Unveiling the Immunogenicity of Ovarian Tumors as the Crucial Catalyst for Therapeutic Success
Abstract
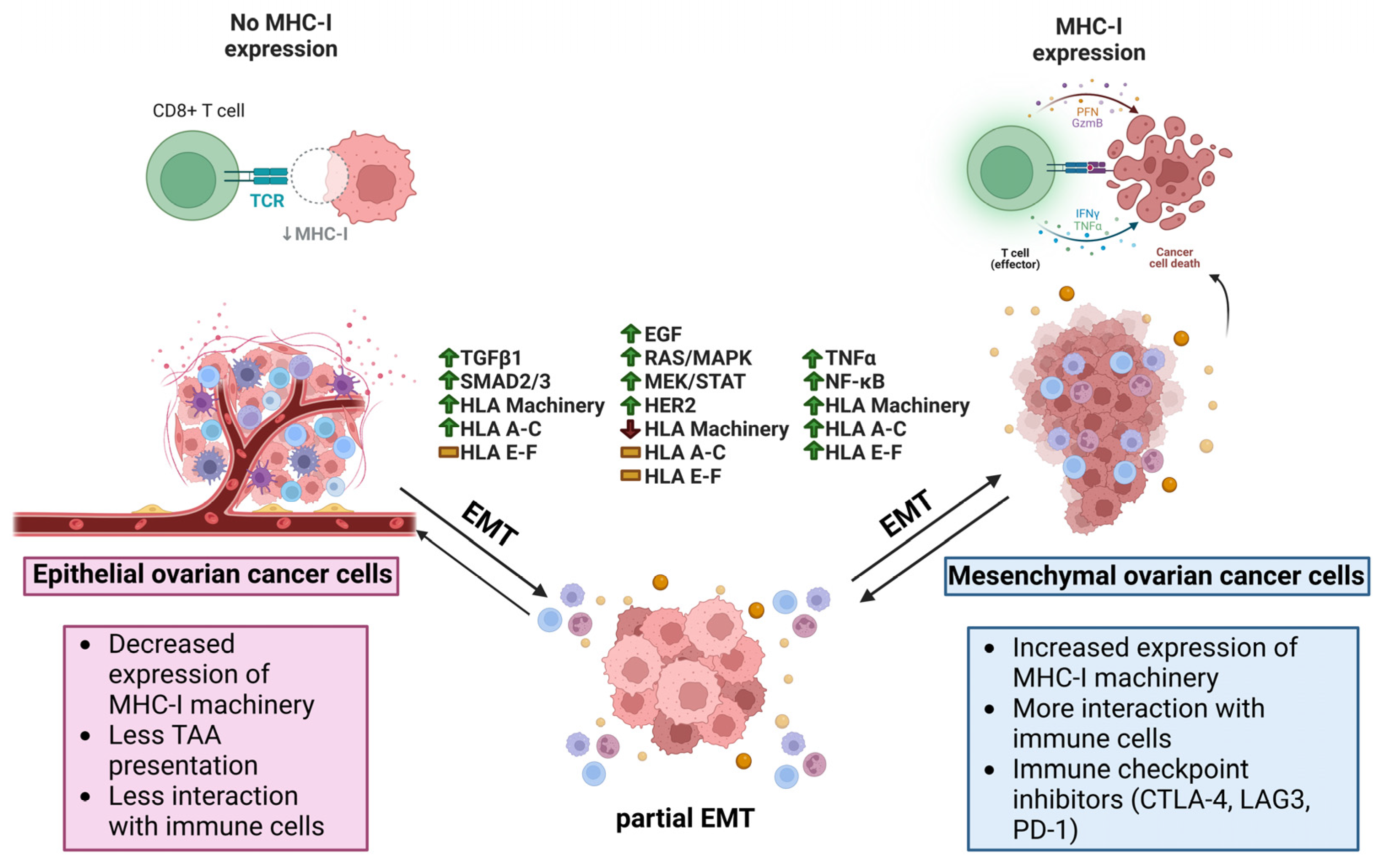
Simple Summary
Abstract
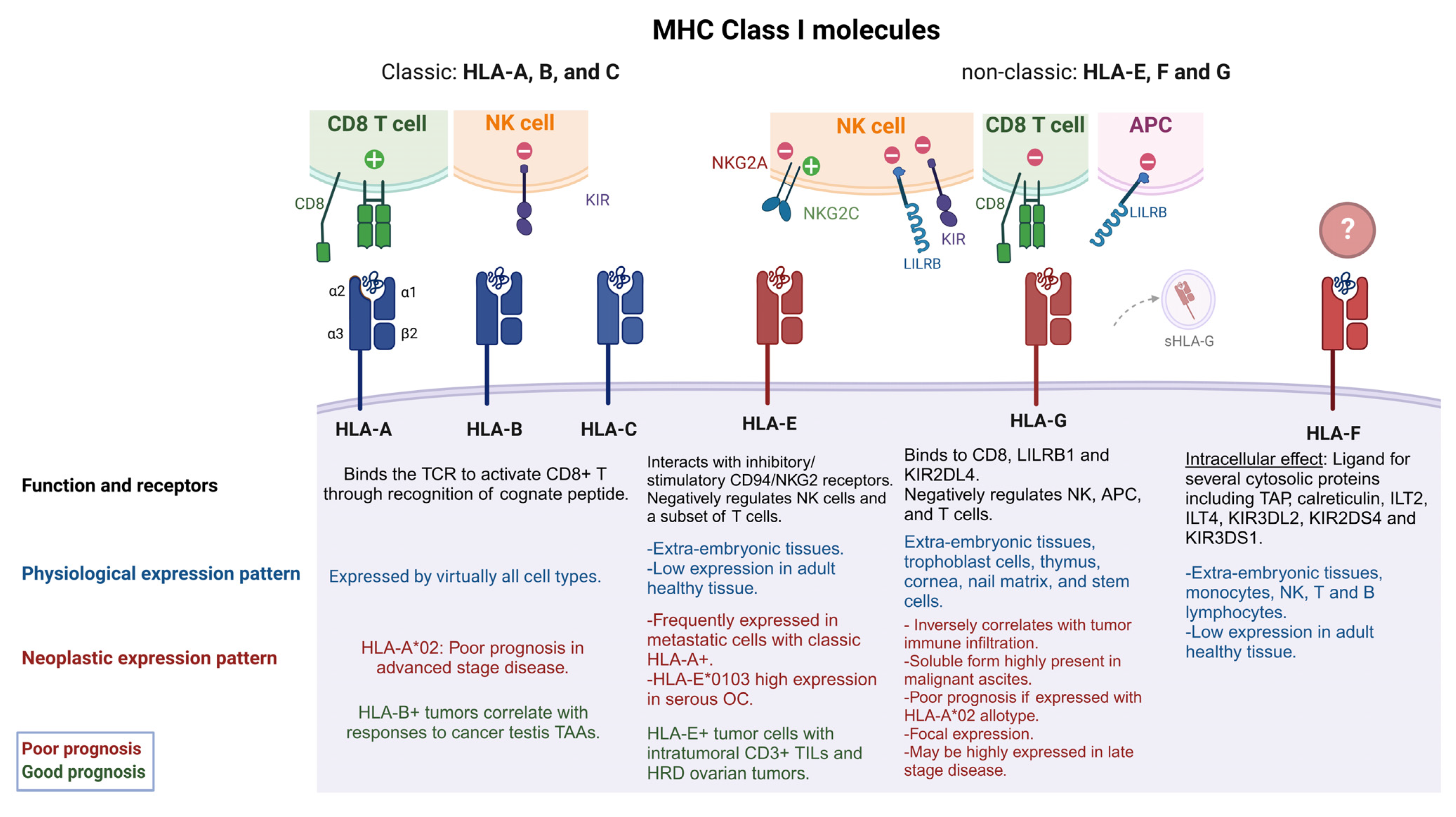
1. Overview of MHC Class I and Class II Molecules
2. Ovarian Cancer Immunogenicity
2.1. Classic HLA Class I
2.2. Non-Classic HLA Class I
2.2.1. HLA-E
2.2.2. HLA-F
2.2.3. HLA-G
2.3. NLRC5, the Master Regulator of MHC Class I Expression
3. Other Tumor Microenvironment Factors Influencing Ovarian Cancer Immunogenicity
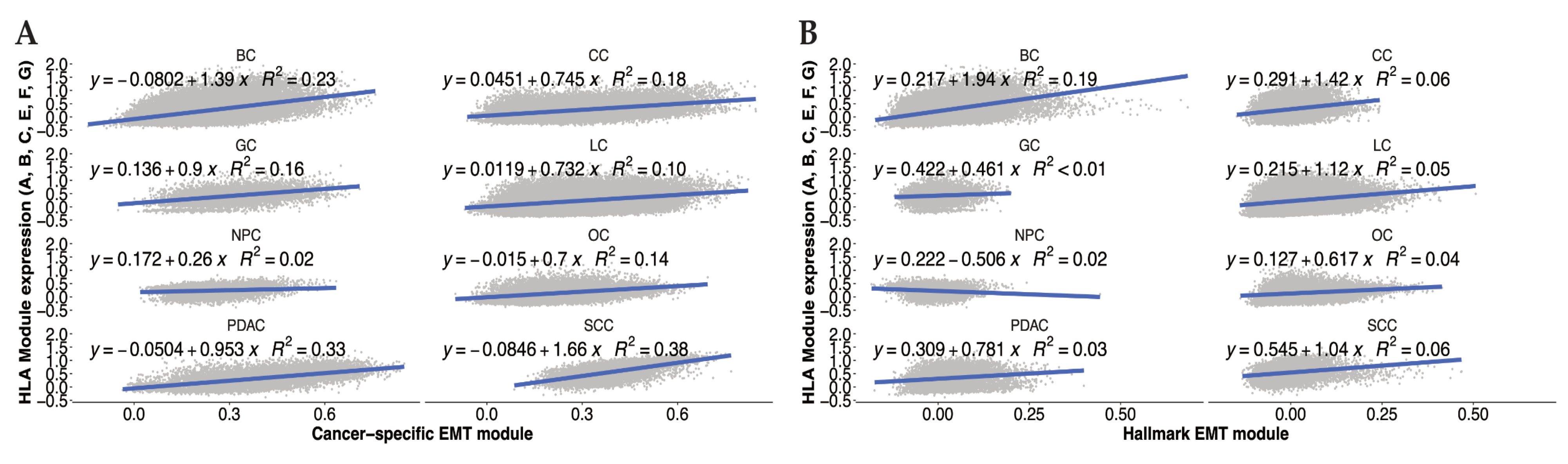
3.1. EMT Effects on HLA Expression in Cancer
3.2. EMT Inducers and HLA I Expression
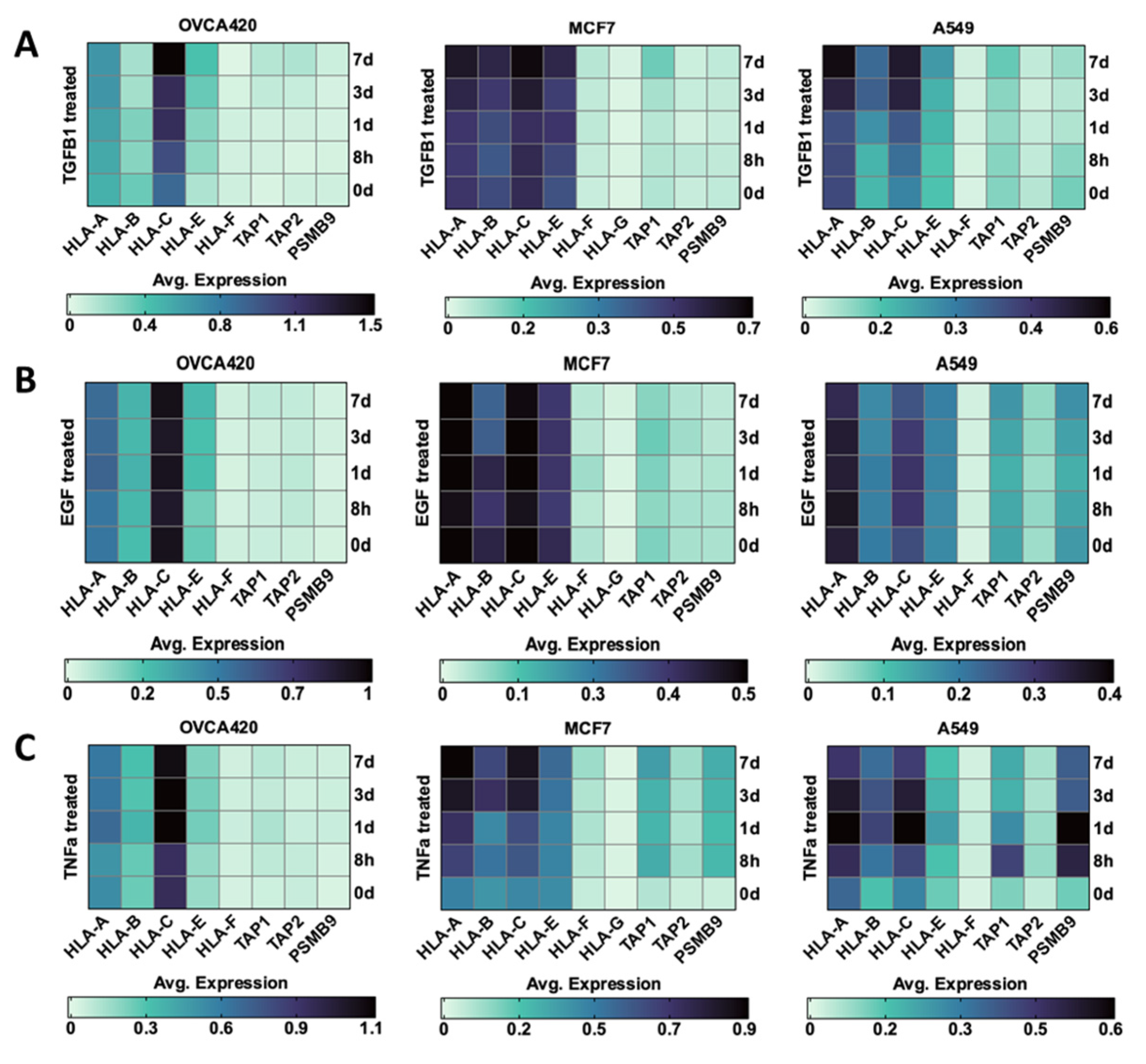
3.2.1. TGF β1
3.2.2. EGF
3.2.3. TNF-α
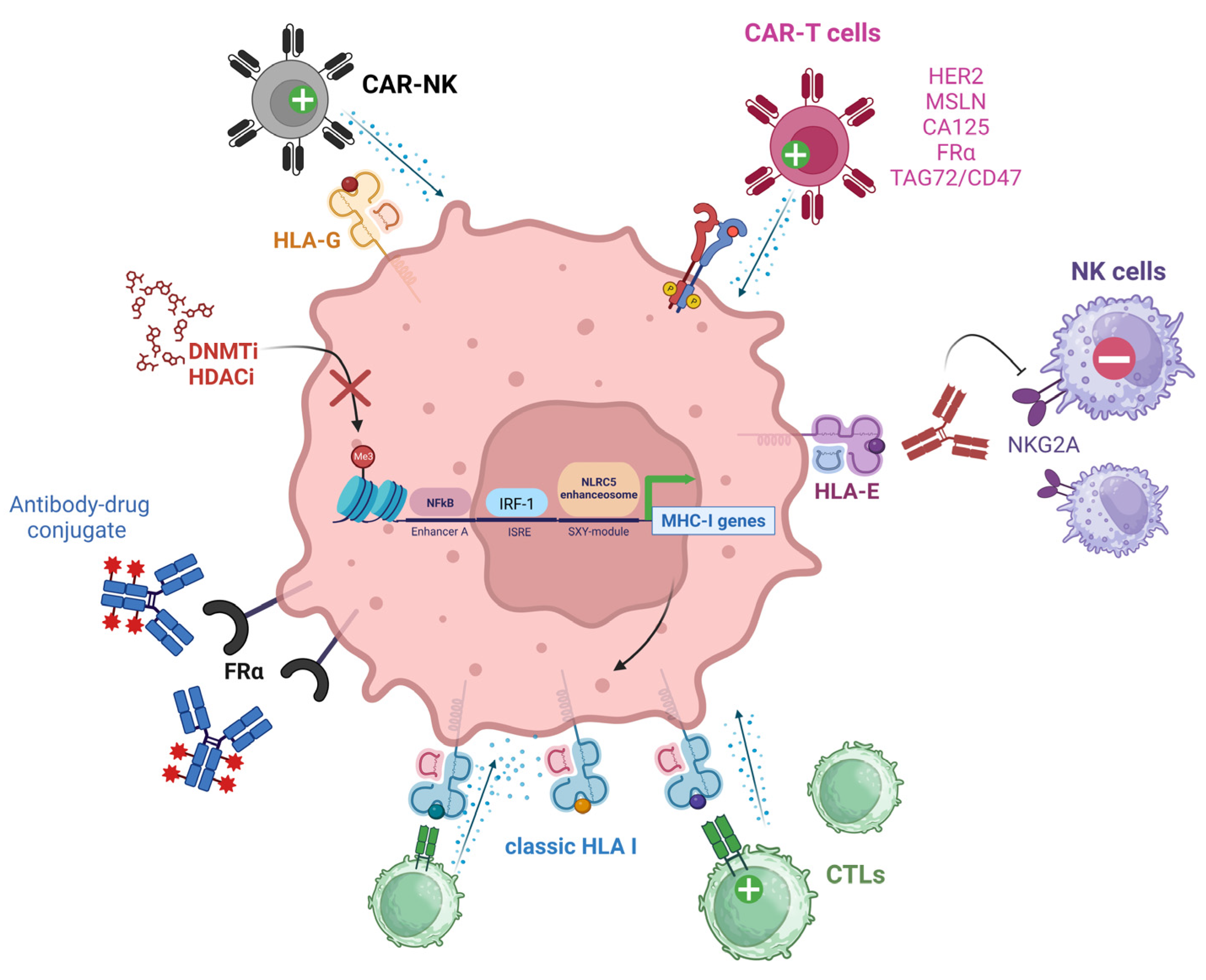
4. Therapeutic Strategies to Overcome the Lack of Immune Recognition in Ovarian Cancer
4.1. Targeting TAAs and HLAs
4.1.1. Chimeric Antigen Receptors
Human Epidermal Growth Factor Receptor 2 (HER2)
Mesothelin
MUC16/CA125
Alpha-Folate Receptor (FRα)
Tumor-Associated Glycoprotein 72 (TAG72)
4.1.2. Targeting MHC I Molecules
HLA-E
HLA-G
4.1.3. Epigenetic Modulation of MHC I Expression
4.2. Therapeutic Advances Targeting EMT in Cancer
4.2.1. TGF β1
4.2.2. EMT-TFs
5. Future Directions and Open Questions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of Abbreviations
| APC | Antigen presenting cells |
| APM | Antigen presenting machinery |
| β2M | β-2 microglobulin |
| CAR-T | Chimeric antigen receptor T (cell) |
| CIITA | MHC class II transactivator |
| CTL | Cytotoxic T cell |
| DC | Dendritic cells |
| DNMTi | DNA methyltransferase inhibitors |
| EGF | Epidermal growth factor |
| EGFR | Epidermal growth factor receptor |
| EMT | Epithelial-mesenchymal transition |
| EMT-TFs | Epithelial-mesenchymal transition transcription factors |
| EOC | Epithelial ovarian cancer |
| HGSC | High-grade serous ovarian cancer |
| HDACi | histone deacetylase inhibitors |
| HLA | Human leukocyte antigens |
| HRD | Homologous recombination deficiency |
| HSDL1 | Hydroxysteroid dehydrogenase–like protein 1 |
| Ig | Immunoglobulin |
| ILT2 | Immunoglobulin-like transcript 2 |
| IFN-γ | Type II interferon |
| IRF-1 | Interferon regulatory factor |
| KIR3DL2 | KIR three Ig domains and long cytoplasmic tail 2 |
| KIR3DS1 | KIR three Ig domains and short cytoplasmic tail 1 |
| KIR3DS4 | KIR three Ig domains and long cytoplasmic tail 4 |
| LSK1 | lysine-specific histone demethylase 1 |
| MHC I | Major histocompatibility complex I |
| NCT | National clinical trial |
| NK | Natural killer |
| NLRC5 | NOD-like receptor family caspase recruitment domain–containing 5 |
| pMHC I | peptide-MHC I complex |
| sHLA | Soluble HLA |
| scFv | single-chain variable fragment |
| PFS | Progression-free survival |
| TAA | Tumor associated antigen |
| TAP | Transporter associated with antigen presentation |
| TCR | T cell receptors |
| TGF β1 | Transforming growth factor beta 1 |
| TGFβR | TGF β receptors |
| TMB | Tumor mutational burden |
| TME | Tumor microenvironment |
| TNFα | Tumor necrosis factor alpha |
| TAG72 | Tumor-associated glycoprotein 72 |
References
- Shiina, T.; Hosomichi, K.; Inoko, H.; Kulski, J.K. The HLA Genomic Loci Map: Expression, Interaction, Diversity and Disease. J. Hum. Genet. 2009, 54, 15–39. [Google Scholar] [CrossRef] [PubMed]
- Garrido, F. HLA Class-I Expression and Cancer Immunotherapy. Adv. Exp. Med. Biol. 2019, 1151, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.; Sato, A. The HLA System. First of Two Parts. N. Engl. J. Med. 2000, 343, 702–709. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.; Sato, A. The HLA System. Second of Two Parts. N. Engl. J. Med. 2000, 343, 782–786. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Tapias, P.; Castiblanco, J.; Anaya, J.-M. HLA Association with Autoimmune Diseases. In Autoimmunity: From Bench to Bedside [Internet]; El Rosario University Press: Bogotá, Colombia, 2013. [Google Scholar]
- D’Souza, M.P.; Adams, E.; Altman, J.D.; Birnbaum, M.E.; Boggiano, C.; Casorati, G.; Chien, Y.; Conley, A.; Eckle, S.B.G.; Früh, K.; et al. Casting a Wider Net: Immunosurveillance by Nonclassical MHC Molecules. PLoS Pathog. 2019, 15, e1007567. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.; Barker, D.J.; Georgiou, X.; Cooper, M.A.; Flicek, P.; Marsh, S.G.E. IPD-IMGT/HLA Database. Nucleic Acids Res. 2020, 48, D948–D955. [Google Scholar] [CrossRef]
- Andersson, E.; Poschke, I.; Villabona, L.; Carlson, J.W.; Lundqvist, A.; Kiessling, R.; Seliger, B.; Masucci, G.V. Non-Classical HLA-Class I Expression in Serous Ovarian Carcinoma: Correlation with the HLA-Genotype, Tumor Infiltrating Immune Cells and Prognosis. Oncoimmunology 2015, 5, e1052213. [Google Scholar] [CrossRef]
- Stern, L.J.; Wiley, D.C. Antigenic Peptide Binding by Class I and Class II Histocompatibility Proteins. Structure 1994, 2, 245–251. [Google Scholar] [CrossRef]
- Chen, Y.; Sidney, J.; Southwood, S.; Cox, A.L.; Sakaguchi, K.; Henderson, R.A.; Appella, E.; Hunt, D.F.; Sette, A.; Engelhard, V.H. Naturally Processed Peptides Longer than Nine Amino Acid Residues Bind to the Class I MHC Molecule HLA-A2.1 with High Affinity and in Different Conformations. J. Immunol. 1994, 152, 2874–2881. [Google Scholar] [CrossRef]
- Rist, M.J.; Theodossis, A.; Croft, N.P.; Neller, M.A.; Welland, A.; Chen, Z.; Sullivan, L.C.; Burrows, J.M.; Miles, J.J.; Brennan, R.M.; et al. HLA Peptide Length Preferences Control CD8+ T Cell Responses. J. Immunol. 2013, 191, 561–571. [Google Scholar] [CrossRef]
- Trolle, T.; McMurtrey, C.P.; Sidney, J.; Bardet, W.; Osborn, S.C.; Kaever, T.; Sette, A.; Hildebrand, W.H.; Nielsen, M.; Peters, B. The Length Distribution of Class I Restricted T Cell Epitopes Is Determined by Both Peptide Supply and MHC Allele Specific Binding Preference. J. Immunol. 2016, 196, 1480–1487. [Google Scholar] [CrossRef]
- Lanier, L.L. NK Cell Recognition. Annu. Rev. Immunol. 2005, 23, 225–274. [Google Scholar] [CrossRef]
- Lanier, L.L.; Phillips, J.H. Inhibitory MHC Class I Receptors on NK Cells and T Cells. Immunol. Today 1996, 17, 86–91. [Google Scholar] [CrossRef]
- Moretta, A.; Bottino, C.; Vitale, M.; Pende, D.; Biassoni, R.; Mingari, M.C.; Moretta, L. Receptors for Hla Class-I Molecules in Human Natural Killer Cells. Annu. Rev. Immunol. 1996, 14, 619–648. [Google Scholar] [CrossRef]
- Braud, V.M.; Allan, D.S.; O’Callaghan, C.A.; Söderström, K.; D’Andrea, A.; Ogg, G.S.; Lazetic, S.; Young, N.T.; Bell, J.I.; Phillips, J.H.; et al. HLA-E Binds to Natural Killer Cell Receptors CD94/NKG2A, B and C. Nature 1998, 391, 795–799. [Google Scholar] [CrossRef]
- López-Botet, M.; Bellón, T. Natural Killer Cell Activation and Inhibition by Receptors for MHC Class I. Curr. Opin. Immunol. 1999, 11, 301–307. [Google Scholar] [CrossRef]
- Moretta, L.; Biassoni, R.; Bottino, C.; Mingari, M.C.; Moretta, A. Human NK-Cell Receptors. Immunol. Today 2000, 21, 420–422. [Google Scholar] [CrossRef]
