Identifying Genetic Mutation Status in Patients with Colorectal Cancer Liver Metastases Using Radiomics-Based Machine-Learning Models
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Data Collection
2.2.1. Genetic Mutation Analysis
2.2.2. Imaging Data
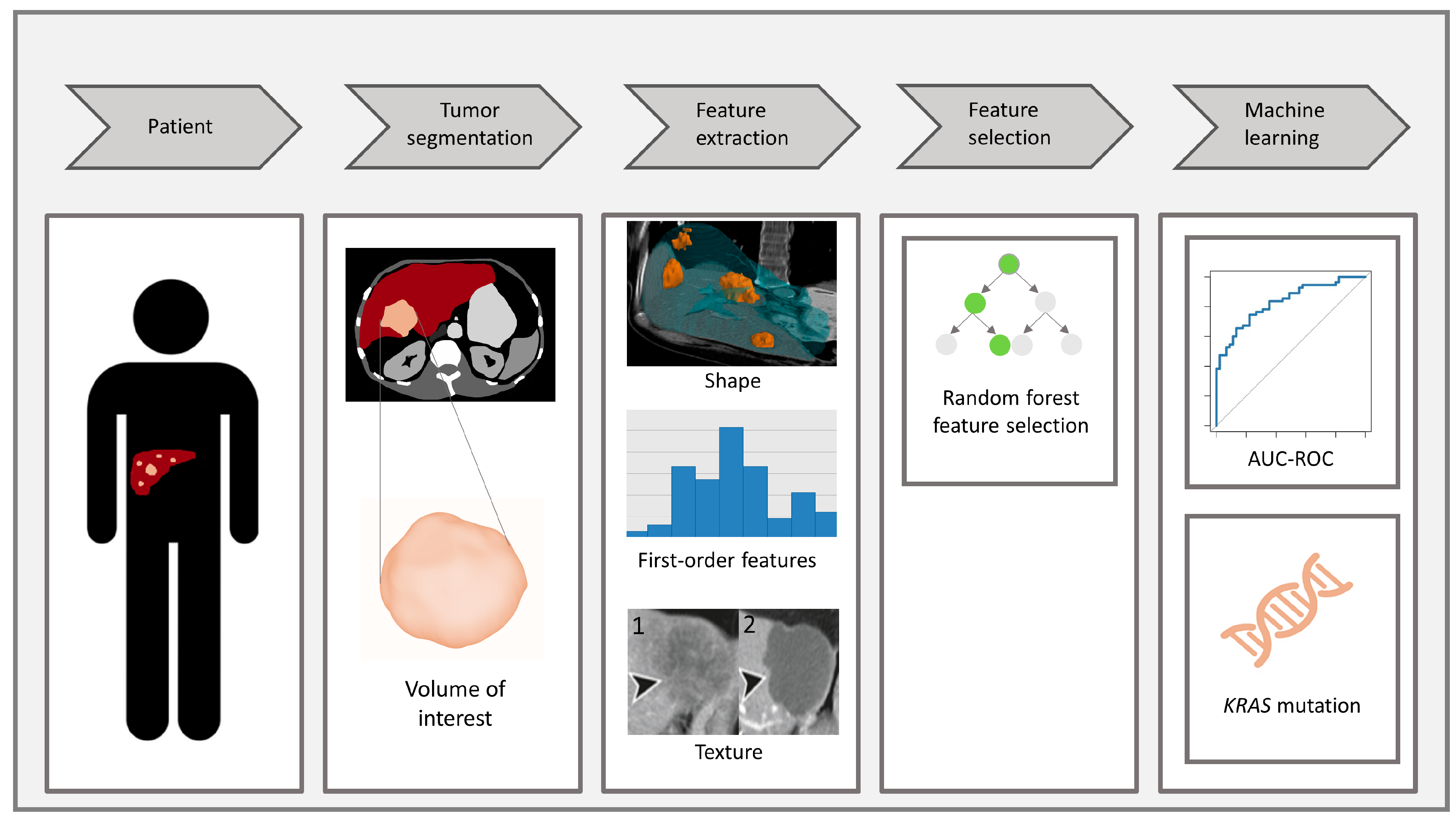
2.3. Radiomics Analysis
2.3.1. Tumor Segmentation
2.3.2. Feature Extraction and Feature Selection
2.3.3. Data Pre-Processing and Modeling
2.4. External Validation
2.5. Statistics
3. Results
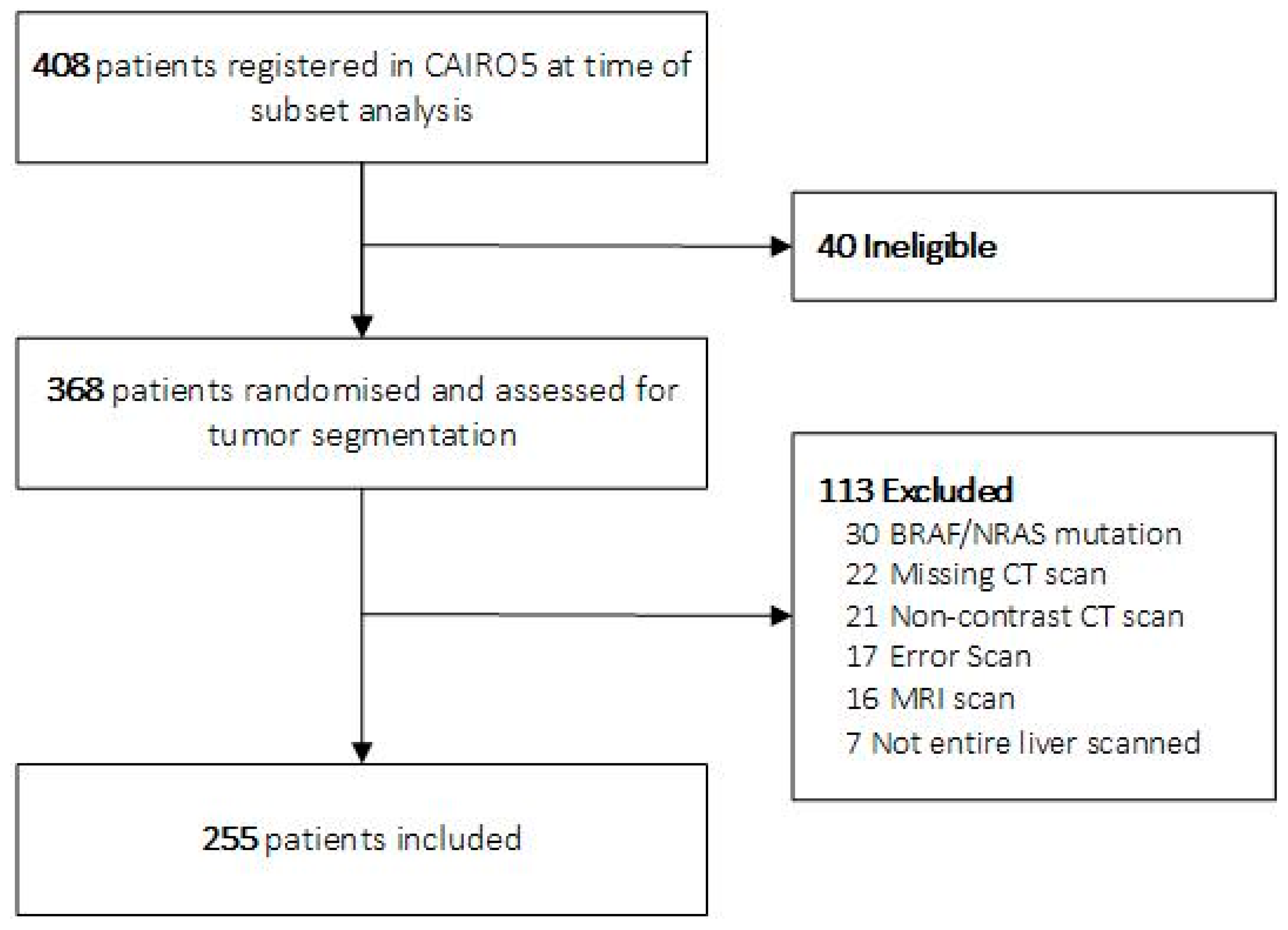
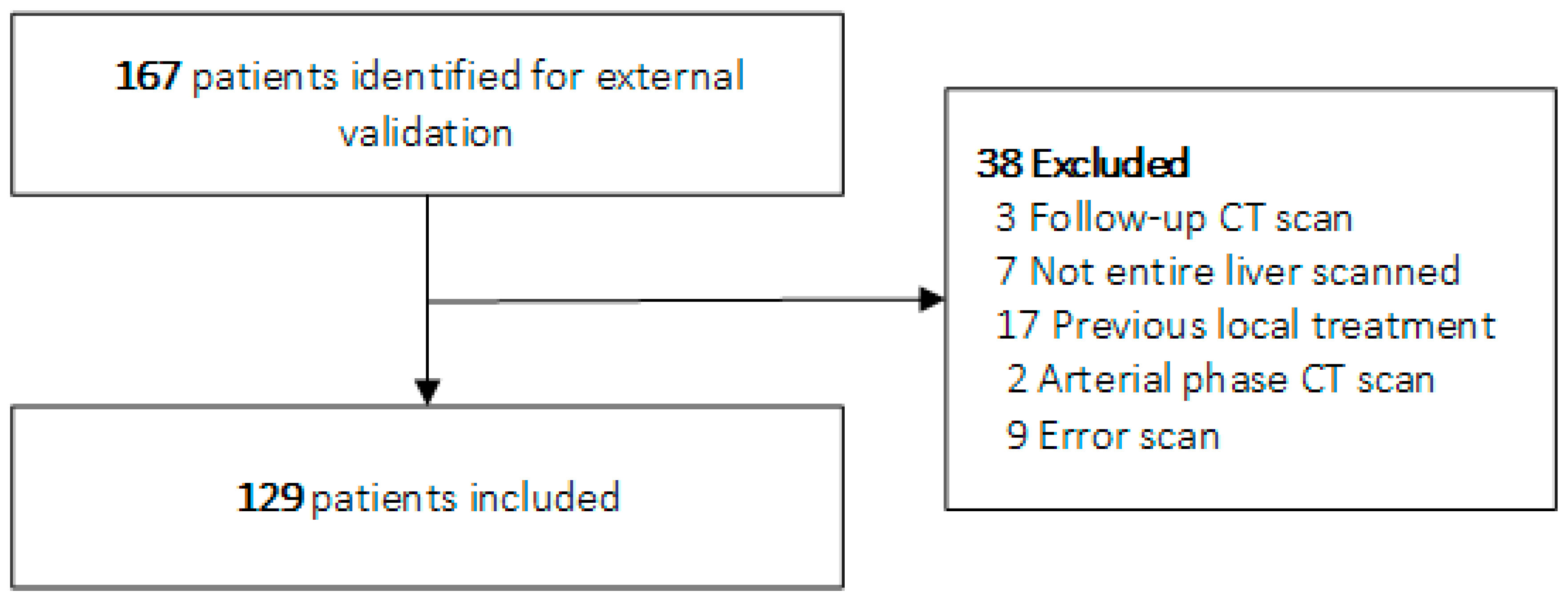
3.1. Study Population
3.2. Feature Selection and Modeling
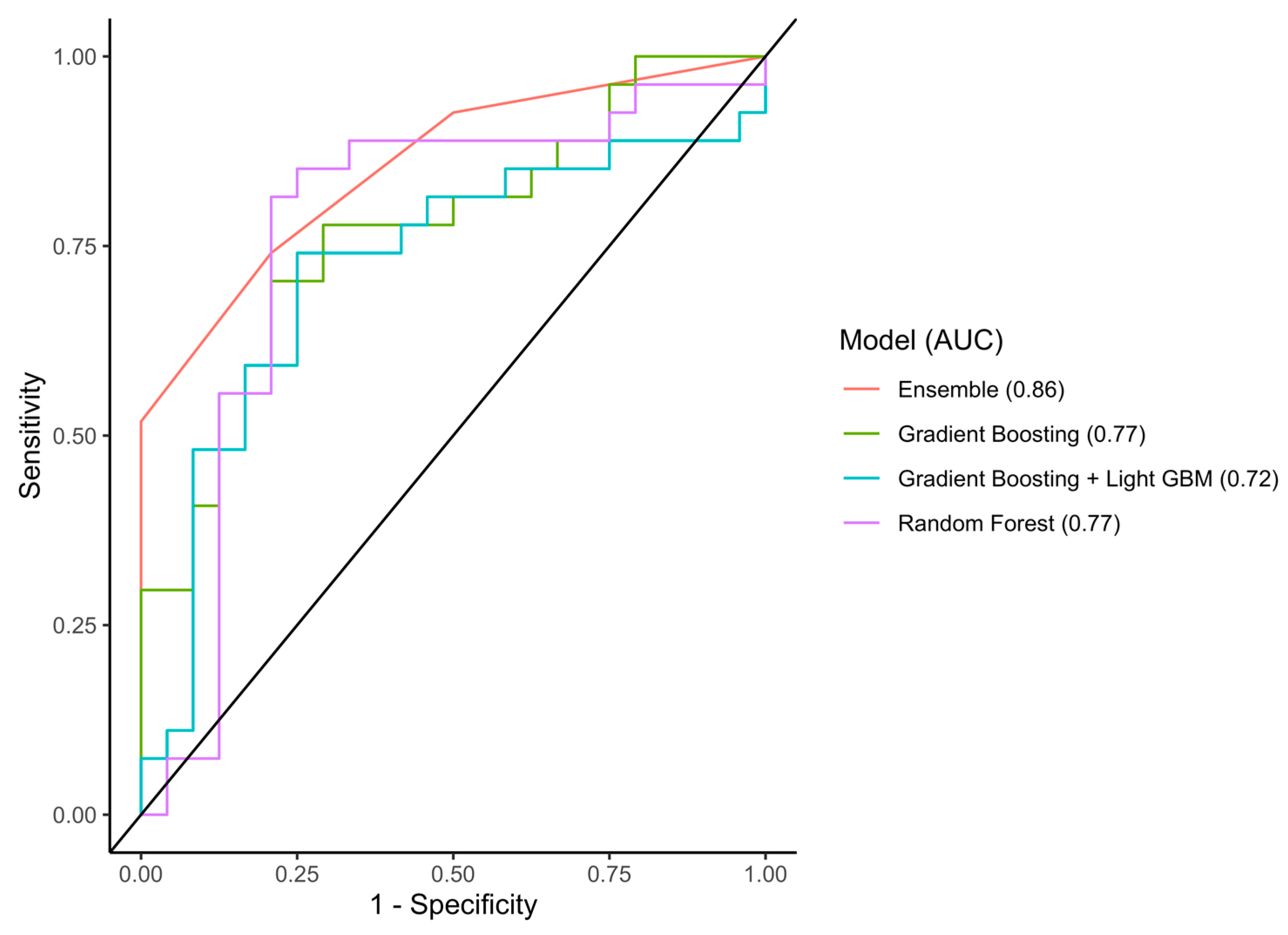
3.3. Performance of the Models
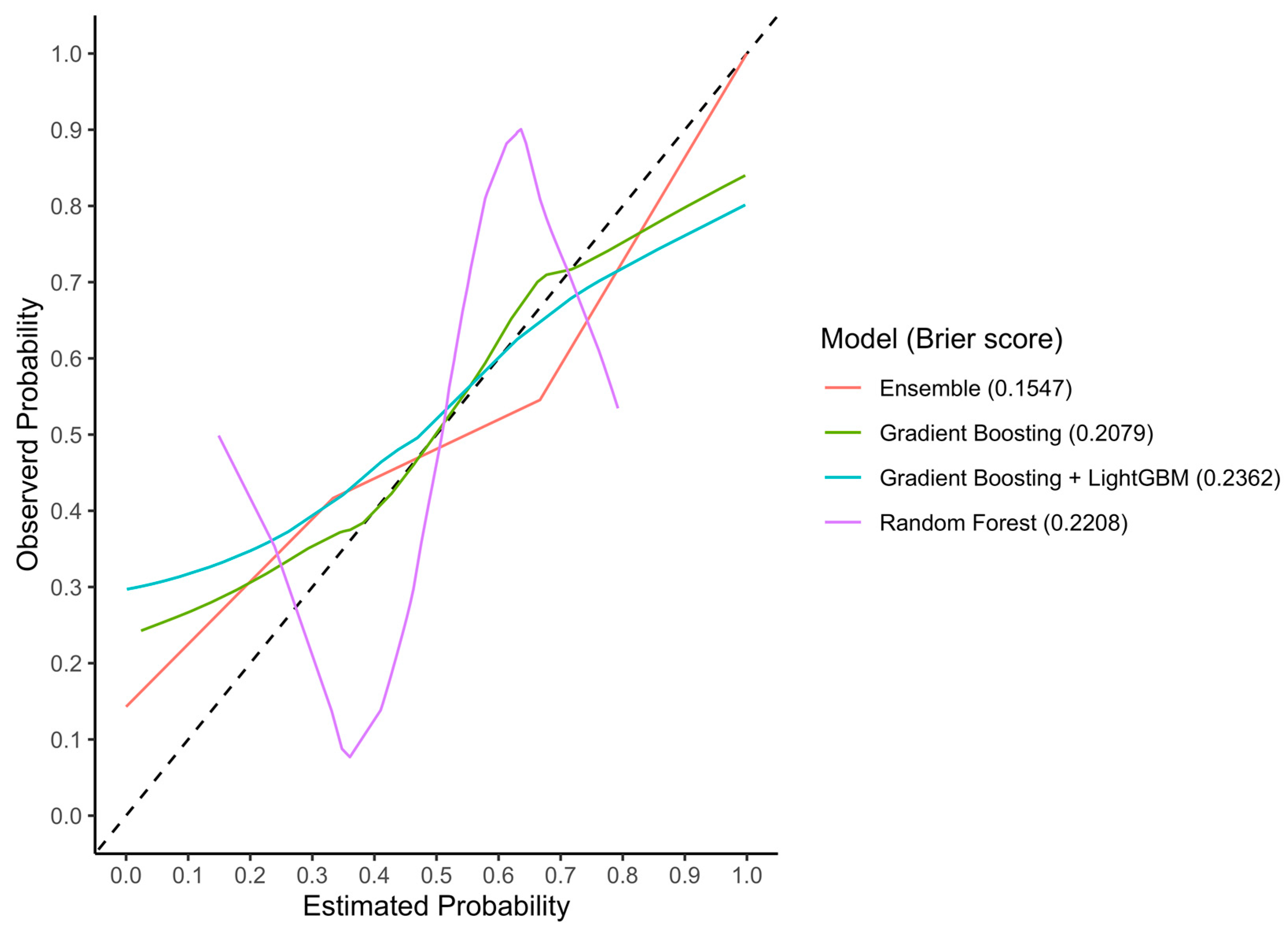
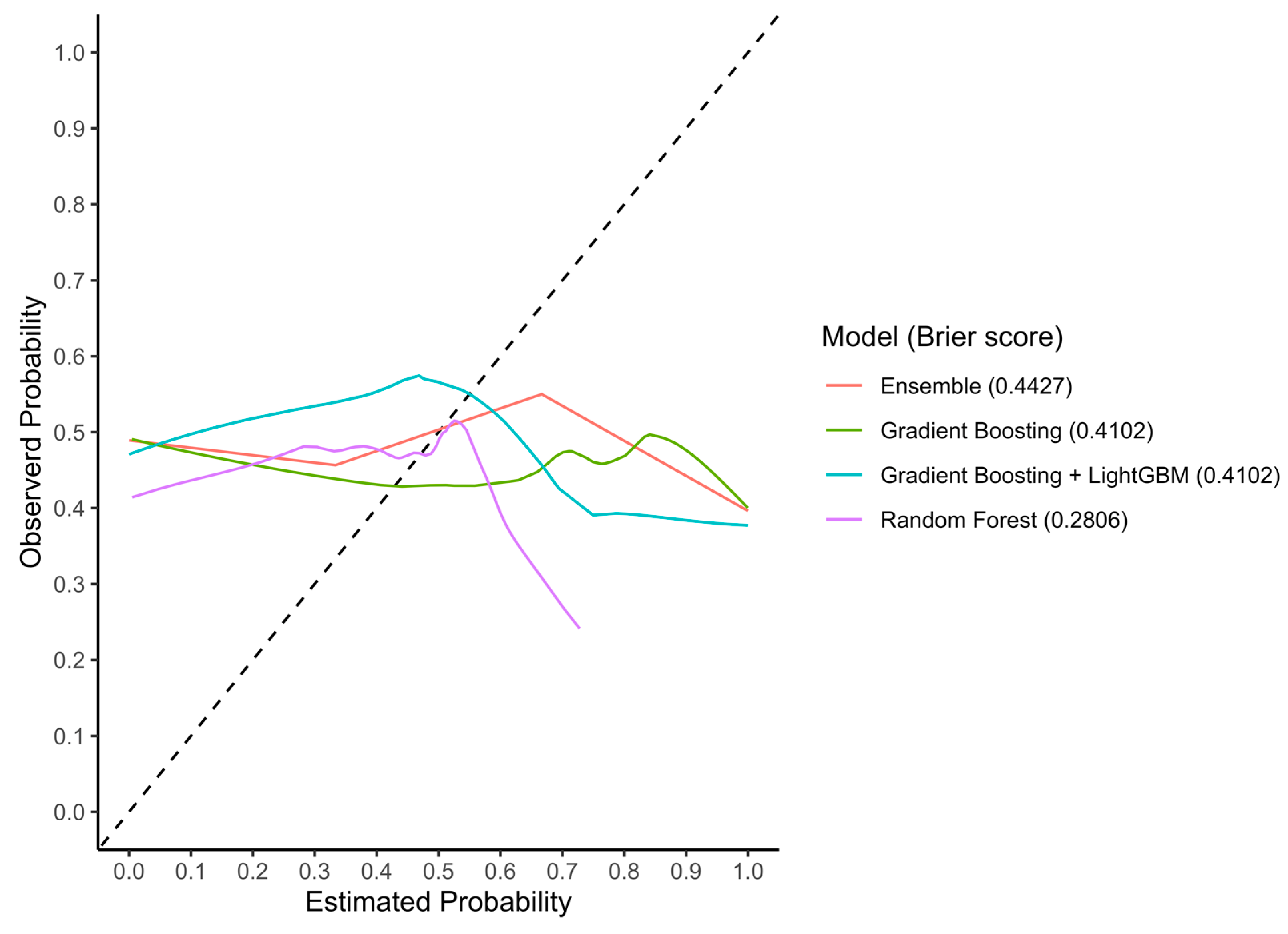
3.3.1. Discrimination Accuracy
3.3.2. Calibration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, E.K.; Adam, R.; Bilchik, A.J.; Jaeck, D.; Vauthey, J.N.; Mahvi, D. Improving resectability of hepatic colorectal metastases: Expert consensus statement. Ann. Surg. Oncol. 2006, 13, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Donadon, M.; Ribero, D.; Morris-Stiff, G.; Abdalla, E.K.; Vauthey, J.N. New paradigm in the management of liver-only metastases from colorectal cancer. Gastrointest. Cancer Res. GCR 2007, 1, 20–27. [Google Scholar] [PubMed]
- Adam, R.; Kitano, Y. Multidisciplinary approach of liver metastases from colorectal cancer. Ann. Gastroenterol. Surg. 2019, 3, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Lam, V.W.; Spiro, C.; Laurence, J.M.; Johnston, E.; Hollands, M.J.; Pleass, H.C.; Richardson, A.J. A systematic review of clinical response and survival outcomes of downsizing systemic chemotherapy and rescue liver surgery in patients with initially unresectable colorectal liver metastases. Ann. Surg. Oncol. 