the level of tumor-infiltrating lymphocytes in patients with triple-negative breast cancer: Usefulness of breast MRI computer-aided detection and diagnosis. J. Magn. Reson. Imaging JMRI 2018, 47, 760–766. [Google Scholar] [CrossRef]
- Braman, N.; Prasanna, P.; Whitney, J.; Singh, S.; Beig, N.; Etesami, M.; Bates, D.D.B.; Gallagher, K.; Bloch, B.N.; Vulchi, M.; et al. Association of Peritumoral Radiomics With Tumor Biology and Pathologic Response to Preoperative Targeted Therapy for HER2 (ERBB2)-Positive Breast Cancer. JAMA Netw. Open 2019, 2, e192561. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.J.; Jin, Z.; Zhang, Y.L.; Liang, Y.S.; Cheng, Z.X.; Chen, L.X.; Liang, Y.Y.; Wei, X.H.; Kong, Q.C.; Guo, Y.; et al. Whole-Lesion Histogram Analysis of the Apparent Diffusion Coefficient as a Quantitative Imaging Biomarker for Assessing the Level of Tumor-Infiltrating Lymphocytes: Value in Molecular Subtypes of Breast Cancer. Front. Oncol. 2020, 10, 611571. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, X.; Teng, X.; Rubin, D.L.; Napel, S.; Daniel, B.L.; Li, R. Magnetic resonance imaging and molecular features associated with tumor-infiltrating lymphocytes in breast cancer. Breast Cancer Res. BCR 2018, 20, 101. [Google Scholar] [CrossRef] [PubMed]
- Murakami, W.; Tozaki, M.; Sasaki, M.; Hida, A.I.; Ohi, Y.; Kubota, K.; Sagara, Y. Correlation between (18)F-FDG uptake on PET/MRI and the level of tumor-infiltrating lymphocytes (TILs) in triple-negative and HER2-positive breast cancer. Eur. J. Radiol. 2020, 123, 108773. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.J.; Kong, Q.C.; Cheng, Z.X.; Liang, Y.S.; Jin, Z.; Chen, L.X.; Hu, W.K.; Liang, Y.Y.; Wei, X.H.; Guo, Y.; et al. Performance of radiomics models for tumour-infiltrating lymphocyte (TIL) prediction in breast cancer: The role of the dynamic contrast-enhanced (DCE) MRI phase. Eur. Radiol. 2022, 32, 864–875. [Google Scholar] [CrossRef] [PubMed]
- Akkus, Z.; Cai, J.; Boonrod, A.; Zeinoddini, A.; Weston, A.D.; Philbrick, K.A.; Erickson, B.J. A Survey of Deep-Learning Applications in Ultrasound: Artificial Intelligence-Powered Ultrasound for Improving Clinical Workflow. J. Am. Coll. Radiol. 2019, 16, 1318–1328. [Google Scholar] [CrossRef]
- Guo, X.; Liu, Z.; Sun, C.; Zhang, L.; Wang, Y.; Li, Z.; Shi, J.; Wu, T.; Cui, H.; Zhang, J.; et al. Deep learning radiomics of ultrasonography: Identifying the risk of axillary non-sentinel lymph node involvement in primary breast cancer. EBioMedicine 2020, 60, 103018. [Google Scholar] [CrossRef]
- Qian, X.; Pei, J.; Zheng, H.; Xie, X.; Yan, L.; Zhang, H.; Han, C.; Gao, X.; Zhang, H.; Zheng, W.; et al. Prospective assessment of breast cancer risk from multimodal multiview ultrasound images via clinically applicable deep learning. Nat. Biomed. Eng. 2021, 5, 522–532. [Google Scholar] [CrossRef]
- Zhou, L.Q.; Wu, X.L.; Huang, S.Y.; Wu, G.G.; Ye, H.R.; Wei, Q.; Bao, L.Y.; Deng, Y.B.; Li, X.R.; Cui, X.W.; et al. Lymph Node Metastasis Prediction from Primary Breast Cancer US Images Using Deep Learning. Radiology 2020, 294, 19–28. [Google Scholar] [CrossRef]
- Gu, J.; Tong, T.; He, C.; Xu, M.; Yang, X.; Tian, J.; Jiang, T.; Wang, K. Deep learning radiomics of ultrasonography can predict response to neoadjuvant chemotherapy in breast cancer at an early stage of treatment: A prospective study. Eur. Radiol. 2022, 32, 2099–2109. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, D.; Tang, S.C.; Luo, X.M.; Chuan, Z.R.; Lv, W.Z.; Jiang, F.; Ni, X.J.; Cui, X.W.; Dietrich, C.F. Deep learning with convolutional neural network in the assessment of breast cancer molecular subtypes based on US images: A multicenter retrospective study. Eur. Radiol. 2021, 31, 3673–3682. [Google Scholar] [CrossRef] [PubMed]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017. [Google Scholar]