- Chicz, R.M.; Urban, R.G.; Lane, W.S.; Gorga, J.C.; Stern, L.J.; Vignali, D.A.; Strominger, J.L. Predominant Naturally Processed Peptides Bound to HLA-DR1 Are Derived from MHC-Related Molecules and Are Heterogeneous in Size. Nature 1992, 358, 764–768. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, T.; Ruiz-Cabello, F.; Garrido, F. Biological Implications of HLA-DR Expression in Tumours. Scand. J. Immunol. 1995, 41, 398–406. [Google Scholar] [CrossRef]
- Cruz-Tapias, P.; Castiblanco, J.; Anaya, J.-M. Major Histocompatibility Complex: Antigen Processing and Presentation; El Rosario University Press: Bogotá, Colombia, 2013. [Google Scholar]
- Daar, A.S.; Fuggle, S.V.; Fabre, J.W.; Ting, A.; Morris, P.J. The Detailed Distribution of MHC Class II Antigens in Normal Human Organs. Transplantation 1984, 38, 293–298. [Google Scholar] [CrossRef]
- Aultman, D.; Adamashvili, I.; Yaturu, K.; Langford, M.; Gelder, F.; Gautreaux, M.; Ghali, G.E.; McDonald, J. Soluble HLA in Human Body Fluids. Hum. Immunol. 1999, 60, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Puppo, F.; Scudeletti, M.; Indiveri, F.; Ferrone, S. Serum HLA Class I Antigens: Markers and Modulators of an Immune Response? Immunol. Today 1995, 16, 124–127. [Google Scholar] [CrossRef] [PubMed]
- Hauptmann, G.; Bahram, S. Genetics of the Central MHC. Curr. Opin. Immunol. 2004, 16, 668–672. [Google Scholar] [CrossRef]
- Horton, R.; Wilming, L.; Rand, V.; Lovering, R.C.; Bruford, E.A.; Khodiyar, V.K.; Lush, M.J.; Povey, S.; Talbot, C.C.; Wright, M.W.; et al. Gene Map of the Extended Human MHC. Nat. Rev. Genet. 2004, 5, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Schott, G.; Garcia-Blanco, M.A. MHC Class III RNA Binding Proteins and Immunity. RNA Biol. 2020, 18, 640–646. [Google Scholar] [CrossRef]
- Aust, S.; Felix, S.; Auer, K.; Bachmayr-Heyda, A.; Kenner, L.; Dekan, S.; Meier, S.M.; Gerner, C.; Grimm, C.; Pils, D. Absence of PD-L1 on Tumor Cells Is Associated with Reduced MHC I Expression and PD-L1 Expression Increases in Recurrent Serous Ovarian Cancer. Sci. Rep. 2017, 7, 42929. [Google Scholar] [CrossRef]
- Dholakia, J.; Scalise, C.B.; Katre, A.A.; Goldsberry, W.N.; Meza-Perez, S.; Randall, T.D.; Norian, L.A.; Novak, L.; Arend, R.C. Sequential Modulation of the Wnt/β-Catenin Signaling Pathway Enhances Tumor-Intrinsic MHC I Expression and Tumor Clearance. Gynecol. Oncol. 2022, 164, 170–180. [Google Scholar] [CrossRef]
- Hamanishi, J.; Mandai, M.; Iwasaki, M.; Okazaki, T.; Tanaka, Y.; Yamaguchi, K.; Higuchi, T.; Yagi, H.; Takakura, K.; Minato, N.; et al. Programmed Cell Death 1 Ligand 1 and Tumor-Infiltrating CD8+ T Lymphocytes Are Prognostic Factors of Human Ovarian Cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 3360–3365. [Google Scholar] [CrossRef]
- Sato, E.; Olson, S.H.; Ahn, J.; Bundy, B.; Nishikawa, H.; Qian, F.; Jungbluth, A.A.; Frosina, D.; Gnjatic, S.; Ambrosone, C.; et al. Intraepithelial CD8+ Tumor-Infiltrating Lymphocytes and a High CD8+/Regulatory T Cell Ratio Are Associated with Favorable Prognosis in Ovarian Cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 18538–18543. [Google Scholar] [CrossRef]
- Vitale, M.; Pelusi, G.; Taroni, B.; Gobbi, G.; Micheloni, C.; Rezzani, R.; Donato, F.; Wang, X.; Ferrone, S. HLA Class I Antigen Down-Regulation in Primary Ovary Carcinoma Lesions: Association with Disease Stage. Clin. Cancer Res. 2005, 11, 67–72. [Google Scholar] [CrossRef]
- Zhang, L.; Conejo-Garcia, J.R.; Katsaros, D.; Gimotty, P.A.; Massobrio, M.; Regnani, G.; Makrigiannakis, A.; Gray, H.; Schlienger, K.; Liebman, M.N.; et al. Intratumoral T Cells, Recurrence, and Survival in Epithelial Ovarian Cancer. N. Engl. J. Med. 2003, 348, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.D.; Warren, R.L.; Gibb, E.A.; Martin, S.D.; Spinelli, J.J.; Nelson, B.H.; Holt, R.A. Neo-Antigens Predicted by Tumor Genome Meta-Analysis Correlate with Increased Patient Survival. Genome Res. 2014, 24, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Wick, D.A.; Webb, J.R.; Nielsen, J.S.; Martin, S.D.; Kroeger, D.R.; Milne, K.; Castellarin, M.; Twumasi-Boateng, K.; Watson, P.H.; Holt, R.A.; et al. Surveillance of the Tumor Mutanome by T Cells during Progression from Primary to Recurrent Ovarian Cancer. Clin. Cancer Res. 2014, 20, 1125–1134. [Google Scholar] [CrossRef]
- Han, L.Y.; Fletcher, M.S.; Urbauer, D.L.; Mueller, P.; Landen, C.N.; Kamat, A.A.; Lin, Y.G.; Merritt, W.M.; Spannuth, W.A.; Deavers, M.T.; et al. HLA Class I Antigen Processing Machinery Component Expression and Intratumoral T-Cell Infiltrate as Independent Prognostic Markers in Ovarian Carcinoma. Clin. Cancer Res. 2008, 14, 3372–3379. [Google Scholar] [CrossRef] [PubMed]
- Santoiemma, P.P.; Reyes, C.; Wang, L.-P.; McLane, M.W.; Feldman, M.D.; Tanyi, J.L.; Powell, D.J. Systematic Evaluation of Multiple Immune Markers Reveals Prognostic Factors in Ovarian Cancer. Gynecol. Oncol. 2016, 143, 120–127. [Google Scholar] [CrossRef]
- Garrido, F.; Algarra, I. MHC Antigens and Tumor Escape from Immune Surveillance. Adv. Cancer Res. 2001, 83, 117–158. [Google Scholar] [CrossRef]
- Garrido, F.; Cabrera, T.; Aptsiauri, N. “Hard” and “Soft” Lesions Underlying the HLA Class I Alterations in Cancer Cells: Implications for Immunotherapy. Int. J. Cancer 2010, 127, 249–256. [Google Scholar] [CrossRef]
- Hobart, M.; Ramassar, V.; Goes, N.; Urmson, J.; Halloran, P.F. The Induction of Class I and II Major Histocompatibility Complex by Allogeneic Stimulation Is Dependent on the Transcription Factor Interferon Regulatory Factor 1 (IRF-1): Observations in IRF-1 Knockout Mice. Transplantation 1996, 62, 1895–1901. [Google Scholar] [CrossRef]
- Meissner, T.B.; Li, A.; Biswas, A.; Lee, K.-H.; Liu, Y.-J.; Bayir, E.; Iliopoulos, D.; van den Elsen, P.J.; Kobayashi, K.S. NLR Family Member NLRC5 Is a Transcriptional Regulator of MHC Class I Genes. Proc. Natl. Acad. Sci. USA 2010, 107, 13794–13799. [Google Scholar] [CrossRef]
- Naumann, M.; Scheidereit, C. Activation of NF-Kappa B in Vivo Is Regulated by Multiple Phosphorylations. EMBO J. 1994, 13, 4597–4607. [Google Scholar] [CrossRef]
- Goodell, V.; Salazar, L.G.; Urban, N.; Drescher, C.W.; Gray, H.; Swensen, R.E.; McIntosh, M.W.; Disis, M.L. Antibody Immunity to the P53 Oncogenic Protein Is a Prognostic Indicator in Ovarian Cancer. J. Clin. Oncol. 2006, 24, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.D.; Brown, S.D.; Wick, D.A.; Nielsen, J.S.; Kroeger, D.R.; Twumasi-Boateng, K.; Holt, R.A.; Nelson, B.H. Low Mutation Burden in Ovarian Cancer May Limit the Utility of Neoantigen-Targeted Vaccines. PLoS ONE 2016, 11, e0155189. [Google Scholar] [CrossRef]
- Luo, N.; Nixon, M.J.; Gonzalez-Ericsson, P.I.; Sanchez, V.; Opalenik, S.R.; Li, H.; Zahnow, C.A.; Nickels, M.L.; Liu, F.; Tantawy, M.N.; et al. DNA Methyltransferase Inhibition Upregulates MHC-I to Potentiate Cytotoxic T Lymphocyte Responses in Breast Cancer. Nat. Commun. 2018, 9, 248. [Google Scholar] [CrossRef]