2012, 19, 1292–1301. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, E.K.; Vauthey, J.N.; Ellis, L.M.; Ellis, V.; Pollock, R.; Broglio, K.R.; Hess, K.; Curley, S.A. Recurrence and outcomes following hepatic resection, radiofrequency ablation, and combined resection/ablation for colorectal liver metastases. Ann. Surg. 2004, 239, 818–825; discussion 825–827. [Google Scholar] [CrossRef] [PubMed]
- Venook, A.P.; Niedzwiecki, D.; Lenz, H.J.; Innocenti, F.; Fruth, B.; Meyerhardt, J.A.; Schrag, D.; Greene, C.; O’Neil, B.H.; Atkins, J.N.; et al. Effect of First-Line Chemotherapy Combined with Cetuximab or Bevacizumab on Overall Survival in Patients With KRAS Wild-Type Advanced or Metastatic Colorectal Cancer: A Randomized Clinical Trial. J. Am. Med. Assoc. 2017, 317, 2392–2401. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Köhne, C.H.; Láng, I.; Folprecht, G.; Nowacki, M.P.; Cascinu, S.; Shchepotin, I.; Maurel, J.; Cunningham, D.; Tejpar, S.; et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: Updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J. Clin. Oncol. 2011, 29, 2011–2019. [Google Scholar] [CrossRef]
- Piawah, S.; Venook, A.P. Targeted therapy for colorectal cancer metastases: A review of current methods of molecularly targeted therapy and the use of tumor biomarkers in the treatment of metastatic colorectal cancer. Cancer 2019, 125, 4139–4147. [Google Scholar] [CrossRef]