- Burstein, H.J.; Curigliano, G.; Loibl, S.; Dubsky, P.; Gnant, M.; Poortmans, P.; Colleoni, M.; Denkert, C.; Piccart-Gebhart, M.; Regan, M.; et al. Estimating the benefits of therapy for early-stage breast cancer: The St. Gallen International Consensus Guidelines for the primary therapy of early breast cancer 2019. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 1541–1557. [Google Scholar] [CrossRef] [PubMed]
- Loi, S. The ESMO clinical practise guidelines for early breast cancer: Diagnosis, treatment and follow-up: On the winding road to personalized medicine. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 1183–1184. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Mayer, A.T.; Li, R. Integrated imaging and molecular analysis to decipher tumor microenvironment in the era of immunotherapy. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar] [CrossRef]
- Fukui, K.; Masumoto, N.; Shiroma, N.; Kanou, A.; Sasada, S.; Emi, A.; Kadoya, T.; Yokozaki, M.; Arihiro, K.; Okada, M. Novel tumor-infiltrating lymphocytes ultrasonography score based on ultrasonic tissue findings predicts tumor-infiltrating lymphocytes in breast cancer. Breast Cancer 2019, 26, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Bian, T.; Wu, Z.; Lin, Q.; Mao, Y.; Wang, H.; Chen, J.; Chen, Q.; Fu, G.; Cui, C.; Su, X. Evaluating Tumor-Infiltrating Lymphocytes in Breast Cancer Using Preoperative MRI-Based Radiomics. J. Magn. Reson. Imaging JMRI 2022, 55, 772–784. [Google Scholar] [CrossRef] [PubMed]
- Su, G.H.; Xiao, Y.; Jiang, L.; Zheng, R.C.; Wang, H.; Chen, Y.; Gu, Y.J.; You, C.; Shao, Z.M. Radiomics features for assessing tumor-infiltrating lymphocytes correlate with molecular traits of triple-negative breast cancer. J. Transl. Med. 2022, 20, 471. [Google Scholar] [CrossRef]
- Shad, H.S.; Rizvee, M.M.; Roza, N.T.; Hoq, S.M.A.; Monirujjaman Khan, M.; Singh, A.; Zaguia, A.; Bourouis, S. Comparative Analysis of Deepfake Image Detection Method Using Convolutional Neural Network. Comput. Intell. Neurosci. 2021, 2021, 3111676. [Google Scholar] [CrossRef]
- Ogawa, N.; Maeda, K.; Ogawa, T.; Haseyama, M. Deterioration Level Estimation Based on Convolutional Neural Network Using Confidence-Aware Attention Mechanism for Infrastructure Inspection. Sensors 2022, 22, 382. [Google Scholar] [CrossRef]
- Jia, Y.; Zhu, Y.; Li, T.; Song, X.; Duan, Y.; Yang, D.; Nie, F. Evaluating Tumor-Infiltrating Lymphocytes in Breast Cancer: The Role of Conventional Ultrasound and Contrast-Enhanced Ultrasound. J. Ultrasound Med. 2022, 9999, 1–12. [Google Scholar] [CrossRef]
- Tamaki, K.; Sasano, H.; Ishida, T.; Ishida, K.; Miyashita, M.; Takeda, M.; Amari, M.; Harada-Shoji, N.; Kawai, M.; Hayase, T.; et al. The correlation between ultrasonographic findings and pathologic features in breast disorders. Jpn. J. Clin. Oncol. 2010, 40, 905–912. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 1–48. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y. Courville A: Convolutional networks. In Deep Learning; MIT Press: Cambridge, MA, USA, 2016; Volume 2016, pp. 330–372. [Google Scholar]
- Glorot, X.; Bordes, A.; Bengio, Y. Deep sparse rectifier neural networks. In Proceedings of the Fourteenth International Conference on Artificial Intelligence and Statistics, Fort Lauderdale, FL, USA, 11–13 April 2011; JMLR Workshop and Conference Proceedings. pp. 315–323. Available online: http://proceedings.mlr.press/v15/glorot11a (accessed on 3 December 2022).