- Taylor, B.C.; Balko, J.M. Mechanisms of MHC-I Downregulation and Role in Immunotherapy Response. Front. Immunol. 2022, 13, 844866. [Google Scholar] [CrossRef] [PubMed]
- Rolland, P.; Deen, S.; Scott, I.; Durrant, L.; Spendlove, I. Human Leukocyte Antigen Class I Antigen Expression Is an Independent Prognostic Factor in Ovarian Cancer. Clin. Cancer Res. 2007, 13, 3591–3596. [Google Scholar] [CrossRef] [PubMed]
- Szender, J.B.; Eng, K.H.; Matsuzaki, J.; Miliotto, A.; Gnjatic, S.; Tsuji, T.; Odunsi, K. HLA Superfamily Assignment Is a Predictor of Immune Response to Cancer Testis Antigens and Survival in Ovarian Cancer. Gynecol. Oncol. 2016, 142, 158–162. [Google Scholar] [CrossRef][Green Version]
- Shukla, S.A.; Rooney, M.S.; Rajasagi, M.; Tiao, G.; Dixon, P.M.; Lawrence, M.S.; Stevens, J.; Lane, W.J.; Dellagatta, J.L.; Steelman, S.; et al. Comprehensive Analysis of Cancer-Associated Somatic Mutations in Class I HLA Genes. Nat. Biotechnol. 2015, 33, 1152–1158. [Google Scholar] [CrossRef]
- Schuster, H.; Peper, J.K.; Bösmüller, H.-C.; Röhle, K.; Backert, L.; Bilich, T.; Ney, B.; Löffler, M.W.; Kowalewski, D.J.; Trautwein, N.; et al. The Immunopeptidomic Landscape of Ovarian Carcinomas. Proc. Natl. Acad. Sci. USA 2017, 114, E9942–E9951. [Google Scholar] [CrossRef]
- Griesinger, L.; Nyarko-Odoom, A.; Martinez, S.A.; Shen, N.W.; Ring, K.L.; Gaughan, E.M.; Mills, A.M. PD-L1 and MHC Class I Expression in High-Grade Ovarian Cancers, Including Platinum-Resistant Recurrences Treated with Checkpoint Inhibitor Therapy. Appl. Immunohistochem. Mol. Morphol. 2023, 31, 197–203. [Google Scholar] [CrossRef]
- Monos, D.S.; Winchester, R.J. The Major Histocompatibility Complex. In Clinical Immunology: Principles and Practice, 5th ed.; Elsevier: Amsterdam, The Netherlands, 2019; Volume 5, pp. 79–92. [Google Scholar]
- Wyatt, R.C.; Lanzoni, G.; Russell, M.A.; Gerling, I.; Richardson, S.J. What the HLA-I!—Classical and Non-Classical HLA Class I and Their Potential Roles in Type 1 Diabetes. Curr. Diab. Rep. 2019, 19, 159. [Google Scholar] [CrossRef]
- Menier, C.; Saez, B.; Horejsi, V.; Martinozzi, S.; Krawice-Radanne, I.; Bruel, S.; Le Danff, C.; Reboul, M.; Hilgert, I.; Rabreau, M.; et al. Characterization of Monoclonal Antibodies Recognizing HLA-G or HLA-E: New Tools to Analyze the Expression of Nonclassical HLA Class I Molecules. Hum. Immunol. 2003, 64, 315–326. [Google Scholar] [CrossRef]
- Marín, R.; Ruiz-Cabello, F.; Pedrinaci, S.; Méndez, R.; Jiménez, P.; Geraghty, D.E.; Garrido, F. Analysis of HLA-E Expression in Human Tumors. Immunogenetics 2003, 54, 767–775. [Google Scholar] [CrossRef]
- Wei, X.; Orr, H.T. Differential Expression of HLA-E, HLA-F, and HLA-G Transcripts in Human Tissue. Hum. Immunol. 1990, 29, 131–142. [Google Scholar] [CrossRef]
- Lee, N.; Llano, M.; Carretero, M.; Ishitani, A.; Navarro, F.; López-Botet, M.; Geraghty, D.E. HLA-E Is a Major Ligand for the Natural Killer Inhibitory Receptor CD94/NKG2A. Proc. Natl. Acad. Sci. USA 1998, 95, 5199–5204. [Google Scholar] [CrossRef]
- Speiser, D.E.; Valmori, D.; Rimoldi, D.; Pittet, M.J.; Liénard, D.; Cerundolo, V.; MacDonald, H.R.; Cerottini, J.C.; Romero, P. CD28-Negative Cytolytic Effector T Cells Frequently Express NK Receptors and Are Present at Variable Proportions in Circulating Lymphocytes from Healthy Donors and Melanoma Patients. Eur. J. Immunol. 1999, 29, 1990–1999. [Google Scholar] [CrossRef]
- Gooden, M.; Lampen, M.; Jordanova, E.S.; Leffers, N.; Trimbos, J.B.; van der Burg, S.H.; Nijman, H.; van Hall, T. HLA-E Expression by Gynecological Cancers Restrains Tumor-Infiltrating CD8+ T Lymphocytes. Proc. Natl. Acad. Sci. USA 2011, 108, 10656–10661. [Google Scholar] [CrossRef]
- Eugène, J.; Jouand, N.; Ducoin, K.; Dansette, D.; Oger, R.; Deleine, C.; Leveque, E.; Meurette, G.; Podevin, J.; Matysiak, T.; et al. The Inhibitory Receptor CD94/NKG2A on CD8+ Tumor-Infiltrating Lymphocytes in Colorectal Cancer: A Promising New Druggable Immune Checkpoint in the Context of HLAE/Β2m Overexpression. Mod. Pathol. 2020, 33, 468–482. [Google Scholar] [CrossRef]
- Fumet, J.-D.; Lardenois, E.; Ray-Coquard, I.; Harter, P.; Joly, F.; Canzler, U.; Truntzer, C.; Tredan, O.; Liebrich, C.; Lortholary, A.; et al. Genomic Instability Is Defined by Specific Tumor Microenvironment in Ovarian Cancer: A Subgroup Analysis of AGO OVAR 12 Trial. Cancers 2022, 14, 1189. [Google Scholar] [CrossRef]
- Li, T.; Chen, Z.J. The CGAS–CGAMP–STING Pathway Connects DNA Damage to Inflammation, Senescence, and Cancer. J. Exp. Med. 2018, 215, 1287–1299. [Google Scholar] [CrossRef]
- Nguyen, S.; Beziat, V.; Dhedin, N.; Kuentz, M.; Vernant, J.P.; Debre, P.; Vieillard, V. HLA-E Upregulation on IFN-γ-Activated AML Blasts Impairs CD94/NKG2A-Dependent NK Cytolysis after Haplo-Mismatched Hematopoietic SCT. Bone Marrow. Transpl. 2009, 43, 693–699. [Google Scholar] [CrossRef]
- Zheng, H.; Lu, R.; Xie, S.; Wen, X.; Wang, H.; Gao, X.; Guo, L. Human Leukocyte Antigen-E Alleles and Expression in Patients with Serous Ovarian Cancer. Cancer Sci. 2015, 106, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Ishitani, A.; Geraghty, D.E. HLA-F Is a Surface Marker on Activated Lymphocytes. Eur. J. Immunol. 2010, 40, 2308–2318. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, S.D.; Biro, P.A.; Holmes, C.H. HLA-F Is a Predominantly Empty, Intracellular, TAP-Associated MHC Class Ib Protein with a Restricted Expression Pattern1. J. Immunol. 2000, 164, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Lepin, E.J.M.; Bastin, J.M.; Allan, D.S.J.; Roncador, G.; Braud, V.M.; Mason, D.Y.; van der Merwe, P.A.; McMichael, A.J.; Bell, J.I.; Powis, S.H.; et al. Functional Characterization of HLA-F and Binding of HLA-F Tetramers to ILT2 and ILT4 Receptors. Eur. J. Immunol. 2000, 30, 3552–3561. [Google Scholar] [CrossRef] [PubMed]
- Goodridge, J.P.; Burian, A.; Lee, N.; Geraghty, D.E. HLA-F and MHC Class I Open Conformers Are Ligands for NK Cell Ig-like Receptors. J. Immunol. 2013, 191, 3553–3562. [Google Scholar] [CrossRef]
- Burian, A.; Wang, K.L.; Finton, K.A.K.; Lee, N.; Ishitani, A.; Strong, R.K.; Geraghty, D.E. HLA-F and MHC-I Open Conformers Bind Natural Killer Cell Ig-Like Receptor KIR3DS1. PLoS ONE 2016, 11, e0163297. [Google Scholar] [CrossRef]
- Goodridge, J.P.; Lee, N.; Burian, A.; Pyo, C.-W.; Tykodi, S.S.; Warren, E.H.; Yee, C.; Riddell, S.R.; Geraghty, D.E. HLA-F and MHC-I Open Conformers Cooperate in a MHC-I Antigen Cross-Presentation Pathway. J. Immunol. 2013, 191, 1567–1577. [Google Scholar] [CrossRef]
- Hrbac, T.; Kopkova, A.; Siegl, F.; Vecera, M.; Ruckova, M.; Kazda, T.; Jancalek, R.; Hendrych, M.; Hermanova, M.; Vybihal, V.; et al. HLA-E and HLA-F Are Overexpressed in Glioblastoma and HLA-E Increased After Exposure to Ionizing Radiation. Cancer Genom. Proteom. 2022, 19, 151–162. [Google Scholar] [CrossRef]
- Fang, W.; Xia, Y. LncRNA HLA-F-AS1 Attenuates the Ovarian Cancer Development by Targeting MiR-21-3p/PEG3 Axis. Anti Cancer Drugs 2022, 33, 671. [Google Scholar] [CrossRef]
