A New HEK293 Cell with CR2 Region of E1A Gene Deletion Prevents the Emergence of Replication-Competent Adenovirus
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Reagents
2.2. Cell Proliferation Assay
2.3. Screening of Monoclonal Cell
2.4. CRISPR/Cas9 Mediated Deletions and Sequencing
2.5. Western-Blot Assay
2.6. E1A Gene Copies Number Tests
2.7. HAdVs and Replication
2.8. HAdVs Titration
2.9. DNA Extraction
2.10. Real-Time PCR
2.11. RCA Test
2.12. Statistical Analysis
3. Results
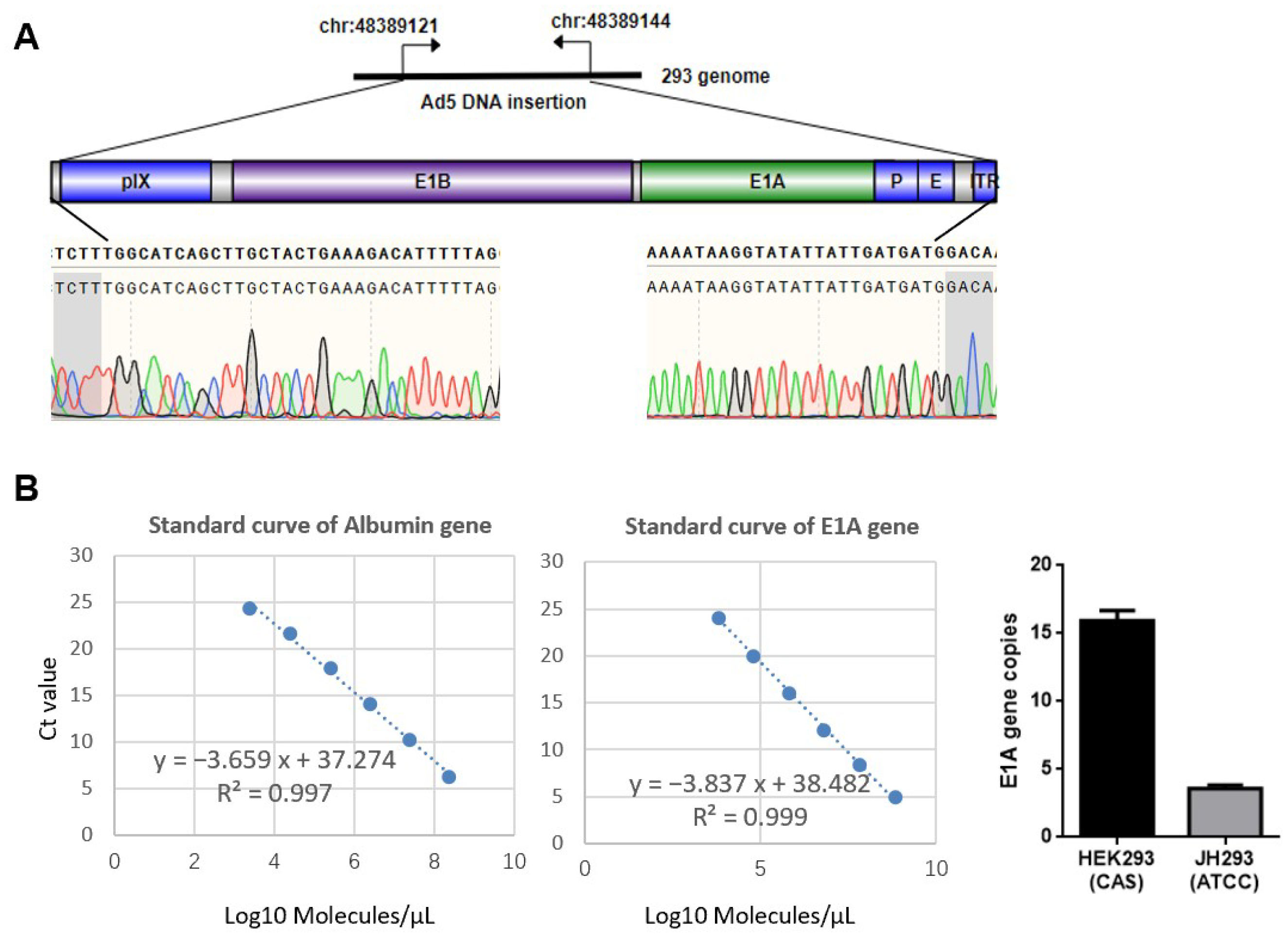
3.1. Different Sources of 293 Cell Have Different Copy Numbers of E1A
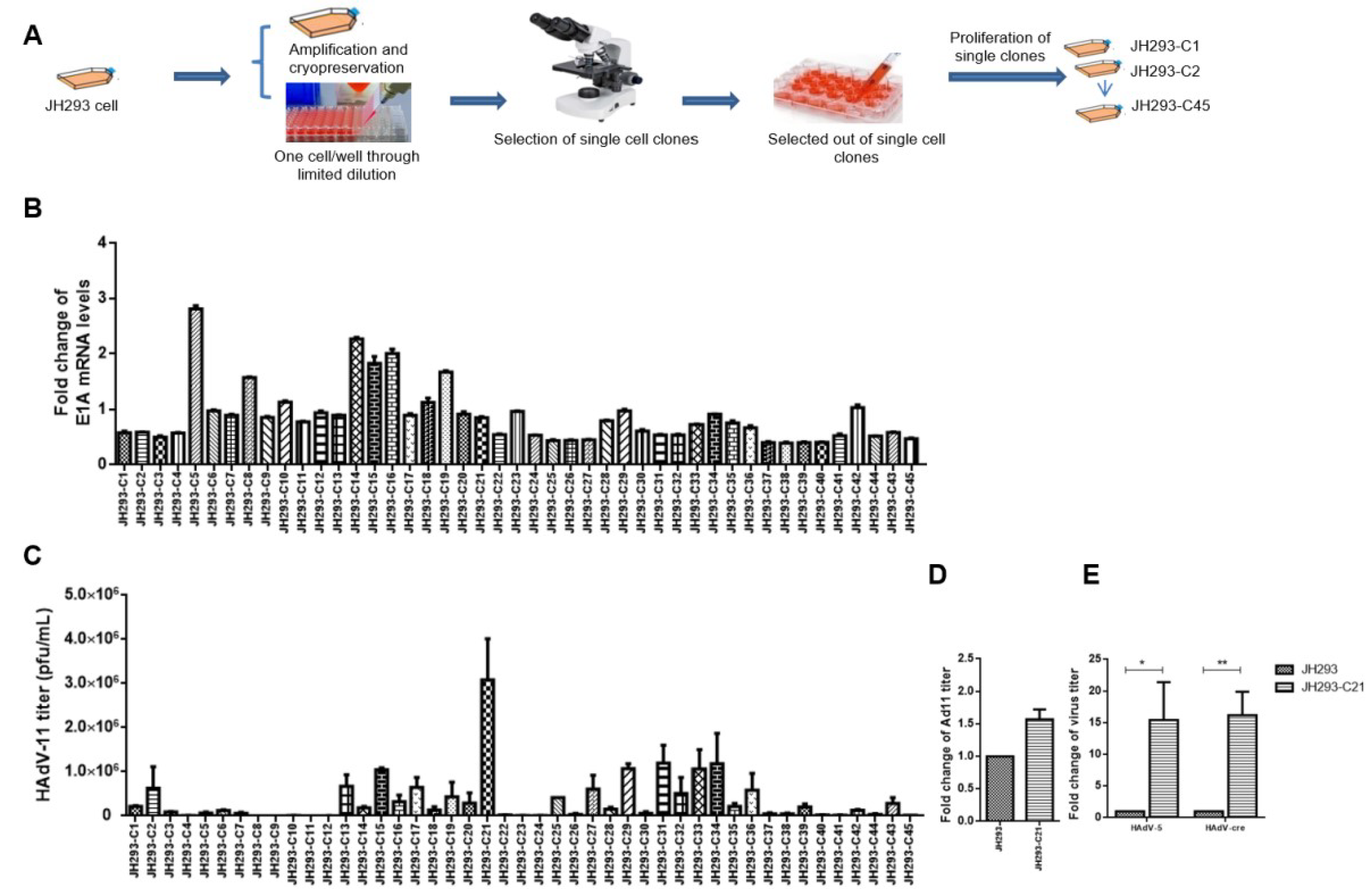
3.2. Selection of JH293 Single Clone JH293-C21 with High AdV Production Ability
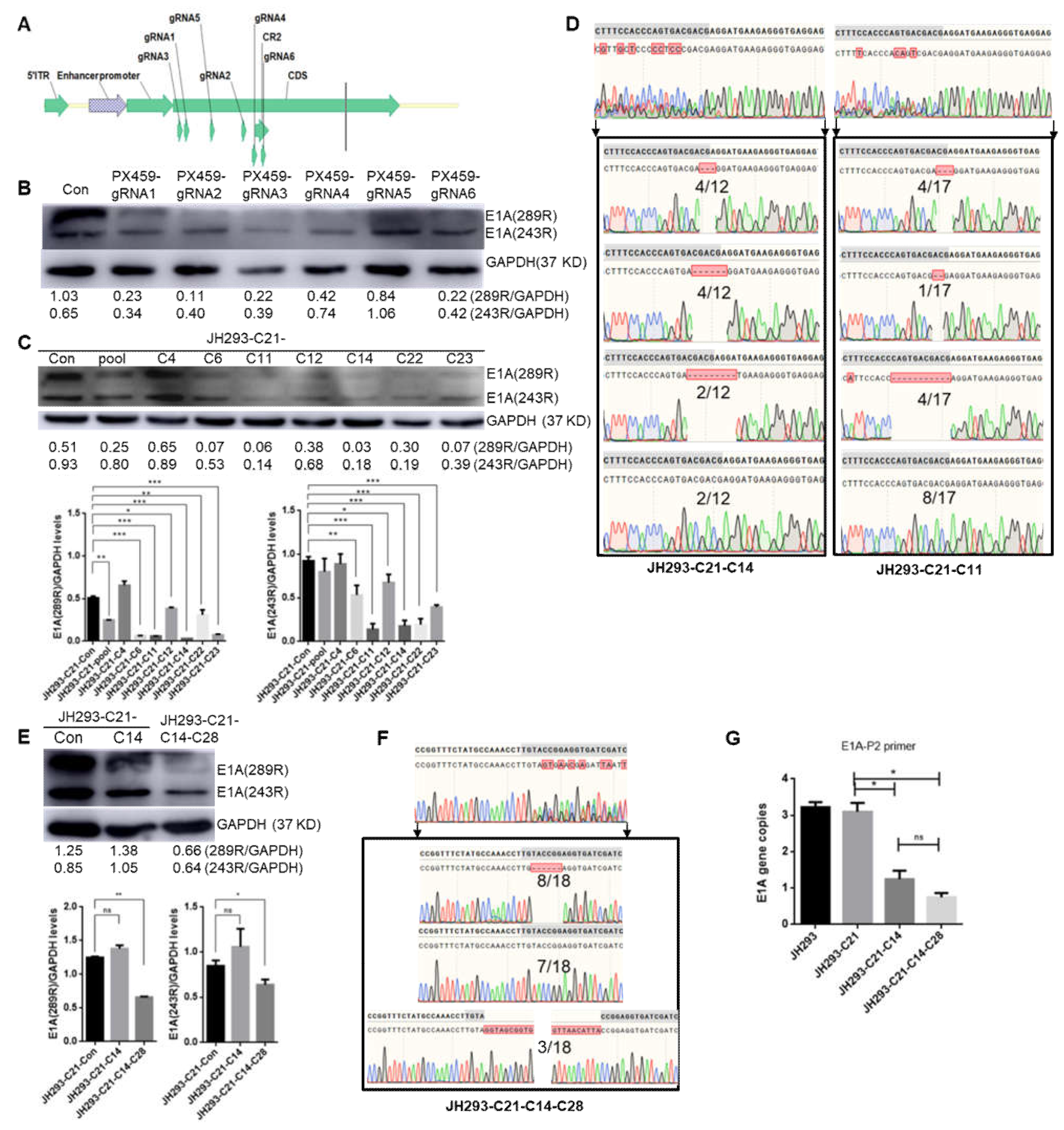
3.3. Construction of the CR2 Region of the E1A Knock-Out Cell Clone JH293-C21-C14-C28
3.4. The JH293-C21-C14-C28 Clone Had Similar Cell Proliferation and AdV Production Ability to JH293-C21 Cells
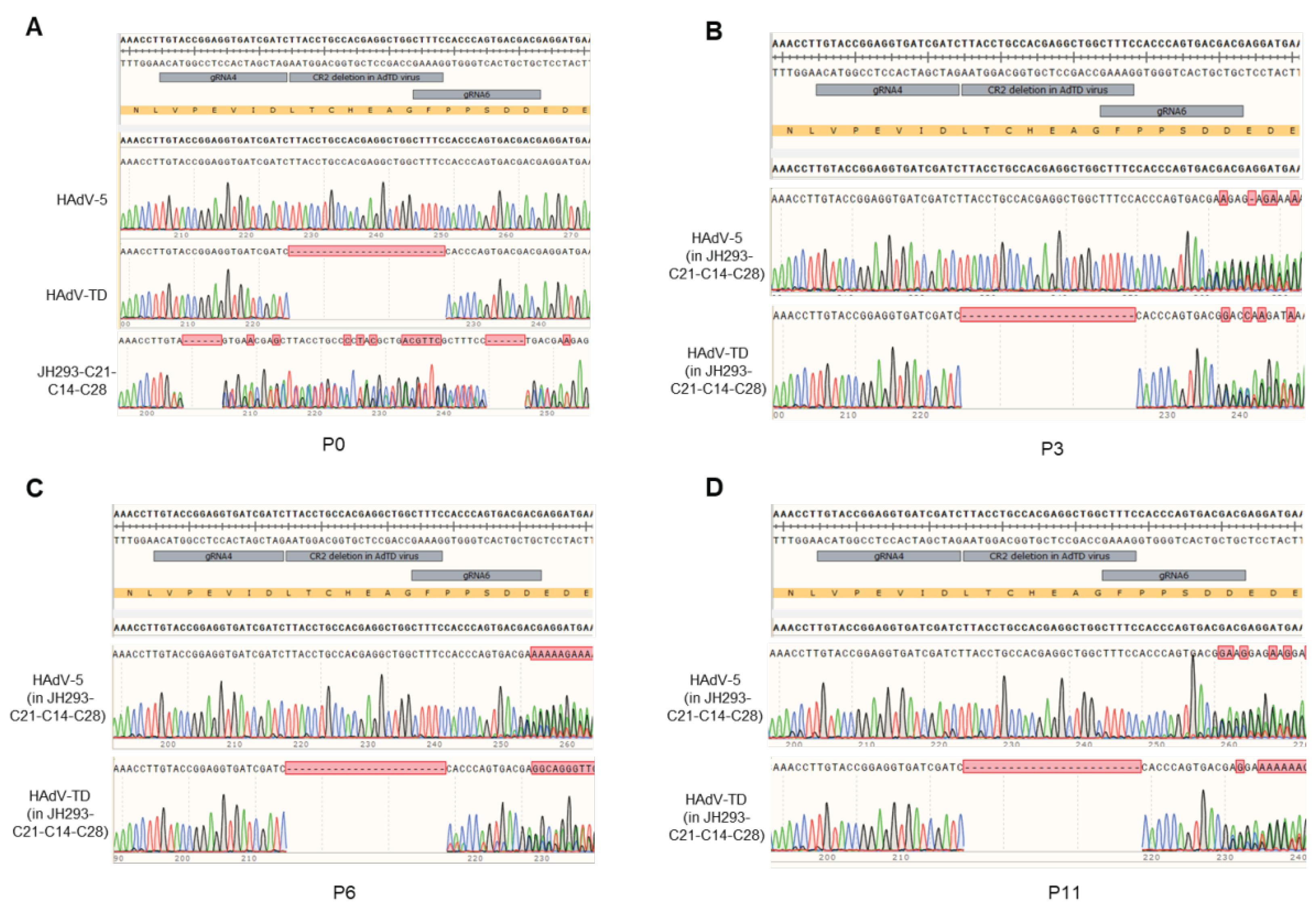
3.5. The JH293-C21-C14-C28 Clone Prevents the Production of RCA in Recombinant AdVs
3.6. No E1A Gene Recombination Occurred in the Production of Oncolytic Adenovirus in the JH293-C21-C14-C28 Cell Line
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rowe, W.P.; Huebner, R.J.; Gilmore, L.K.; Parrott, R.H.; Ward, T.G. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc. Soc. Exp. Biol. Med. 1953, 84, 570–573. [Google Scholar] [CrossRef] [PubMed]
- Mese, K.; Bunz, O.; Schellhorn, S.; Volkwein, W.; Jung, D.; Gao, J.; Zhang, W.; Baiker, A.; Ehrhardt, A. Identification of novel human adenovirus candidates using the coxsackievirus and adenovirus receptor for cell entry. Virol. J. 2020, 17, 52. [Google Scholar] [CrossRef]