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning, Lille, France, 7–9 July 2015; Volume 37, pp. 448–456. Available online: http://proceedings.mlr.press/v37/ioffe15.html (accessed on 3 December 2022).
- Lin, M.; Chen, Q.; Yan, S. Network in network. arXiv 2013, arXiv:131244002013. [Google Scholar]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Bahdanau, D.; Cho, K.; Bengio, Y. Neural machine translation by jointly learning to align and translate. arXiv 2014, arXiv:140904732014. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition: 2016, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. Available online: https://openaccess.thecvf.com/content_cvpr_2016/html/He_Deep_Residual_Learning_CVPR_2016_paper.html (accessed on 3 December 2022).
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:140915562014. [Google Scholar]
- Howard, A.; Sandler, M.; Chu, G.; Chen, L.-C.; Chen, B.; Tan, M.; Wang, W.; Zhu, Y.; Pang, R.; Vasudevan, V. Searching for mobilenetv3. In Proceedings of the IEEE/CVF International Conference on Computer Vsion: 2019, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 1314–1324. Available online: https://openaccess.thecvf.com/content_ICCV_2019/html/Howard_Searching_for_MobileNetV3_ICCV_2019_paper.html (accessed on 3 December 2022).
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010119292020. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition: 2017, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. Available online: https://openaccess.thecvf.com/content_cvpr_2017/html/Huang_Densely_Connected_Convolutional_CVPR_2017_paper.html (accessed on 3 December 2022).
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition: 2018, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. Available online: https://openaccess.thecvf.com/content_cvpr_2018/html/Hu_Squeeze-and-Excitation_Networks_CVPR_2018_paper.html (accessed on 3 December 2022).
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV): 2018, Munich, Germany, 8–14 September 2018; pp. 3–19. Available online: https://openaccess.thecvf.com/content_ECCV_2018/html/Sanghyun_Woo_Convolutional_Block_Attention_ECCV_2018_paper.html (accessed on 3 December 2022).
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:141269802014. [Google Scholar]
- Paszke, A.; Gross, S.; Chintala, S.; Chanan, G.; Yang, E.; DeVito, Z.; Lin, Z.; Desmaison, A.; Antiga, L.; Lerer, A. Automatic Differentiation in Pytorch. 2017. Available online: https://openreview.net/forum?id=BJJsrmfCZ (accessed on 3 December 2022).