- Kovats, S.; Main, E.K.; Librach, C.; Stubblebine, M.; Fisher, S.J.; DeMars, R. A Class I Antigen, HLA-G, Expressed in Human Trophoblasts. Science 1990, 248, 220–223. [Google Scholar] [CrossRef]
- Cirulli, V.; Zalatan, J.; McMaster, M.; Prinsen, R.; Salomon, D.R.; Ricordi, C.; Torbett, B.E.; Meda, P.; Crisa, L. The Class I HLA Repertoire of Pancreatic Islets Comprises the Nonclassical Class Ib Antigen HLA-G. Diabetes 2006, 55, 1214–1222. [Google Scholar] [CrossRef]
- Le Discorde, M.; Moreau, P.; Sabatier, P.; Legeais, J.-M.; Carosella, E.D. Expression of HLA-G in Human Cornea, an Immune-Privileged Tissue. Hum. Immunol. 2003, 64, 1039–1044. [Google Scholar] [CrossRef]
- Menier, C.; Rabreau, M.; Challier, J.-C.; Le Discorde, M.; Carosella, E.D.; Rouas-Freiss, N. Erythroblasts Secrete the Nonclassical HLA-G Molecule from Primitive to Definitive Hematopoiesis. Blood 2004, 104, 3153–3160. [Google Scholar] [CrossRef]
- Crisa, L.; McMaster, M.T.; Ishii, J.K.; Fisher, S.J.; Salomon, D.R. Identification of a Thymic Epithelial Cell Subset Sharing Expression of the Class Ib HLA-G Molecule with Fetal Trophoblasts. J. Exp. Med. 1997, 186, 289–298. [Google Scholar] [CrossRef]
- Rouas-Freiss, N.; Moreau, P.; LeMaoult, J.; Carosella, E.D. The Dual Role of HLA-G in Cancer. J. Immunol. Res. 2014, 2014, 359748. [Google Scholar] [CrossRef]
- Barbaro, G.; Inversetti, A.; Cristodoro, M.; Ticconi, C.; Scambia, G.; Di Simone, N. HLA-G and Recurrent Pregnancy Loss. Int. J. Mol. Sci. 2023, 24, 2557. [Google Scholar] [CrossRef] [PubMed]
- Contini, P.; Ghio, M.; Poggi, A.; Filaci, G.; Indiveri, F.; Ferrone, S.; Puppo, F. Soluble HLA-A,-B,-C and -G Molecules Induce Apoptosis in T and NK CD8+ Cells and Inhibit Cytotoxic T Cell Activity through CD8 Ligation. Eur. J. Immunol. 2003, 33, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Kleinberg, L.; Flørenes, V.A.; Skrede, M.; Dong, H.P.; Nielsen, S.; McMaster, M.T.; Nesland, J.M.; Shih, I.-M.; Davidson, B. Expression of HLA-G in Malignant Mesothelioma and Clinically Aggressive Breast Carcinoma. Virchows Arch. 2006, 449, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Rouas-Freiss, N.; Moreau, P.; Ferrone, S.; Carosella, E.D. HLA-G Proteins in Cancer: Do They Provide Tumor Cells with an Escape Mechanism? Cancer Res. 2005, 65, 10139–10144. [Google Scholar] [CrossRef] [PubMed]
- Carosella, E.D.; Favier, B.; Rouas-Freiss, N.; Moreau, P.; Lemaoult, J. Beyond the Increasing Complexity of the Immunomodulatory HLA-G Molecule. Blood 2008, 111, 4862–4870. [Google Scholar] [CrossRef]
- Lin, A.; Zhang, X.; Xu, H.-H.; Xu, D.-P.; Ruan, Y.-Y.; Yan, W.-H. HLA-G Expression Is Associated with Metastasis and Poor Survival in the Balb/c Nu/Nu Murine Tumor Model with Ovarian Cancer. Int. J. Cancer 2012, 131, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Kanai, T.; Fujii, T.; Unno, N.; Yamashita, T.; Hyodo, H.; Miki, A.; Hamai, Y.; Kozuma, S.; Taketani, Y. Human Leukocyte Antigen-G-Expressing Cells Differently Modulate the Release of Cytokines from Mononuclear Cells Present in the Decidua versus Peripheral Blood. Am. J. Reprod. Immunol. 2001, 45, 94–99. [Google Scholar] [CrossRef]
- Kanai, T.; Fujii, T.; Kozuma, S.; Yamashita, T.; Miki, A.; Kikuchi, A.; Taketani, Y. Soluble HLA-G Influences the Release of Cytokines from Allogeneic Peripheral Blood Mononuclear Cells in Culture. Mol. Hum. Reprod. 2001, 7, 195–200. [Google Scholar] [CrossRef]
- Babay, W.; Ben Yahia, H.; Boujelbene, N.; Zidi, N.; Laaribi, A.B.; Kacem, D.; Ben Ghorbel, R.; Boudabous, A.; Ouzari, H.-I.; Rizzo, R.; et al. Clinicopathologic Significance of HLA-G and HLA-E Molecules in Tunisian Patients with Ovarian Carcinoma. Hum. Immunol. 2018, 79, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Singer, G.; Rebmann, V.; Chen, Y.-C.; Liu, H.-T.; Ali, S.Z.; Reinsberg, J.; McMaster, M.T.; Pfeiffer, K.; Chan, D.W.; Wardelmann, E.; et al. HLA-G Is a Potential Tumor Marker in Malignant Ascites. Clin. Cancer Res. 2003, 9, 4460–4464. [Google Scholar] [PubMed]
- Babay, W.; Boujelbene, N.; Ben Yahia, H.; Bortolotti, D.; Zemni, I.; Ouzari, H.-I.; Chelbi, H.; Mezlini, A.; Rizzo, R.; Zidi, I. Prognostic Significance of High Circulating SHLA-G in Ovarian Carcinoma. HLA 2021, 98, 357–365. [Google Scholar] [CrossRef]
- McCormick, J.; Whitley, G.S.J.; Le Bouteiller, P.; Cartwright, J.E. Soluble HLA-G Regulates Motility and Invasion of the Trophoblast-Derived Cell Line SGHPL-4. Hum. Reprod. 2009, 24, 1339–1345. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rutten, M.J.; Dijk, F.; Savci-Heijink, C.D.; Buist, M.R.; Kenter, G.G.; van de Vijver, M.J.; Jordanova, E.S. HLA-G Expression Is an Independent Predictor for Improved Survival in High Grade Ovarian Carcinomas. J. Immunol. Res. 2014, 2014, 274584. [Google Scholar] [CrossRef]
- Menier, C.; Prevot, S.; Carosella, E.D.; Rouas-Freiss, N. Human Leukocyte Antigen-G Is Expressed in Advanced-Stage Ovarian Carcinoma of High-Grade Histology. Hum. Immunol. 2009, 70, 1006–1009. [Google Scholar] [CrossRef]
- Downs, I.; Vijayan, S.; Sidiq, T.; Kobayashi, K.S. CITA/NLRC5: A Critical Transcriptional Regulator of MHC Class I Gene Expression. Biofactors 2016, 42, 349–357. [Google Scholar] [CrossRef]
- Staehli, F.; Ludigs, K.; Heinz, L.X.; Seguín-Estévez, Q.; Ferrero, I.; Braun, M.; Schroder, K.; Rebsamen, M.; Tardivel, A.; Mattmann, C.; et al. NLRC5 Deficiency Selectively Impairs MHC Class I- Dependent Lymphocyte Killing by Cytotoxic T Cells. J. Immunol. 2012, 188, 3820–3828. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Wang, Y.; Chen, F.; Huang, Y.; Zhu, S.; Leng, Q.; Wang, H.; Shi, Y.; Qian, Y. NLRC5 Regulates MHC Class I Antigen Presentation in Host Defense against Intracellular Pathogens. Cell Res. 2012, 22, 836–847. [Google Scholar] [CrossRef] [PubMed]
- Yoshihama, S.; Roszik, J.; Downs, I.; Meissner, T.B.; Vijayan, S.; Chapuy, B.; Sidiq, T.; Shipp, M.A.; Lizee, G.A.; Kobayashi, K.S. NLRC5/MHC Class I Transactivator Is a Target for Immune Evasion in Cancer. Proc. Natl. Acad. Sci. USA 2016, 113, 5999–6004. [Google Scholar] [CrossRef] [PubMed]
- Yoshihama, S.; Vijayan, S.; Sidiq, T.; Kobayashi, K.S. NLRC5/CITA: A Key Player in Cancer Immune Surveillance. Trends Cancer 2017, 3, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, G.M.; Galpin, K.J.C.; McCloskey, C.W.; Vanderhyden, B.C. The Tumor Microenvironment of Epithelial Ovarian Cancer and Its Influence on Response to Immunotherapy. Cancers 2018, 10, E242. [Google Scholar] [CrossRef]
- Bubeník, J. Tumour MHC Class I Downregulation and Immunotherapy (Review). Oncol. Rep. 2003, 10, 2005–2008. [Google Scholar] [CrossRef]
- Norell, H.; Carlsten, M.; Ohlum, T.; Malmberg, K.-J.; Masucci, G.; Schedvins, K.; Altermann, W.; Handke, D.; Atkins, D.; Seliger, B.; et al. Frequent Loss of HLA-A2 Expression in Metastasizing Ovarian Carcinomas Associated with Genomic Haplotype Loss and HLA-A2-Restricted HER-2/Neu-Specific Immunity. Cancer Res. 2006, 66, 6387–6394. [Google Scholar] [CrossRef]