- Davison, A.J.; Benkő, M.; Harrach, B. Genetic content and evolution of adenoviruses. J. Gen. Virol. 2003, 84 Pt 11, 2895–2908. [Google Scholar] [CrossRef]
- Russell, W.C. Adenoviruses: Update on structure and function. J. Gen. Virol. 2009, 90 Pt 1, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Bányai, K.; Martella, V.; Meleg, E.; Kisfali, P.; Péterfi, Z.; Benkö, M.; Melegh, B.; Szucs, G. Searching for HAdV-52, the putative gastroenteritis-associated human adenovirus serotype in Southern Hungary. New Microbiol. 2009, 32, 185–188. [Google Scholar] [PubMed]
- Yang, C.; Zhu, C.; Qian, Y.; Deng, J.; Zhang, B.; Zhu, R.; Wang, F.; Sun, Y.; Chen, D.; Guo, Q.; et al. Application of Human Adenovirus Genotyping by Phylogenetic Analysis in an Outbreak to Identify Nosocomial Infection. Virol. Sin. 2021, 36, 393–401. [Google Scholar] [CrossRef]
- Kaján, G.L.; Affranio, I.; Tóthné Bistyák, A.; Kecskeméti, S.; Benkő, M. An emerging new fowl adenovirus genotype. Heliyon 2019, 5, e01732. [Google Scholar] [CrossRef]
- Ghebremedhin, B. Human adenovirus: Viral pathogen with increasing importance. Eur. J. Microbiol. Immunol. 2014, 4, 26–33. [Google Scholar] [CrossRef]
- MacNeil, K.M.; Dodge, M.J.; Evans, A.M.; Tessier, T.M.; Weinberg, J.B.; Mymryk, J.S. Adenoviruses in medicine: Innocuous pathogen, predator, or partner. Trends Mol. Med. 2023, 29, 4–19. [Google Scholar] [CrossRef]
- Tseha, S.T. Role of Adenoviruses in Cancer Therapy. Front. Oncol. 2022, 12, 772659. [Google Scholar] [CrossRef] [PubMed]