- Hajian-Tilaki, K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Casp. J. Intern. Med. 2013, 4, 627. [Google Scholar]
- Wong, H.B.; Lim, G.H. Measures of diagnostic accuracy: Sensitivity, specificity, PPV and NPV. Proc. Singap. Healthc. 2011, 20, 316–318. [Google Scholar] [CrossRef]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 1–13. [Google Scholar] [CrossRef] [PubMed]


| Characteristics | Training Cohort (n = 298) | IV Cohort (n = 98) | P1 | EV Cohort (n = 98) | P2 |
|---|---|---|---|---|---|
| Age, years, mean ± SD | 53.1 ± 11.9 | 52.4 ± 11.8 | 0.76 | 54.1 ± 10.7 | 0.13 |
| US size, cm mean ± SD | 2.38 ± 0.97 | 2.44 ± 0.91 | 0.50 | 2.50 ± 1.01 | 0.57 |
| Ki67 | 0.32 | ||||
| ≤20 | 103 (34.6%) | 28 (28.6%) | 36 (36.7%) | 0.72 | |
| >20 | 195 (65.4%) | 70 (71.4%) | 62 (63.3%) | ||
| ER | 0.61 | 0.30 | |||
| Positive | 207 (69.5%) | 71 (72.4%) | 74 (75.5%) | ||
| Negative | 91 (30.5%) | 27 (27.6%) | 24 (24.5%) | ||
| PR | 0.82 | 0.52 | |||
| Positive | 166 (55.7%) | 53 (54.1%) | 55 (56.1%) | ||
| Negative | 132 (44.3%) | 45 (45.9%) | 43 (43.9%) | ||
| HER2 | 0.52 | 0.72 | |||
| Positive | 172 (57.7%) | 57 (58.2%) | 59 (60.2%) | ||
| Negative | 126 (42.3%) | 41 (41.8%) | 39 (39.8%) | ||
| Molecular subtype | 0.63 | 0.99 | |||
| HR+ and HER2− | 101 (33.9%) | 33 (33.7%) | 32 (32.7%) | ||
| HR+ and HER2+ | 138 (46.3%) | 42 (42.9%) | 46 (46.9%) | ||
| HER2+ | 34 (11.4%) | 16 (16.3%) | 12 (12.2%) | ||
| Triple-negative | 25 (8.4%) | 7 (7.1%) | 8 (8.2%) | ||
| Histological grade | 0.24 | 0.21 | |||
| 1 | 14 (4.7%) | 5 (5.1%) | 6 (5.1%) | ||
| 2 | 280(94.0%) | 89 (90.8%) | 88 (89.8%) | ||
| 3 | 4 (1.3%) | 4 (4.1%) | 4 (4.1%) | ||
| Tumor type | 0.43 | 0.68 | |||
| Invasive ductal carcinoma | 283 (95.0%) | 91 (92.9%) | 92 (93.9%) | ||
| Others | 15 (5.0%) | 7 (7.1%) | 6 (6.1%) |
| ResNet50 | DenseNet121 | Attention-Based DenseNet121 | Mobilenet_v3 | Vision Transformer | |
|---|---|---|---|---|---|
| IV cohort (n = 98) | |||||
| AUC | 0.906 [0.831, 0.956] | 0.919 [0.847, 0.965] | 0.922 [0.850, 0.967] | 0.885 [0.805, 0.941] | 0.907 [0.832, 0.957] |
| ACC(%) | 79.5 [71.5, 87.6] | 82.8 [75.0, 90.2] | 83.6 [76.3, 91.0] | 79.6 [71.5, 87.6] | 84.7 [77.4, 91.9] |
| SENS(%) | 87.0 [77.8, 96.2] | 87.0 [77.8, 96.2] | 85.2 [75.5, 94.9] | 79.6 [68.6, 90.6] | 87.0 [77.8, 96.2] |
| SPEC(%) | 70.4 [56.5, 84.3] | 77.2 [64.5, 90.0] | 81.8 [70.1, 93.5] | 79.5 [67.3, 91.8] | 81.8 [70.1, 93.5] |
| PPV(%) | 78.3 [67.6, 88.9] | 82.4 [72.3, 92.5] | 85.2 [75.5, 94.9] | 82.7 [72.2, 93.2] | 85.5 [75.9, 95.0] |
| NPV(%) | 81.5 [68.8, 82.4] | 82.9 [71.0, 94.8] | 81.8 [70.1, 93.5] | 76.1 [63.4, 88.7] | 83.7 [72.4, 95.1] |
| F1 score | 0.824 [0.754, 0.895] | 0.846 [0.779, 0.915] | 0.851 [0.784, 0.919] | 0.811 [0.736, 0.887] | 0.862 [0.797, 0.927] |
| EV cohort (n = 98) | |||||
| AUC | 0.858 [0.774, 0.921] | 0.867 [0.784, 0.927] | 0.873 [0.791, 0.932] | 0.888 [0.808, 0.947] | 0.878 [0.796, 0.935] |