- Adam, R.; de Gramont, A.; Figueras, J.; Kokudo, N.; Kunstlinger, F.; Loyer, E.; Poston, G.; Rougier, P.; Rubbia-Brandt, L.; Sobrero, A.; et al. Managing synchronous liver metastases from colorectal cancer: A multidisciplinary international consensus. Cancer Treat. Rev. 2015, 41, 729–741. [Google Scholar] [CrossRef]
- Yamashita, S.; Chun, Y.S.; Kopetz, S.E.; Vauthey, J.N. Biomarkers in colorectal liver metastases. Br. J. Surg. 2018, 105, 618–627. [Google Scholar] [CrossRef] [PubMed]
- Peeters, M.; Oliner, K.S.; Price, T.J.; Cervantes, A.; Sobrero, A.F.; Ducreux, M.; Hotko, Y.; André, T.; Chan, E.; Lordick, F.; et al. Analysis of KRAS/NRAS Mutations in a Phase III Study of Panitumumab with FOLFIRI Compared with FOLFIRI Alone as Second-line Treatment for Metastatic Colorectal Cancer. Clin. Cancer Res. An Off. J. Am. Assoc. Cancer Res. 2015, 21, 5469–5479. [Google Scholar] [CrossRef] [PubMed]
- Eklöf, V.; Wikberg, M.L.; Edin, S.; Dahlin, A.M.; Jonsson, B.A.; Öberg, Å.; Rutegård, J.; Palmqvist, R. The prognostic role of KRAS, BRAF, PIK3CA and PTEN in colorectal cancer. Br. J. Cancer 2013, 108, 2153–2163. [Google Scholar] [CrossRef] [PubMed]
- Peeters, M.; Kafatos, G.; Taylor, A.; Gastanaga, V.M.; Oliner, K.S.; Hechmati, G.; Terwey, J.H.; Van Krieken, J.H. Prevalence of RAS mutations and individual variation patterns among patients with metastatic colorectal cancer: A pooled analysis of randomised controlled trials. Eur. J. Cancer 2015, 51, 1704–1713. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; Van Krieken, J.H.; Aderka, D.; Aguilar, E.A.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann. Oncol. 2016, 27, 1386–1422. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.; Even, A.J.