- Shimono, Y.; Ugalde, M.Z.; Cho, R.W.; Lobo, N.; Dalerba, P.; Qian, D.; Diehn, M.; Liu, H.; Panula, S.P.; Chiao, E.; et al. Down-Regulation of MiRNA-200c Links Breast Cancer Stem Cells with Normal Stem Cells. Cell 2009, 138, 592–603. [Google Scholar] [CrossRef]
- Yang, M.-H.; Hsu, D.S.-S.; Wang, H.-W.; Wang, H.-J.; Lan, H.-Y.; Yang, W.-H.; Huang, C.-H.; Kao, S.-Y.; Tzeng, C.-H.; Tai, S.-K.; et al. Bmi1 Is Essential in Twist1-Induced Epithelial–Mesenchymal Transition. Nat. Cell. Biol. 2010, 12, 982–992. [Google Scholar] [CrossRef] [PubMed]
- Dongre, A.; Weinberg, R.A. New Insights into the Mechanisms of Epithelial–Mesenchymal Transition and Implications for Cancer. Nat. Rev. Mol. Cell. Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Weinberg, R.A. EMT and Cancer: More Than Meets the Eye. Dev. Cell 2019, 49, 313–316. [Google Scholar] [CrossRef]
- Chen, X.-H.; Liu, Z.-C.; Zhang, G.; Wei, W.; Wang, X.-X.; Wang, H.; Ke, H.-P.; Zhang, F.; Wang, H.-S.; Cai, S.-H.; et al. TGF-β and EGF Induced HLA-I Downregulation Is Associated with Epithelial-Mesenchymal Transition (EMT) through Upregulation of Snail in Prostate Cancer Cells. Mol. Immunol. 2015, 65, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Dongre, A.; Rashidian, M.; Reinhardt, F.; Bagnato, A.; Keckesova, Z.; Ploegh, H.L.; Weinberg, R.A. Epithelial-to-Mesenchymal Transition Contributes to Immunosuppression in Breast Carcinomas. Cancer Res. 2017, 77, 3982–3989. [Google Scholar] [CrossRef]
- Pires, P.R.L.; Xavier, P.L.P.; Fukumasu, H. Abstract B180: Effects of EMT Process under MHC Class I and TAP1 Gene Expression Related to Antigen Presentation. Cancer Immunol. Res. 2019, 7, B180. [Google Scholar] [CrossRef]
- Porter, R.L.; Sun, S.; Flores, M.N.; Berzolla, E.; You, E.; Phillips, I.E.; Kc, N.; Desai, N.; Tai, E.C.; Szabolcs, A.; et al. Satellite Repeat RNA Expression in Epithelial Ovarian Cancer Associates with a Tumor-Immunosuppressive Phenotype. J. Clin. Invest. 2022, 132. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.C.; Sankpal, N.V.; Gillanders, W.E. Functional Implications of the Dynamic Regulation of EpCAM during Epithelial-to-Mesenchymal Transition. Biomolecules 2021, 11, 956. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Zhang, Z.; ten Dijke, P. Harnessing Epithelial-Mesenchymal Plasticity to Boost Cancer Immunotherapy. Cell Mol. Immunol. 2023, 20, 318–340. [Google Scholar] [CrossRef]
- Zingg, D.; Debbache, J.; Schaefer, S.M.; Tuncer, E.; Frommel, S.C.; Cheng, P.; Arenas-Ramirez, N.; Haeusel, J.; Zhang, Y.; Bonalli, M.; et al. The Epigenetic Modifier EZH2 Controls Melanoma Growth and Metastasis through Silencing of Distinct Tumour Suppressors. Nat. Commun. 2015, 6, 6051. [Google Scholar] [CrossRef]
- Kleer, C.G.; Cao, Q.; Varambally, S.; Shen, R.; Ota, I.; Tomlins, S.A.; Ghosh, D.; Sewalt, R.G.A.B.; Otte, A.P.; Hayes, D.F.; et al. EZH2 Is a Marker of Aggressive Breast Cancer and Promotes Neoplastic Transformation of Breast Epithelial Cells. Proc. Natl. Acad. Sci. USA 2003, 100, 11606–11611. [Google Scholar] [CrossRef]
- Collett, K.; Eide, G.E.; Arnes, J.; Stefansson, I.M.; Eide, J.; Braaten, A.; Aas, T.; Otte, A.P.; Akslen, L.A. Expression of Enhancer of Zeste Homologue 2 Is Significantly Associated with Increased Tumor Cell Proliferation and Is a Marker of Aggressive Breast Cancer. Clin. Cancer Res. 2006, 12, 1168–1174. [Google Scholar] [CrossRef]
- Zhou, L.; Mudianto, T.; Ma, X.; Riley, R.; Uppaluri, R. Targeting EZH2 Enhances Antigen Presentation, Antitumor Immunity and Circumvents Anti-PD-1 Resistance in Head and Neck Cancer. Clin. Cancer Res. 2020, 26, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.; Xu, H.; Ma, L.; Li, C.; Qin, W.; Chen, X.; Yi, M.; Sun, L.; Liu, B.; Yuan, X. Nintedanib Enhances the Efficacy of PD-L1 Blockade by Upregulating MHC-I and PD-L1 Expression in Tumor Cells. Theranostics 2022, 12, 747–766. [Google Scholar] [CrossRef] [PubMed]
- Josson, S.; Nomura, T.; Lin, J.-T.; Huang, W.-C.; Wu, D.; Zhau, H.E.; Zayzafoon, M.; Weizmann, M.N.; Gururajan, M.; Chung, L.W.K. Β2-Microglobulin Induces Epithelial to Mesenchymal Transition and Confers Cancer Lethality and Bone Metastasis in Human Cancer Cells. Cancer Res. 2011, 71, 2600–2610. [Google Scholar] [CrossRef] [PubMed]
- Chockley, P.J.; Keshamouni, V.G. Immunological Consequences of Epithelial-Mesenchymal Transition in Tumor Progression. J. Immunol. 2016, 197, 691–698. [Google Scholar] [CrossRef]
- Lazaridou, M.-F.; Gonschorek, E.; Massa, C.; Friedrich, M.; Handke, D.; Mueller, A.; Jasinski-Bergner, S.; Dummer, R.; Koelblinger, P.; Seliger, B. Identification of MiR-200a-5p Targeting the Peptide Transporter TAP1 and Its Association with the Clinical Outcome of Melanoma Patients. OncoImmunology 2020, 9, 1774323. [Google Scholar] [CrossRef] [PubMed]
- Camp, F.A.; Brunetti, T.M.; Williams, M.M.; Christenson, J.L.; Sreekanth, V.; Costello, J.C.; Hay, Z.L.Z.; Kedl, R.M.; Richer, J.K.; Slansky, J.E. Antigens Expressed by Breast Cancer Cells Undergoing EMT Stimulate Cytotoxic CD8+ T Cell Immunity. Cancers 2022, 14, 4397. [Google Scholar] [CrossRef]
- Cavallari, I.; Ciccarese, F.; Sharova, E.; Urso, L.; Raimondi, V.; Silic-Benussi, M.; D’Agostino, D.M.; Ciminale, V. The MiR-200 Family of MicroRNAs: Fine Tuners of Epithelial-Mesenchymal Transition and Circulating Cancer Biomarkers. Cancers 2021, 13, 5874. [Google Scholar] [CrossRef]
- Cook, D.P.; Vanderhyden, B.C. Transcriptional Census of Epithelial-Mesenchymal Plasticity in Cancer. Sci. Adv. 2022, 8, eabi7640. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) Hallmark Gene Set Collection. Cell. Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J. TGFβ in Cancer. Cell 2008, 134, 215–230. [Google Scholar] [CrossRef]
- Scheel, C.; Eaton, E.N.; Li, S.H.-J.; Chaffer, C.L.; Reinhardt, F.; Kah, K.-J.; Bell, G.; Guo, W.; Rubin, J.; Richardson, A.L.; et al. Paracrine and Autocrine Signals Induce and Maintain Mesenchymal and Stem Cell States in the Breast. Cell 2011, 145, 926–940. [Google Scholar] [CrossRef] [PubMed]
- Larocca, C.; Cohen, J.R.; Fernando, R.I.; Huang, B.; Hamilton, D.H.; Palena, C. An Autocrine Loop between TGF-Β1 and the Transcription Factor Brachyury Controls the Transition of Human Carcinoma Cells into a Mesenchymal Phenotype. Mol. Cancer Ther. 2013, 12, 1805–1815. [Google Scholar] [CrossRef] [PubMed]
- Valcourt, U.; Kowanetz, M.; Niimi, H.; Heldin, C.-H.; Moustakas, A. TGF-β and the Smad Signaling Pathway Support Transcriptomic Reprogramming during Epithelial-Mesenchymal Cell Transition. Mol. Biol. Cell 2005, 16, 1987–2002. [Google Scholar] [CrossRef] [PubMed]
- Dodagatta-Marri, E.; Meyer, D.S.; Reeves, M.Q.; Paniagua, R.; To, M.D.; Binnewies, M.; Broz, M.L.; Mori, H.; Wu, D.; Adoumie, M.; et al. α-PD-1 Therapy Elevates Treg/Th Balance and Increases Tumor Cell PSmad3 That Are Both Targeted by α-TGFβ Antibody to Promote Durable Rejection and Immunity in Squamous Cell Carcinomas. J. Immunother. Cancer. 2019, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- Soundararajan, R.; Fradette, J.J.; Konen, J.M.; Moulder, S.; Zhang, X.; Gibbons, D.L.; Varadarajan, N.; Wistuba, I.I.; Tripathy, D.; Bernatchez, C.; et al. Targeting the Interplay between Epithelial-to-Mesenchymal-Transition and the Immune System for Effective Immunotherapy. Cancers 2019, 11, 714. [Google Scholar] [CrossRef]