- Lukashev, A.N.; Zamyatnin, A.A., Jr. Viral Vectors for Gene Therapy: Current State and Clinical Perspectives. Biochemistry 2016, 81, 700–708. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.M.; Lai, Y.K.; Rakoczy, P.E. Adenovirus and adeno-associated virus vectors. DNA Cell Biol. 2002, 21, 895–913. [Google Scholar] [CrossRef] [PubMed]
- Ikoma, T.; Takahashi, T.; Nagano, S.; Li, Y.M.; Ohno, Y.; Ando, K.; Fujiwara, T.; Fujiwara, H.; Kosai, K. A definitive role of RhoC in metastasis of orthotopic lung cancer in mice. Clin. Cancer Res. 2004, 10, 1192–1200. [Google Scholar] [CrossRef] [PubMed]
- Kalesnykas, G.; Kokki, E.; Alasaarela, L.; Lesch, H.P.; Tuulos, T.; Kinnunen, K.; Uusitalo, H.; Airenne, K.; Yla-Herttuala, S. Comparative Study of Adeno-associated Virus, Adenovirus, Bacu lovirus and Lentivirus Vectors for Gene Therapy of the Eyes. Curr. Gene Ther. 2017, 17, 235–247. [Google Scholar] [CrossRef]
- Lion, T. Adenovirus infections in immunocompetent and immunocompromised patients. Clin. Microbiol. Rev. 2014, 27, 441–462. [Google Scholar] [CrossRef] [PubMed]
- Leen, A.M.; Rooney, C.M. Adenovirus as an emerging pathogen in immunocompromised patients. Br. J. Haematol. 2005, 128, 135–144. [Google Scholar] [CrossRef]
- Pham, T.T.; Burchette, J.L., Jr.; Hale, L.P. Fatal disseminated adenovirus infections in immunocompromised patients. Am. J. Clin. Pathol. 2003, 120, 575–583. [Google Scholar] [CrossRef]
- Li, S.; Ou, M.; Wang, G.; Tang, L. Application of conditionally replicating adenoviruses in tumor early diagnosis technology, gene-radiation therapy and chemotherapy. Appl. Microbiol. Biotechnol. 2016, 100, 8325–8335. [Google Scholar] [CrossRef]