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Wesdorp, N.J.; Hellingman, T.; Jansma, E.P.; van Waesberghe, J.T.M.; Boellaard, R.; Punt, C.J.A.; Huiskens, J.; Kazemier, G. Advanced analytics and artificial intelligence in gastrointestinal cancer: A systematic review of radiomics predicting response to treatment. Eur. J. Nucl. Med. Mol. Imaging 2020, 48, 1785–1794. [Google Scholar] [CrossRef]
- Wesdorp, N.J.; van Goor, V.J.; Kemna, R.; Jansma, E.P.; van Waesberghe, J.H.T.M.; Swijnenburg, R.J.; Punt, C.J.A.; Huiskens, J.; Kazemier, G. Advanced image analytics predicting clinical outcomes in patients with colorectal liver metastases: A systematic review of the literature. Surg. Oncol. 2021, 38, 101578. [Google Scholar] [CrossRef]
- Staal, F.C.R.; van der Reijd, D.J.; Taghavi, M.; Lambregts, D.M.J.; Beets-Tan, R.G.H.; Maas, M. Radiomics for the Prediction of Treatment Outcome and Survival in Patients with Colorectal Cancer: A Systematic Review. Clin. Color. Cancer 2021, 20, 52–71. [Google Scholar] [CrossRef]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Badic, B.; Tixier, F.; Cheze Le Rest, C.; Hatt, M.; Visvikis, D. Radiogenomics in Colorectal Cancer. Cancers 2021, 13, 973. [Google Scholar] [CrossRef]
- Shi, R.; Chen, W.; Yang, B.; Qu, J.; Cheng, Y.; Zhu, Z.; Gao, Y.; Wang, Q.; Liu, Y.; Li, Z.; et al. Prediction of KRAS, NRAS and BRAF status in colorectal cancer patients with liver metastasis using a deep artificial neural network based on radiomics and semantic features. Am. J. Cancer Res. 2020, 10, 4513–4526. [Google Scholar] [PubMed]
- Brunsell, T.H.; Sveen, A.; Bjornbeth, B.A.; Rosok, B.I.; Danielsen, S.A.; Brudvik, K.W.; Berg, K.C.; Johannessen, B.; Cengija, V.; Abildgaard, A.; et al. High Concordance and Negative Prognostic Impact of RAS/BRAF/PIK3CA Mutations in Multiple Resected Colorectal Liver Metastases. Clin. Color. Cancer 2020, 19, e26–e47. [Google Scholar] [CrossRef] [PubMed]