- erglund, A.K.; Hinson, A.L.; Schnabel, L.V. TGF-β downregulates antigen processing and presentation genes and MHC I surface expression through a Smad3-dependent mechanism. bioRxiv 2023. [Google Scholar] [CrossRef]
- Cook, D.P.; Vanderhyden, B.C. Context Specificity of the EMT Transcriptional Response. Nat. Commun. 2020, 11, 2142. [Google Scholar] [CrossRef]
- Machado, C.D.; Telles, P.D.; Nascimento, I.L. Immunological Characteristics of Mesenchymal Stem Cells. Rev. Bras. Hematol. Hemoter. 2013, 35, 62–67. [Google Scholar] [CrossRef]
- Apavaloaei, A.; Hesnard, L.; Hardy, M.-P.; Benabdallah, B.; Ehx, G.; Thériault, C.; Laverdure, J.-P.; Durette, C.; Lanoix, J.; Courcelles, M.; et al. Induced Pluripotent Stem Cells Display a Distinct Set of MHC I-Associated Peptides Shared by Human Cancers. Cell Rep. 2022, 40. [Google Scholar] [CrossRef]
- Bertone, S.; Schiavetti, F.; Bellomo, R.; Vitale, C.; Ponte, M.; Moretta, L.; Mingari, M.C. Transforming Growth Factor-Beta-Induced Expression of CD94/NKG2A Inhibitory Receptors in Human T Lymphocytes. Eur. J. Immunol. 1999, 29, 23–29. [Google Scholar] [CrossRef]
- Gunturi, A.; Berg, R.E.; Crossley, E.; Murray, S.; Forman, J. The Role of TCR Stimulation and TGF-Beta in Controlling the Expression of CD94/NKG2A Receptors on CD8 T Cells. Eur. J. Immunol. 2005, 35, 766–775. [Google Scholar] [CrossRef]
- Gooden, M.J.M.; van Hall, T. Infiltrating CTLs Are Bothered by HLA-E on Tumors. Oncoimmunology 2012, 1, 92–93. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, B.; Wu, Y.; Ge, Z.; Zhang, X.; Yan, Y.; Xie, Y. NLRC5 Deficiency Ameliorates Cardiac Fibrosis in Diabetic Cardiomyopathy by Regulating EndMT through Smad2/3 Signaling Pathway. Biochem. Biophys. Res. Commun. 2020, 528, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Ni, M.; Li, X.; Meng, X.; Huang, C.; Li, J. NLRC5 Regulates TGF-Β1-Induced Proliferation and Activation of Hepatic Stellate Cells during Hepatic Fibrosis. Int. J. Biochem. Cell Biol. 2016, 70, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Romieu-Mourez, R.; François, M.; Boivin, M.-N.; Stagg, J.; Galipeau, J. Regulation of MHC Class II Expression and Antigen Processing in Murine and Human Mesenchymal Stromal Cells by IFN-γ, TGF-β, and Cell Density1. J. Immunol. 2007, 179, 1549–1558. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Tang, L.; Letterio, J.J.; Benveniste, E.N. The Smad3 Protein Is Involved in TGF-β Inhibition of Class II Transactivator and Class II MHC Expression1. J. Immunol. 2001, 167, 311–319. [Google Scholar] [CrossRef]
- Uribe, M.L.; Marrocco, I.; Yarden, Y. EGFR in Cancer: Signaling Mechanisms, Drugs, and Acquired Resistance. Cancers 2021, 13, 2748. [Google Scholar] [CrossRef] [PubMed]
- Brea, E.J.; Oh, C.Y.; Manchado, E.; Budhu, S.; Gejman, R.S.; Mo, G.; Mondello, P.; Han, J.E.; Jarvis, C.A.; Ulmert, D.; et al. Kinase Regulation of Human MHC Class I Molecule Expression on Cancer Cells. Cancer Immunol. Res. 2016, 4, 936–947. [Google Scholar] [CrossRef]
- Loi, S.; Dushyanthen, S.; Beavis, P.A.; Salgado, R.; Denkert, C.; Savas, P.; Combs, S.; Rimm, D.L.; Giltnane, J.M.; Estrada, M.V.; et al. RAS/MAPK Activation Is Associated with Reduced Tumor-Infiltrating Lymphocytes in Triple-Negative Breast Cancer: Therapeutic Cooperation between MEK and PD-1/PD-L1 Immune Checkpoint Inhibitors. Clin. Cancer Res. 2016, 22, 1499–1509. [Google Scholar] [CrossRef]
- Franklin, D.A.; James, J.L.; Axelrod, M.L.; Balko, J.M. MEK Inhibition Activates STAT Signaling to Increase Breast Cancer Immunogenicity via MHC-I Expression. Cancer Drug Resist. 2020, 3, 603–612. [Google Scholar] [CrossRef]
- Inoue, M.; Mimura, K.; Izawa, S.; Shiraishi, K.; Inoue, A.; Shiba, S.; Watanabe, M.; Maruyama, T.; Kawaguchi, Y.; Inoue, S.; et al. Expression of MHC Class I on Breast Cancer Cells Correlates Inversely with HER2 Expression. Oncoimmunology 2012, 1, 1104–1110. [Google Scholar] [CrossRef]
- Velásquez, L.N.; Milillo, M.A.; Delpino, M.V.; Trotta, A.; Mercogliano, M.F.; Pozner, R.G.; Schillaci, R.; Elizalde, P.V.; Giambartolomei, G.H.; Barrionuevo, P. Inhibition of MHC-I by Brucella Abortus Is an Early Event during Infection and Involves EGFR Pathway. Immunol. Cell Biol. 2017, 95, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Pollack, B.P.; Sapkota, B.; Cartee, T.V. Epidermal Growth Factor Receptor Inhibition Augments the Expression of MHC Class I and II Genes. Clin. Cancer Res. 2011, 17, 4400–4413. [Google Scholar] [CrossRef] [PubMed]
- Wieduwilt, M.J.; Moasser, M.M. The Epidermal Growth Factor Receptor Family: Biology Driving Targeted Therapeutics. Cell Mol. Life Sci. 2008, 65, 1566–1584. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Hiroki, K.; Yamashita, Y. The Role of Epidermal Growth Factor Receptor in Cancer Metastasis and Microenvironment. Biomed. Res. Int. 2013, 2013, 546318. [Google Scholar] [CrossRef] [PubMed]
- Cornel, A.M.; Mimpen, I.L.; Nierkens, S. MHC Class I Downregulation in Cancer: Underlying Mechanisms and Potential Targets for Cancer Immunotherapy. Cancers 2020, 12, 1760. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Kerner, Z.J.; Hong, H.; Sun, J. Targeted Cancer Therapy with Tumor Necrosis Factor-Alpha. Biochem. Insights 2008, 2008, 15–21. [Google Scholar] [CrossRef]
- Lorenzi, S.; Forloni, M.; Cifaldi, L.; Antonucci, C.; Citti, A.; Boldrini, R.; Pezzullo, M.; Castellano, A.; Russo, V.; van der Bruggen, P.; et al. IRF1 and NF-KB Restore MHC Class I-Restricted Tumor Antigen Processing and Presentation to Cytotoxic T Cells in Aggressive Neuroblastoma. PLoS ONE 2012, 7, e46928. [Google Scholar] [CrossRef]
- Forloni, M.; Albini, S.; Limongi, M.Z.; Cifaldi, L.; Boldrini, R.; Nicotra, M.R.; Giannini, G.; Natali, P.G.; Giacomini, P.; Fruci, D. NF-ΚB, and Not MYCN, Regulates MHC Class I and Endoplasmic Reticulum Aminopeptidases in Human Neuroblastoma Cells. Cancer Res. 2010, 70, 916–924. [Google Scholar] [CrossRef]
- Nishio, H.; Yaguchi, T.; Sugiyama, J.; Sumimoto, H.; Umezawa, K.; Iwata, T.; Susumu, N.; Fujii, T.; Kawamura, N.; Kobayashi, A.; et al. Immunosuppression through Constitutively Activated NF-ΚB Signalling in Human Ovarian Cancer and Its Reversal by an NF-ΚB Inhibitor. Br. J. Cancer 2014, 110, 2965–2974. [Google Scholar] [CrossRef]
- Sadelain, M.; Rivière, I.; Riddell, S. Therapeutic T Cell Engineering. Nature 2017, 545, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.; Wickman, E.; DeRenzo, C.; Gottschalk, S. CAR T Cell Therapy for Solid Tumors: Bright Future or Dark Reality? Mol. Ther. 2020, 28, 2320–2339. [Google Scholar] [CrossRef] [PubMed]
- ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ (accessed on 3 November 2023).