- Peter, M.; Kühnel, F. Oncolytic Adenovirus in Cancer Immunotherapy. Cancers 2020, 12, 3354. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yao, Q.M.; Li, J.L.; Chang, Y.; Li, T.; Han, W.L.; Wu, H.P.; Li, L.F.; Qian, Q.J.; Ruan, G.R. Synergistic antitumor activity of triple-regulated oncolytic adenovirus with VSTM1 and daunorubicin in leukemic cells. Apoptosis 2016, 21, 1179–1190. [Google Scholar] [CrossRef]
- Wang, Z.X.; Germino, F.J. Mutational analysis of the conserved region 2 site of adenovirus E1A and its effect on binding to the retinoblastoma gene product: Use of the “double-tagging” assay. Proc. Natl. Acad. Sci. USA 1995, 92, 4631–4635. [Google Scholar] [CrossRef]
- Jiang, H.; Alemany, R.; Gomez-Manzano, C.; Medrano, D.R.; Lemoine, M.G.; Olson, M.V.; Alonso, M.M.; Lee, O.H.; Conrad, C.C.; Yung, W.K.; et al. Downmodulation of E1A protein expression as a novel strategy to design cancer-selective adenoviruses. Neoplasia 2005, 7, 723–729. [Google Scholar] [CrossRef]
- Abudoureyimu, M.; Lai, Y.; Tian, C.; Wang, T.; Wang, R.; Chu, X. Oncolytic Adenovirus-A Nova for Gene-Targeted Oncolytic Viral Therapy in HCC. Front. Oncol. 2019, 9, 1182. [Google Scholar] [CrossRef] [PubMed]
- Lochmüller, H.; Jani, A.; Huard, J.; Prescott, S.; Simoneau, M.; Massie, B.; Karpati, G.; Acsadi, G. Emergence of early region 1-containing replication-competent adenovirus in stocks of replication-defective adenovirus recombinants (delta E1 + delta E3) during multiple passages in 293 cells. Hum. Gene Ther. 1994, 5, 1485–1491. [Google Scholar] [CrossRef] [PubMed]