- Loes, I.M.; Immervoll, H.; Sorbye, H.; Angelsen, J.H.; Horn, A.; Knappskog, S.; Lønning, P.E. Impact of KRAS, BRAF, PIK3CA, TP53 status and intraindividual mutation heterogeneity on outcome after liver resection for colorectal cancer metastases. Int. J. Cancer 2016, 139, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.L.; Zhao, J.X.; Zhao, L.P.; Tian, J.H.; Huang, G. Current status and quality of radiomic studies for predicting KRAS mutations in colorectal cancer patients: A systematic review and meta-analysis. Eur. J. Radiol. 2023, 158, 110640. [Google Scholar] [CrossRef]
- Huiskens, J.; van Gulik, T.M.; van Lienden, K.P.; Engelbrecht, M.R.; Meijer, G.A.; van Grieken, N.C.; Schriek, J.; Keijser, A.; Mol, L.; Molenaar, I.Q.; et al. Treatment strategies in colorectal cancer patients with initially unresectable liver-only metastases, a study protocol of the randomised phase 3 CAIRO5 study of the Dutch Colorectal Cancer Group (DCCG). BMC Cancer 2015, 15, 365. [Google Scholar] [CrossRef] [PubMed]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104. [Google Scholar] [CrossRef]
- Brudvik, K.W.; Mise, Y.; Chung, M.H.; Chun, Y.S.; Kopetz, S.E.; Passot, G.; Conrad, C.; Maru, D.M.; Aloia, T.A.; Vauthey, J.N. RAS Mutation Predicts Positive Resection Margins and Narrower Resection Margins in Patients Undergoing Resection of Colorectal Liver Metastases. Ann. Surg. Oncol. 2016, 23, 2635–2643. [Google Scholar] [CrossRef]