- Zhang, X.-W.; Wu, Y.-S.; Xu, T.-M.; Cui, M.-H. CAR-T Cells in the Treatment of Ovarian Cancer: A Promising Cell Therapy. Biomolecules 2023, 13, 465. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.-Y.; Bang, Y.-J. HER2-Targeted Therapies — a Role beyond Breast Cancer. Nat. Rev. Clin. Oncol. 2020, 17, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Olvera, A.; Dueñas-González, A.; Gallardo-Rincón, D.; Candelaria, M.; De la Garza-Salazar, J. Prognostic, Predictive and Therapeutic Implications of HER2 in Invasive Epithelial Ovarian Cancer. Cancer Treat Rev. 2006, 32, 180–190. [Google Scholar] [CrossRef]
- Sun, M.; Shi, H.; Liu, C.; Liu, J.; Liu, X.; Sun, Y. Construction and Evaluation of a Novel Humanized HER2-Specific Chimeric Receptor. Breast Cancer Res. 2014, 16, R61. [Google Scholar] [CrossRef]
- Chang, K.; Pastan, I. Molecular Cloning of Mesothelin, a Differentiation Antigen Present on Mesothelium, Mesotheliomas, and Ovarian Cancers. Proc. Natl. Acad. Sci. USA 1996, 93, 136–140. [Google Scholar] [CrossRef]
- Ho, M.; Hassan, R.; Zhang, J.; Wang, Q.-C.; Onda, M.; Bera, T.; Pastan, I. Humoral Immune Response to Mesothelin in Mesothelioma and Ovarian Cancer Patients. Clin. Cancer Res. 2005, 11, 3814–3820. [Google Scholar] [CrossRef]
- Beatty, G.L.; Haas, A.R.; Maus, M.V.; Torigian, D.A.; Soulen, M.C.; Plesa, G.; Chew, A.; Zhao, Y.; Levine, B.L.; Albelda, S.M.; et al. Mesothelin-Specific Chimeric Antigen Receptor MRNA-Engineered T Cells Induce Anti-Tumor Activity in Solid Malignancies. Cancer Immunol. Res. 2014, 2, 112–120. [Google Scholar] [CrossRef]
- Rao, T.D.; Tian, H.; Ma, X.; Yan, X.; Thapi, S.; Schultz, N.; Rosales, N.; Monette, S.; Wang, A.; Hyman, D.M.; et al. Expression of the Carboxy-Terminal Portion of MUC16/CA125 Induces Transformation and Tumor Invasion. PLoS ONE 2015, 10, e0126633. [Google Scholar] [CrossRef]
- Felder, M.; Kapur, A.; Gonzalez-Bosquet, J.; Horibata, S.; Heintz, J.; Albrecht, R.; Fass, L.; Kaur, J.; Hu, K.; Shojaei, H.; et al. MUC16 (CA125): Tumor Biomarker to Cancer Therapy, a Work in Progress. Mol. Cancer 2014, 13, 129. [Google Scholar] [CrossRef] [PubMed]
- Chekmasova, A.A.; Rao, T.D.; Nikhamin, Y.; Park, K.J.; Levine, D.A.; Spriggs, D.R.; Brentjens, R.J. Successful Eradication of Established Peritoneal Ovarian Tumors in SCID-Beige Mice Following Adoptive Transfer of T Cells Genetically Targeted to the MUC16 Antigen. Clin. Cancer Res. 2010, 16, 3594–3606. [Google Scholar] [CrossRef] [PubMed]
- Parker, N.; Turk, M.J.; Westrick, E.; Lewis, J.D.; Low, P.S.; Leamon, C.P. Folate Receptor Expression in Carcinomas and Normal Tissues Determined by a Quantitative Radioligand Binding Assay. Anal. Biochem. 2005, 338, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Kalli, K.R.; Oberg, A.L.; Keeney, G.L.; Christianson, T.J.H.; Low, P.S.; Knutson, K.L.; Hartmann, L.C. Folate Receptor Alpha as a Tumor Target in Epithelial Ovarian Cancer. Gynecol. Oncol. 2008, 108, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Knutson, K.L.; Krco, C.J.; Erskine, C.L.; Goodman, K.; Kelemen, L.E.; Wettstein, P.J.; Low, P.S.; Hartmann, L.C.; Kalli, K.R. T-Cell Immunity to the Folate Receptor Alpha Is Prevalent in Women with Breast or Ovarian Cancer. J. Clin. Oncol. 2006, 24, 4254–4261. [Google Scholar] [CrossRef] [PubMed]
- Kershaw, M.H.; Westwood, J.A.; Parker, L.L.; Wang, G.; Eshhar, Z.; Mavroukakis, S.A.; White, D.E.; Wunderlich, J.R.; Canevari, S.; Rogers-Freezer, L.; et al. A Phase I Study on Adoptive Immunotherapy Using Gene-Modified T Cells for Ovarian Cancer. Clin. Cancer Res. 2006, 12, 6106–6115. [Google Scholar] [CrossRef] [PubMed]
- Ebel, W.; Routhier, E.L.; Foley, B.; Jacob, S.; McDonough, J.M.; Patel, R.K.; Turchin, H.A.; Chao, Q.; Kline, J.B.; Old, L.J.; et al. Preclinical Evaluation of MORAb-003, a Humanized Monoclonal Antibody Antagonizing Folate Receptor-Alpha. Cancer Immun. 2007, 7, 6. [Google Scholar] [PubMed]
- Shimizu, T.; Fujiwara, Y.; Yonemori, K.; Koyama, T.; Sato, J.; Tamura, K.; Shimomura, A.; Ikezawa, H.; Nomoto, M.; Furuuchi, K.; et al. First-in-Human Phase 1 Study of MORAb-202, an Antibody–Drug Conjugate Comprising Farletuzumab Linked to Eribulin Mesylate, in Patients with Folate Receptor-α–Positive Advanced Solid Tumors. Clin. Cancer Res. 2021, 27, 3905–3915. [Google Scholar] [CrossRef] [PubMed]
- Matulonis, U.A.; Lorusso, D.; Oaknin, A.; Pignata, S.; Dean, A.; Denys, H.; Colombo, N.; Van Gorp, T.; Konner, J.A.; Marin, M.R.; et al. Efficacy and Safety of Mirvetuximab Soravtansine in Patients with Platinum-Resistant Ovarian Cancer with High Folate Receptor Alpha Expression: Results from the SORAYA Study. JCO 2023, 41, 2436–2445. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.N.; Martin, L.P.; O’Malley, D.M.; Matulonis, U.A.; Konner, J.A.; Perez, R.P.; Bauer, T.M.; Ruiz-Soto, R.; Birrer, M.J. Safety and Activity of Mirvetuximab Soravtansine (IMGN853), a Folate Receptor Alpha-Targeting Antibody-Drug Conjugate, in Platinum-Resistant Ovarian, Fallopian Tube, or Primary Peritoneal Cancer: A Phase I Expansion Study. J. Clin. Oncol. 2017, 35, 1112–1118. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, D.M.; Matulonis, U.A.; Birrer, M.J.; Castro, C.M.; Gilbert, L.; Vergote, I.; Martin, L.P.; Mantia-Smaldone, G.M.; Martin, A.G.; Bratos, R.; et al. Phase Ib Study of Mirvetuximab Soravtansine, a Folate Receptor Alpha (FRα)-Targeting Antibody-Drug Conjugate (ADC), in Combination with Bevacizumab in Patients with Platinum-Resistant Ovarian Cancer. Gynecol. Oncol. 2020, 157, 379–385. [Google Scholar] [CrossRef]
- Vergote, I.; Armstrong, D.; Scambia, G.; Teneriello, M.; Sehouli, J.; Schweizer, C.; Weil, S.C.; Bamias, A.; Fujiwara, K.; Ochiai, K.; et al. A Randomized, Double-Blind, Placebo-Controlled, Phase III Study to Assess Efficacy and Safety of Weekly Farletuzumab in Combination with Carboplatin and Taxane in Patients with Ovarian Cancer in First Platinum-Sensitive Relapse. J. Clin. Oncol. 2016, 34, 2271–2278. [Google Scholar] [CrossRef]
- Shu, R.; Evtimov, V.J.; Hammett, M.V.; Nguyen, N.-Y.N.; Zhuang, J.; Hudson, P.J.; Howard, M.C.; Pupovac, A.; Trounson, A.O.; Boyd, R.L. Engineered CAR-T Cells Targeting TAG-72 and CD47 in Ovarian Cancer. Mol. Ther. Oncolytics. 2021, 20, 325–341. [Google Scholar] [CrossRef]
- Johnson, V.G.; Schlom, J.; Paterson, A.J.; Bennett, J.; Magnani, J.L.; Colcher, D. Analysis of a Human Tumor-Associated Glycoprotein (TAG-72) Identified by Monoclonal Antibody B72.3. Cancer Res. 1986, 46, 850–857. [Google Scholar]
- Chauhan, S.C.; Vinayek, N.; Maher, D.M.; Bell, M.C.; Dunham, K.A.; Koch, M.D.; Lio, Y.; Jaggi, M. Combined Staining of TAG-72, MUC1, and CA125 Improves Labeling Sensitivity in Ovarian Cancer: Antigens for Multi-Targeted Antibody-Guided Therapy. J. Histochem. Cytochem. 2007, 55, 867–875. [Google Scholar] [CrossRef] [PubMed]
- Ponnusamy, M.P.; Venkatraman, G.; Singh, A.P.; Chauhan, S.C.; Johansson, S.L.; Jain, M.; Smith, L.; Davis, J.S.; Remmenga, S.W.; Batra, S.K. Expression of TAG-72 in Ovarian Cancer and Its Correlation with Tumor Stage and Patient Prognosis. Cancer Lett. 2007, 251, 247–257. [Google Scholar] [CrossRef]
- Jaiswal, S.; Jamieson, C.H.M.; Pang, W.W.; Park, C.Y.; Chao, M.P.; Majeti, R.; Traver, D.; van Rooijen, N.; Weissman, I.L. CD47 Is Upregulated on Circulating Hematopoietic Stem Cells and Leukemia Cells to Avoid Phagocytosis. Cell 2009, 138, 271–285. [Google Scholar] [CrossRef]
- Tinker, A.V.; Hirte, H.W.; Provencher, D.; Butler, M.; Ritter, H.; Tu, D.; Azim, H.A.; Paralejas, P.; Grenier, N.; Hahn, S.-A.; et al. Dose-Ranging and Cohort-Expansion Study of Monalizumab (IPH2201) in Patients with Advanced Gynecologic Malignancies: A Trial of the Canadian Cancer Trials Group (CCTG): IND221. Clin. Cancer Res. 2019, 25, 6052–6060. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Oaknin, A.; Sanchez-Simon, I.; Salgado, A.C.; Patel, S.P.; Oza, A.; Das, M.; Kourtesis, P.; Ascierto, M.L.; Diamond, J.R. 518 Phase 1B Trial of Monalizumab (NKG2A Inhibitor) plus Durvalumab: Safety and Efficacy in Patients with Metastatic Ovarian, Cervical, and Microsatellite-Stable Endometrial Cancers. Int. J. Gynecol. Cancer 2020, 30. [Google Scholar] [CrossRef]
- Jan, C.-I.; Huang, S.-W.; Canoll, P.; Bruce, J.N.; Lin, Y.-C.; Pan, C.-M.; Lu, H.-M.; Chiu, S.-C.; Cho, D.-Y. Targeting Human Leukocyte Antigen G with Chimeric Antigen Receptors of Natural Killer Cells Convert Immunosuppression to Ablate Solid Tumors. J. Immunother. Cancer 2021, 9, e003050. [Google Scholar] [CrossRef]
- Stone, M.L.; Chiappinelli, K.B.; Li, H.; Murphy, L.M.; Travers, M.E.; Topper, M.J.; Mathios, D.; Lim, M.; Shih, I.-M.; Wang, T.-L.; et al. Epigenetic Therapy Activates Type I Interferon Signaling in Murine Ovarian Cancer to Reduce Immunosuppression and Tumor Burden. Proc. Natl. Acad. Sci. USA 2017, 114, E10981–E10990. [Google Scholar] [CrossRef] [PubMed]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via DsRNA Including Endogenous Retroviruses. Cell 2015, 162, 974–986. [Google Scholar] [CrossRef] [PubMed]
- Stone, M.L.; Chiappinelli, K.B.; Li, H.; Murphy, L.M.; Travers, M.E.; Topper, M.J.; Mathios, D.; Lim, M.; Shih, I.-M.; Wang, T.-L.; et al. Reply to Haffner et al.: DNA Hypomethylation Renders Tumors More Immunogenic. Proc. Natl. Acad. Sci. USA 2018, 115, E8583–E8584. [Google Scholar] [CrossRef] [PubMed]
- Moufarrij, S.; Srivastava, A.; Gomez, S.; Hadley, M.; Palmer, E.; Austin, P.T.; Chisholm, S.; Diab, N.; Roche, K.; Yu, A.; et al. Combining DNMT and HDAC6 Inhibitors Increases Anti-Tumor Immune Signaling and Decreases Tumor Burden in Ovarian Cancer. Sci. Rep. 2020, 10, 3470. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.B.; Meza-Perez, S.; Londoño, A.; Katre, A.; Peabody, J.E.; Smith, H.J.; Forero, A.; Norian, L.A.; Straughn, J.M.; Buchsbaum, D.J.; et al. Epigenetic Modifiers Upregulate MHC II and Impede Ovarian Cancer Tumor Growth. Oncotarget 2017, 8, 44159–44170. [Google Scholar] [CrossRef] [PubMed]