- Fallaux, F.J.; Bout, A.; van der Velde, I.; van den Wollenberg, D.J.; Hehir, K.M.; Keegan, J.; Auger, C.; Cramer, S.J.; van Ormondt, H.; van der Eb, A.J.; et al. New helper cells and matched early region 1-deleted adenovirus vectors prevent generation of replication-competent adenoviruses. Hum. Gene Ther. 1998, 9, 1909–1917. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.P.; Engdahl, R.K.; Wilson, J.M. A cell line for high-yield production of E1-deleted adenovirus vectors without the emergence of replication-competent virus. Hum. Gene Ther. 2000, 11, 213–219. [Google Scholar] [CrossRef]
- Barczak, W.; Suchorska, W.; Rubiś, B.; Kulcenty, K. Universal real-time PCR-based assay for lentiviral titration. Mol. Biotechnol. 2015, 57, 195–200. [Google Scholar] [CrossRef]
- Wang, Y.; Hallden, G.; Hill, R.; Anand, A.; Liu, T.C.; Francis, J.; Brooks, G.; Lemoine, N.; Kirn, D. E3 gene manipulations affect oncolytic adenovirus activity in immunocompetent tumor models. Nat. Biotechnol. 2003, 21, 1328–1335. [Google Scholar] [CrossRef]
- Wang, P.; Li, X.; Wang, J.; Gao, D.; Li, Y.; Li, H.; Chu, Y.; Zhang, Z.; Liu, H.; Jiang, G.; et al. Re-designing Interleukin-12 to enhance its safety and potential as an anti-tumor immunotherapeutic agent. Nat. Commun. 2017, 8, 1395. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty percent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Ishii-Watabe, A.; Uchida, E.; Iwata, A.; Nagata, R.; Satoh, K.; Fan, K.; Murata, M.; Mizuguchi, H.; Kawasaki, N.; Kawanishi, T.; et al. Detection of replication-competent adenoviruses spiked into recombinant adenovirus vector products by infectivity PCR. Mol. Ther. 2003, 8, 1009–1016. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.C.; Boone, M.; Meuris, L.; Lemmens, I.; Van Roy, N.; Soete, A.; Reumers, J.; Moisse, M.; Plaisance, S.; Drmanac, R.; et al. Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nat. Commun. 2014, 5, 4767. [Google Scholar] [CrossRef] [PubMed]