- Segal, E.; Sirlin, C.B.; Ooi, C.; Adler, A.S.; Gollub, J.; Chen, X.; Chan, B.K.; Matcuk, G.R.; Barry, C.T.; Chang, H.Y.; et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat. Biotechnol. 2007, 25, 675–680. [Google Scholar] [CrossRef]
- Taguchi, N.; Oda, S.; Yokota, Y.; Yamamura, S.; Imuta, M.; Tsuchigame, T.; Nagayama, Y.; Kidoh, M.; Nakaura, T.; Shiraishi, S.; et al. CT texture analysis for the prediction of KRAS mutation status in colorectal cancer via a machine learning approach. Eur. J. Radiol. 2019, 118, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Dong, D.; Fang, M.; Zhu, Y.; Zang, Y.; Liu, Z.; Zhang, H.; Ying, J.; Zhao, X.; Tian, J. Can CT-based radiomics signature predict KRAS/NRAS/BRAF mutations in colorectal cancer? Eur. Radiol. 2018, 28, 2058–2067. [Google Scholar] [CrossRef] [PubMed]
- van de Sande, D.; van Genderen, M.E.; Huiskens, J.; Gommers, D.; van Bommel, J. Moving from bytes to bedside: A systematic review on the use of artificial intelligence in the intensive care unit. Intensive Care Med. 2021, 47, 750–760. [Google Scholar] [CrossRef] [PubMed]
- van de Sande, D.; Van Genderen, M.E.; Smit, J.M.; Huiskens, J.; Visser, J.J.; Veen, R.E.R.; Van Unen, E.; Hilgers, O.; Gommers, D.; van Bommel, J. Developing, implementing and governing artificial intelligence in medicine: A step-by-step approach to prevent an artificial intelligence winter. BMJ Health Care Inform. 2022, 29, e100495. [Google Scholar] [CrossRef]
- Marshall, C.; Thirion, P.; Mihai, A.; Armstrong, J.G.; Cournane, S.; Hickey, D.; McClean, B.; Quinn, J. Interobserver Variability of Gross Tumor Volume Delineation for Colorectal Liver Metastases Using Computed Tomography and Magnetic Resonance Imaging. Adv. Radiat. Oncol. 2023, 8, 101020. [Google Scholar] [CrossRef]