- Von Hoff, D.D.; Rasco, D.W.; Heath, E.I.; Munster, P.N.; Schellens, J.H.M.; Isambert, N.; Tourneau, C.L.; O’Neil, B.; Mathijssen, R.H.J.; Lopez-Martin, J.A.; et al. Phase I Study of CC-486 Alone and in Combination with Carboplatin or Nab-Paclitaxel in Patients with Relapsed or Refractory Solid Tumors. Clin. Cancer Res. 2018, 24, 4072–4080. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.; Loo Yau, H.; Chakravarthy, A.; Wang, B.; Shen, S.Y.; Ettayebi, I.; Ishak, C.A.; Bedard, P.L.; Abdul Razak, A.; R Hansen, A.; et al. An Open-Label, Phase II Multicohort Study of an Oral Hypomethylating Agent CC-486 and Durvalumab in Advanced Solid Tumors. J. Immunother. Cancer 2020, 8, e000883. [Google Scholar] [CrossRef] [PubMed]
- Davis, F.M.; Stewart, T.A.; Thompson, E.W.; Monteith, G.R. Targeting EMT in Cancer: Opportunities for Pharmacological Intervention. Trends Pharmacol. Sci. 2014, 35, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Jonckheere, S.; Adams, J.; De Groote, D.; Campbell, K.; Berx, G.; Goossens, S. Epithelial-Mesenchymal Transition (EMT) as a Therapeutic Target. Cells Tissues Organs. 2021, 211, 157–182. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Sun, T. Editorial: Epithelial-Mesenchymal Transition (EMT) as a Therapeutic Target in Cancer. Front. Oncol. 2023, 13. [Google Scholar]
- Huang, Y.; Hong, W.; Wei, X. The Molecular Mechanisms and Therapeutic Strategies of EMT in Tumor Progression and Metastasis. J. Hematol. Oncol. 2022, 15, 129. [Google Scholar] [CrossRef]
- Serrano-Gomez, S.J.; Maziveyi, M.; Alahari, S.K. Regulation of Epithelial-Mesenchymal Transition through Epigenetic and Post-Translational Modifications. Mol. Cancer 2016, 15. [Google Scholar] [CrossRef]
- Wu, S.-M.; Jan, Y.-J.; Tsai, S.-C.; Pan, H.-C.; Shen, C.-C.; Yang, C.-N.; Lee, S.-H.; Liu, S.-H.; Shen, L.-W.; Chiu, C.-S.; et al. Targeting Histone Deacetylase-3 Blocked Epithelial-Mesenchymal Plasticity and Metastatic Dissemination in Gastric Cancer. Cell Biol. Toxicol. 2023, 39, 1873–1896. [Google Scholar] [CrossRef]
- Du, B.; Shim, J.S. Targeting Epithelial–Mesenchymal Transition (EMT) to Overcome Drug Resistance in Cancer. Molecules 2016, 21, 965. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Settleman, J. EMT, Cancer Stem Cells and Drug Resistance: An Emerging Axis of Evil in the War on Cancer. Oncogene 2010, 29, 4741–4751. [Google Scholar] [CrossRef] [PubMed]
- Bedard, P.L.; Hernando-Calvo, A.; Carvajal, R.D.; Morris, V.K.; Paik, P.K.; Zandberg, D.P.; Kaczmar, J.M.; Niculescu, L.; Bohr, D.; Reiners, R.; et al. A Phase 1 Trial of the Bifunctional EGFR/TGFβ Fusion Protein BCA101 Alone and in Combination with Pembrolizumab in Patients with Advanced Solid Tumors. JCO 2022, 40, 2513. [Google Scholar] [CrossRef]
- Kim, C.; Liu, S.V.; Crawford, J.; Torres, T.; Chen, V.; Thompson, J.; Tan, M.; Esposito, G.; Subramaniam, D.S.; Giaccone, G. A Phase I Trial of Dasatinib and Osimertinib in TKI Naïve Patients with Advanced EGFR-Mutant Non-Small-Cell Lung Cancer. Front. Oncol. 2021, 11, 728155. [Google Scholar] [CrossRef] [PubMed]
- Morrison, J.; Thoma, C.; Goodall, R.J.; Lyons, T.J.; Gaitskell, K.; Wiggans, A.J.; Bryant, A. Epidermal Growth Factor Receptor Blockers for the Treatment of Ovarian Cancer. Cochrane Database Syst. Rev. 2018, 2018, CD007927. [Google Scholar] [CrossRef] [PubMed]
- Subbiah, V.; Dumbrava, E.I.; Jiang, Y.; Thein, K.Z.; Naing, A.; Hong, D.S.; Fu, S.; Piha-Paul, S.A.; Tsimberidou, A.M.; Janku, F.; et al. Dual EGFR Blockade with Cetuximab and Erlotinib Combined with Anti-VEGF Antibody Bevacizumab in Advanced Solid Tumors: A Phase 1 Dose Escalation Triplet Combination Trial. Exp. Hematol. Oncol. 2020, 9, 7. [Google Scholar] [CrossRef]
- Seo, J.; Ha, J.; Kang, E.; Cho, S. The Role of Epithelial–Mesenchymal Transition-Regulating Transcription Factors in Anti-Cancer Drug Resistance. Arch. Pharm. Res. 2021, 44, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Wawruszak, A.; Kalafut, J.; Okon, E.; Czapinski, J.; Halasa, M.; Przybyszewska, A.; Miziak, P.; Okla, K.; Rivero-Muller, A.; Stepulak, A. Histone Deacetylase Inhibitors and Phenotypical Transformation of Cancer Cells. Cancers 2019, 11, 148. [Google Scholar] [CrossRef] [PubMed]
- Boulding, T.; McCuaig, R.D.; Tan, A.; Hardy, K.; Wu, F.; Dunn, J.; Kalimutho, M.; Sutton, C.R.; Forwood, J.K.; Bert, A.G.; et al. LSD1 Activation Promotes Inducible EMT Programs and Modulates the Tumour Microenvironment in Breast Cancer. Sci. Rep. 2018, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- Martín-Otal, C.; Lasarte-Cia, A.; Serrano, D.; Casares, N.; Conde, E.; Navarro, F.; Sánchez-Moreno, I.; Gorraiz, M.; Sarrión, P.; Calvo, A.; et al. Targeting the Extra Domain A of Fibronectin for Cancer Therapy with CAR-T Cells. J. Immunother. Cancer 2022, 10, e004479. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Lu, Z.-R. Targeting Fibronectin for Cancer Imaging and Therapy. J. Mater. Chem. B Mater. Biol. Med. 2017, 5, 639–654. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Xie, Q.; Liu, Y.; Gao, Y.; Qu, Z.; Mo, L.; Xu, Y.; Chen, R.; Shi, L. A Small Vimentin-Binding Molecule Blocks Cancer Exosome Release and Reduces Cancer Cell Mobility. Front. Pharmacol. 2021, 12, 627394. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Ye, M.; Zhou, J.; Wang, Z.; Zhu, X. Targeting E-Cadherin Expression with Small Molecules for Digestive Cancer Treatment. Am. J. Transl. Res. 2019, 11, 3932–3944. [Google Scholar] [PubMed]
- Mrozik, K.M.; Blaschuk, O.W.; Cheong, C.M.; Zannettino, A.C.W.; Vandyke, K. N-Cadherin in Cancer Metastasis, Its Emerging Role in Haematological Malignancies and Potential as a Therapeutic Target in Cancer. BMC Cancer 2018, 18, 939. [Google Scholar] [CrossRef] [PubMed]
- Dausset, J. Iso-leuco-anticorps. Acta Haematol. 1958, 20, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Payne, R.; Rolfs, M.R. Fetomaternal Leukocyte Incompatibility. J. Clin. Invest. 1958, 37, 1756–1763. [Google Scholar] [CrossRef] [PubMed]
- Van Rood, J.J.; Eernisse, J.G.; Van Leeuwen, A. Leucocyte Antibodies in Sera from Pregnant Women. Nature 1958, 181, 1735–1736. [Google Scholar] [CrossRef] [PubMed]
- Thorsby, E. A Short History of HLA. Tissue Antigens. 2009, 74, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Harndahl, M.; Rasmussen, M.; Roder, G.; Dalgaard Pedersen, I.; Sørensen, M.; Nielsen, M.; Buus, S. Peptide-MHC Class I Stability Is a Better Predictor than Peptide Affinity of CTL Immunogenicity. Eur. J. Immunol. 2012, 42, 1405–1416. [Google Scholar] [CrossRef] [PubMed]
- Micheletti, F.; Bazzaro, M.; Canella, A.; Marastoni, M.; Traniello, S.; Gavioli, R. The lifespan of major histocompatibility complex class I/peptide complexes determines the efficiency of cytotoxic T-lymphocyte responses. Immunology 1999, 96, 411–415. [Google Scholar] [CrossRef]
- Bassani-Sternberg, M.; Pletscher-Frankild, S.; Jensen, L.J.; Mann, M. Mass Spectrometry of Human Leukocyte Antigen Class I Peptidomes Reveals Strong Effects of Protein Abundance and Turnover on Antigen Presentation*[S]. Mol. Cell. Proteom. 2015, 14, 658–673. [Google Scholar] [CrossRef]
- Kraya, A.A.; Maxwell, K.N.; Eiva, M.A.; Wubbenhorst, B.; Pluta, J.; Feldman, M.; Nayak, A.; Powell, D.J.; Domchek, S.M.; Vonderheide, R.H.; et al. PTEN Loss and BRCA1 Promoter Hypermethylation Negatively Predict for Immunogenicity in BRCA-Deficient Ovarian Cancer. JCO Precis. Oncol. 2022, 6, e2100159. [Google Scholar] [CrossRef] [PubMed]
- Chowell, D.; Morris, L.G.T.; Grigg, C.M.; Weber, J.K.; Samstein, R.M.; Makarov, V.; Kuo, F.; Kendall, S.M.; Requena, D.; Riaz, N.; et al. Patient HLA Class I Genotype Influences Cancer Response to Checkpoint Blockade Immunotherapy. Science 2018, 359, 582–587. [Google Scholar] [CrossRef]
- Brunekreeft, K.L.; Paijens, S.T.; Wouters, M.C.A.; Komdeur, F.L.; Eggink, F.A.; Lubbers, J.M.; Workel, H.H.; Van Der Slikke, E.C.; Pröpper, N.E.J.; Leffers, N.; et al. Deep Immune Profiling of Ovarian Tumors Identifies Minimal MHC-I Expression after Neoadjuvant Chemotherapy as Negatively Associated with T-Cell-Dependent Outcome. Oncoimmunology 2020, 9, 1760705. [Google Scholar] [CrossRef]
- Stevenson, J.P.; Kindler, H.L.; Papasavvas, E.; Sun, J.; Jacobs-Small, M.; Hull, J.; Schwed, D.; Ranganathan, A.; Newick, K.; Heitjan, D.F.; et al. Immunological Effects of the TGFβ-Blocking Antibody GC1008 in Malignant Pleural Mesothelioma Patients. Oncoimmunology 2013, 2, e26218. [Google Scholar] [CrossRef]
- O’Reilly, E.M.; Golan, T.; Ikeda, M.; Milella, M.; Taieb, J.; Wainberg, Z.A.; Wang, L.-W.; Gyambibi, N.; Lopez-Martin, E.M.; Xu, K.; et al. Phase III Study (DaNIS-2) of the Anti–TGF-β Monoclonal Antibody (MAb) NIS793 with Nab-Paclitaxel/Gemcitabine (NG) versus NG Alone in Patients (Pts) with First-Line Metastatic Pancreatic Ductal Adenocarcinoma (MPDAC). JCO 2022, 40, TPS4193. [Google Scholar] [CrossRef]
- Pesch, A.M.; Pierce, L.J.; Speers, C.W. Modulating the Radiation Response for Improved Outcomes in Breast Cancer. JCO Precis. Oncol. 2021. [Google Scholar] [CrossRef]
- Kaczmar, J.M.; Zandberg, D.P.; Wong, D.J.L.; Yilmaz, E.; Sherman, E.J.; Hernando-Calvo, A.; Sacco, A.G.; Chung, C.H.; Bohr, D.; Reiners, R.; et al. Dose Expansion Results of the Bifunctional EGFR/TGFβ Inhibitor BCA101 with Pembrolizumab in Patients with Recurrent, Metastatic Head and Neck Squamous Cell Carcinoma. J. Clin. Oncol. 2023. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodriguez, G.M.; Yakubovich, E.; Vanderhyden, B.C. Unveiling the Immunogenicity of Ovarian Tumors as the Crucial Catalyst for Therapeutic Success. Cancers 2023, 15, 5694. https://doi.org/10.3390/cancers15235694
Rodriguez GM, Yakubovich E, Vanderhyden BC. Unveiling the Immunogenicity of Ovarian Tumors as the Crucial Catalyst for Therapeutic Success. Cancers. 2023; 15(23):5694. https://doi.org/10.3390/cancers15235694
Chicago/Turabian StyleRodriguez, Galaxia M., Edward Yakubovich, and Barbara C. Vanderhyden. 2023. "Unveiling the Immunogenicity of Ovarian Tumors as the Crucial Catalyst for Therapeutic Success" Cancers 15, no. 23: 5694. https://doi.org/10.3390/cancers15235694
APA StyleRodriguez, G. M., Yakubovich, E., & Vanderhyden, B. C. (2023). Unveiling the Immunogenicity of Ovarian Tumors as the Crucial Catalyst for Therapeutic Success. Cancers, 15(23), 5694. https://doi.org/10.3390/cancers15235694