| ACC(%) | 81.6 [73.8, 89.3] | 81.6 [75.0, 90.2] | 79.5 [71.5, 87.6] | 79.6 [71.5, 87.7] | 82.7 [75.1, 90.2] |
| SENS(%) | 85.1 [75.4, 94.8] | 85.1 [75.4, 94.8] | 90.7 [82.8, 98.6] | 75.9 [64.3, 87.5] | 85.2 [75.5, 94.9] |
| SPEC(%) | 77.2 [64.5, 90.0] | 79.5 [67.2, 91.8] | 65.9 [51.5, 80.3] | 84.1 [73.0, 95.2] | 79.5 [67.3, 91.8] |
| PPV(%) | 82.1 [71.8, 92.3] | 83.6 [73.6, 93.6] | 76.6 [65.9, 87.1] | 85.4 [75.2, 95.7] | 83.6 [73.6, 93.6] |
| NPV(%) | 80.9 [68.7, 93.1] | 81.3 [69.4, 93.3] | 85.3 [72.9, 97.6] | 74.0 [61.5, 86.5] | 81.3 [69.4, 93.4] |
| F1 score | 0.836 [0.766, 0.906] | 0.844 [0.775, 0.912] | 0.830 [0.762, 0.898] | 0.803 [0.726, 0.881] | 0.844 [0.775, 0.913] |
| ResNet50 (Truth) | DenseNet121 (Truth) | Att DenseNet121 (Truth) | MobileNet_v3 (Truth) | Vision Transformer (Truth) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Prediction | High | Low | High | Low | High | Low | High | Low | ||
| IV cohort | ||||||||||
| High | 31 | 7 | 34 | 7 | 36 | 8 | 35 | 11 | 36 | 7 |
| Low | 13 | 47 | 10 | 47 | 8 | 46 | 9 | 43 | 8 | 47 |
| EV cohort | ||||||||||
| High | 34 | 8 | 35 | 8 | 29 | 5 | 37 | 13 | 35 | 8 |
| Low | 10 | 46 | 9 | 46 | 15 | 49 | 7 | 41 | 9 | 46 |
| Molecular Subtypes | ACC | SENS | SPEC | PPV | NPV |
|---|---|---|---|---|---|
| HR+ and HER2− | 78.1% | 75.0% | 83.3% | 88.2% | 66.7% |
| HR+ and HER2+ | 78.3% | 75.0% | 85.7% | 66.7% | 60.0% |
| ER-, PR- and HER2+ | 83.3% | 80.0% | 85.7% | 80.0% | 85.7% |
| Triple-negative | 87.5% | 75.0% | 100.0% | 100.0% | 80.0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, Y.; Wu, R.; Lu, X.; Duan, Y.; Zhu, Y.; Ma, Y.; Nie, F. Deep Learning with Transformer or Convolutional Neural Network in the Assessment of Tumor-Infiltrating Lymphocytes (TILs) in Breast Cancer Based on US Images: A Dual-Center Retrospective Study. Cancers 2023, 15, 838. https://doi.org/10.3390/cancers15030838
Jia Y, Wu R, Lu X, Duan Y, Zhu Y, Ma Y, Nie F. Deep Learning with Transformer or Convolutional Neural Network in the Assessment of Tumor-Infiltrating Lymphocytes (TILs) in Breast Cancer Based on US Images: A Dual-Center Retrospective Study. Cancers. 2023; 15(3):838. https://doi.org/10.3390/cancers15030838
Chicago/Turabian StyleJia, Yingying, Ruichao Wu, Xiangyu Lu, Ying Duan, Yangyang Zhu, Yide Ma, and Fang Nie. 2023. "Deep Learning with Transformer or Convolutional Neural Network in the Assessment of Tumor-Infiltrating Lymphocytes (TILs) in Breast Cancer Based on US Images: A Dual-Center Retrospective Study" Cancers 15, no. 3: 838. https://doi.org/10.3390/cancers15030838
APA StyleJia, Y., Wu, R., Lu, X., Duan, Y., Zhu, Y., Ma, Y., & Nie, F. (2023). Deep Learning with Transformer or Convolutional Neural Network in the Assessment of Tumor-Infiltrating Lymphocytes (TILs) in Breast Cancer Based on US Images: A Dual-Center Retrospective Study. Cancers, 15(3), 838. https://doi.org/10.3390/cancers15030838