- van ‘t Erve, I.; Wesdorp, N.J.; Medina, J.E.; Ferreira, L.; Leal, A.; Huiskens, J.; Bolhuis, K.; van Waesberghe, J.H.T.; Swijnenburg, R.J.; van den Broek, D.; et al. KRAS A146 Mutations Are Associated with Distinct Clinical Behavior in Patients With Colorectal Liver Metastases. JCO Precis. Oncol. 2021, PO2100223. [Google Scholar] [CrossRef]

| Baseline Characteristics | Discovery Cohort (n = 255) | External Validation Set (n = 129) | p-Value |
|---|---|---|---|
| Age (years) | 62 (55–70) | 62 (53–69) | 0.607 |
| Sex | 0.068 | ||
| Male | 170 (66.7) | 73 (56.6) | |
| Female | 85 (33.3) | 56 (43.4) | |
| KRAS mutational status | 0.149 | ||
| Mutation | 136 (53.3) | 58 (45.0) | |
| Wild-type | 119 (46.7) | 71 (55.0) | |
| Number of liver metastases | 12 (7–23) | 5 (2–11) | <0.0001 |
| Machine-Learning Classifier | Cohort | AUC (95% CI) | Sensitivity | Specificity | Accuracy |
|---|---|---|---|---|---|
| Random Forest | Train | 0.97 (0.95–0.99) | 0.82 | 0.95 | 0.89 |
| Test | 0.77 (0.62–0.93) | 0.85 | 0.75 | 0.80 | |
| External validation | 0.54 (0.44–0.64) | 0.44 | 0.54 | 0.50 | |
| Gradient Boosting | Train | 0.96 (0.94–1.00) | 0.95 | 0.98 | 0.98 |
| Test | 0.77 (0.64–0.90) | 0.67 | 0.79 | 0.73 | |
| External validation | 0.52 (0.42–0.62) | 0.46 | 0.57 | 0.52 | |
| Gradient Boosting (LightGBM) | Train | 0.98 (0.97–1.00) | 0.98 | 0.99 | 0.98 |
| Test | 0.72 (0.57–0.87) | 0.75 | 0.67 | 0.71 | |
| External validation | 0.56 (0.46–0.67) | 0.28 | 0.59 | 0.42 | |
| Ensemble | Train | 0.97 (0.94–1.00) | 0.97 | 0.93 | 0.95 |
| Test | 0.86 (0.76–0.95) | 0.93 | 0.58 | 0.77 | |