- Bylund, L.; Kytölä, S.; Lui, W.O.; Larsson, C.; Weber, G. Analysis of the cytogenetic stability of the human embryonal kidney cell line 293 by cytogenetic and STR profiling approaches. Cytogenet. Genome Res. 2004, 106, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Wold, W.S.; Toth, K. Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr. Gene Ther. 2013, 13, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Nishikawaji, Y.; Kawakami, H.; Kosai, K.I. Adenovirus Biology, Recombinant Adenovirus, and Adenovirus Usage in Gene Therapy. Viruses 2021, 13, 2502. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.C.; Guan, X.H.; Li, Y.H.; Huang, J.Y.; Jiang, T.; Hou, L.H.; Li, J.X.; Yang, B.F.; Wang, L.; Wang, W.J.; et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 2020, 396, 479–488. [Google Scholar] [CrossRef]
- Matsuda, K.; Migueles, S.A.; Huang, J.; Bolkhovitinov, L.; Stuccio, S.; Griesman, T.; Pullano, A.A.; Kang, B.H.; Ishida, E.; Zimmerman, M.; et al. A replication-competent adenovirus-vectored influenza vaccine induces durable systemic and mucosal immunity. J. Clin. Investig. 2021, 131, e140794. [Google Scholar] [CrossRef]
- Fares, J.; Ahmed, A.U.; Ulasov, I.V.; Sonabend, A.M.; Miska, J.; Lee-Chang, C.; Balyasnikova, I.V.; Chandler, J.P.; Portnow, J.; Tate, M.C.; et al. Neural stem cell delivery of an oncolytic adenovirus in newly diagnosed malignant glioma: A first-in-human, phase 1, dose-escalation trial. Lancet Oncol. 2021, 22, 1103–1114. [Google Scholar] [CrossRef]


| Mutation Type | ||||
|---|---|---|---|---|
| 2#1 | Clones of T vector | Change of gRNA6 situation | Change of gRNA4 situation besides gRNA6 situation | percentage |
| 2#2 | T1/T5/T8/T9/T11/T14/T20 | 6 bases deletion | No change | 7/18 |
| 2#3 | T2/T3/T4/T7/T10/T12/T15/T17 | 9 bases deletion | 6 bases deletion | 8/18 |
| Total of E1A mutation: 18/18 | T6/T13/T18 | 9 bases deletion | 194 bases insertion | 3/18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lian, X.; Zhao, X.; Zhong, J.; Zhang, C.; Chu, Y.; Wang, Y.; Lu, S.; Wang, Z. A New HEK293 Cell with CR2 Region of E1A Gene Deletion Prevents the Emergence of Replication-Competent Adenovirus. Cancers 2023, 15, 5713. https://doi.org/10.3390/cancers15245713
Lian X, Zhao X, Zhong J, Zhang C, Chu Y, Wang Y, Lu S, Wang Z. A New HEK293 Cell with CR2 Region of E1A Gene Deletion Prevents the Emergence of Replication-Competent Adenovirus. Cancers. 2023; 15(24):5713. https://doi.org/10.3390/cancers15245713
Chicago/Turabian StyleLian, Xueqi, Xiaoyan Zhao, Jingjing Zhong, Chenglin Zhang, Yongchao Chu, Yaohe Wang, Shuangshuang Lu, and Zhimin Wang. 2023. "A New HEK293 Cell with CR2 Region of E1A Gene Deletion Prevents the Emergence of Replication-Competent Adenovirus" Cancers 15, no. 24: 5713. https://doi.org/10.3390/cancers15245713
APA StyleLian, X., Zhao, X., Zhong, J., Zhang, C., Chu, Y., Wang, Y., Lu, S., & Wang, Z. (2023). A New HEK293 Cell with CR2 Region of E1A Gene Deletion Prevents the Emergence of Replication-Competent Adenovirus. Cancers, 15(24), 5713. https://doi.org/10.3390/cancers15245713