| External validation | 0.47 (0.37–0.56) | 0.30 | 0.74 | 0.50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wesdorp, N.; Zeeuw, M.; van der Meulen, D.; van ‘t Erve, I.; Bodalal, Z.; Roor, J.; van Waesberghe, J.H.; Moos, S.; van den Bergh, J.; Nota, I.; et al. Identifying Genetic Mutation Status in Patients with Colorectal Cancer Liver Metastases Using Radiomics-Based Machine-Learning Models. Cancers 2023, 15, 5648. https://doi.org/10.3390/cancers15235648
Wesdorp N, Zeeuw M, van der Meulen D, van ‘t Erve I, Bodalal Z, Roor J, van Waesberghe JH, Moos S, van den Bergh J, Nota I, et al. Identifying Genetic Mutation Status in Patients with Colorectal Cancer Liver Metastases Using Radiomics-Based Machine-Learning Models. Cancers. 2023; 15(23):5648. https://doi.org/10.3390/cancers15235648
Chicago/Turabian StyleWesdorp, Nina, Michiel Zeeuw, Delanie van der Meulen, Iris van ‘t Erve, Zuhir Bodalal, Joran Roor, Jan Hein van Waesberghe, Shira Moos, Janneke van den Bergh, Irene Nota, and et al. 2023. "Identifying Genetic Mutation Status in Patients with Colorectal Cancer Liver Metastases Using Radiomics-Based Machine-Learning Models" Cancers 15, no. 23: 5648. https://doi.org/10.3390/cancers15235648
APA StyleWesdorp, N., Zeeuw, M., van der Meulen, D., van ‘t Erve, I., Bodalal, Z., Roor, J., van Waesberghe, J. H., Moos, S., van den Bergh, J., Nota, I., van Dieren, S., Stoker, J., Meijer, G., Swijnenburg, R.-J., Punt, C., Huiskens, J., Beets-Tan, R., Fijneman, R., Marquering, H., ... on behalf of the Dutch Colorectal Cancer Group Liver Expert Panel. (2023). Identifying Genetic Mutation Status in Patients with Colorectal Cancer Liver Metastases Using Radiomics-Based Machine-Learning Models. Cancers, 15(23), 5648. https://doi.org/10.3390/cancers15235648






