Enhanced Risk Stratification in Early-Stage Endometrial Cancer: Integrating POLE through Droplet Digital PCR and L1CAM
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Immunohistochemistry for p53, MMR Proteins, and L1CAM
2.3. Droplet Digital PCR Assay to Detect POLE Mutation
2.4. Microsatellite Instability Test Using PNA Probe-Mediated Real-Time PCR
2.5. Statistical Analysis
3. Results
3.1. Patient Characteristics
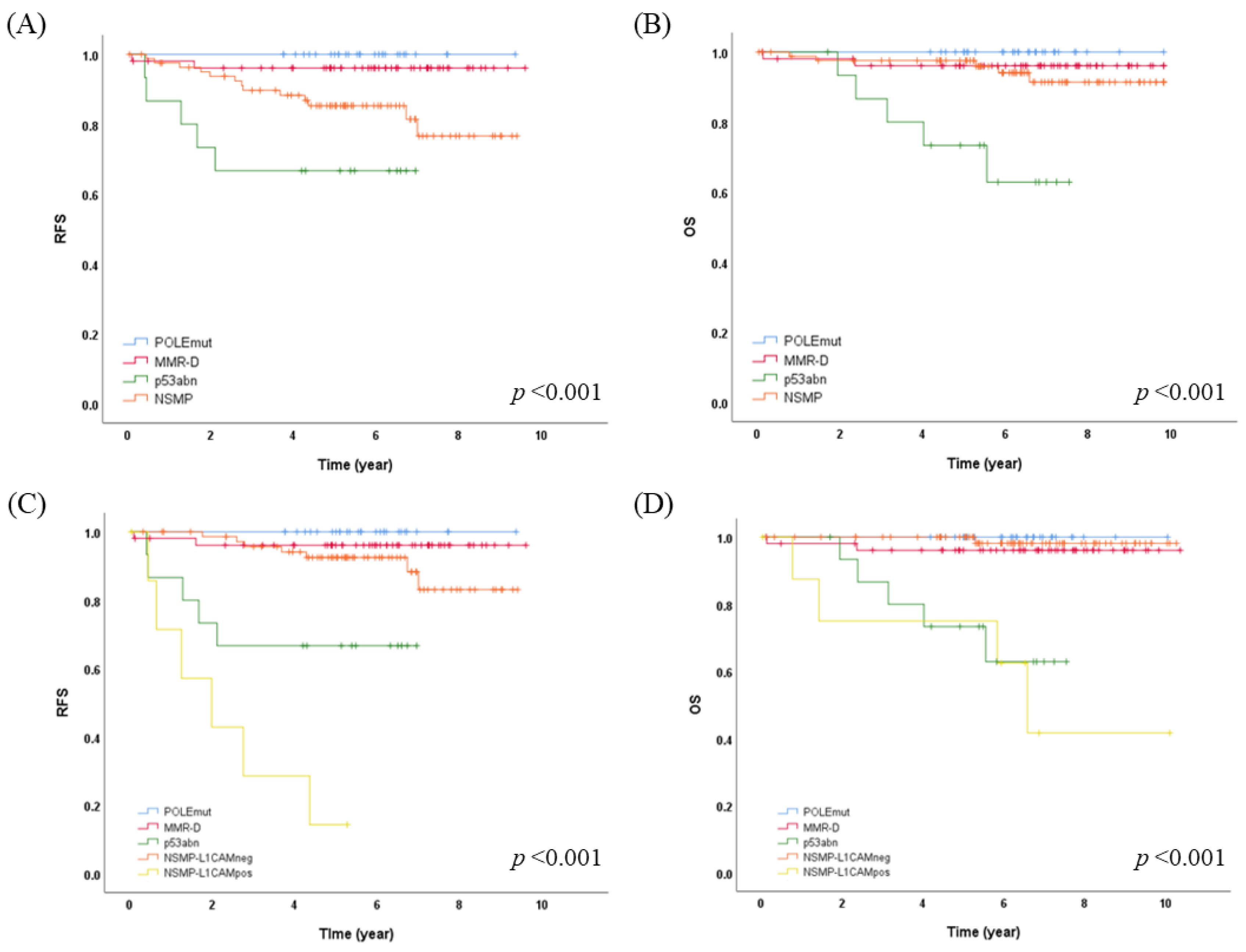
3.2. Molecular Classification Using Surrogate Markers and Its Clinical Significance
3.3. L1CAM Expression and Its Impact on Prognosis and Molecular Classification
3.4. Enhanced Risk Stratification in Early-Stage EC by Integrating Molecular L1CAM Classification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Morice, P.; Leary, A.; Creutzberg, C.; Abu-Rustum, N.; Darai, E. Endometrial cancer. Lancet 2016, 387, 1094–1108. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Ha, H.I.; Chang, H.K.; Park, S.J.; Lim, J.; Won, Y.J.; Lim, M.C. The incidence and survival of cervical, ovarian, and endometrial cancer in Korea, 1999–2017: Korea Central Cancer Registry. Obstet. Gynecol. Sci. 2021, 64, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Won, Y.J.; Lee, J.J.; Jung, K.W.; Kong, H.J.; Im, J.S.; Seo, H.G.; Community of Population-Based Regional Cancer, R. Cancer Statistics in Korea: Incidence, Mortality, Survival, and Prevalence in 2018. Cancer Res. Treat. 2021, 53, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar]
- McAlpine, J.; Leon-Castillo, A.; Bosse, T. The rise of a novel classification system for endometrial carcinoma; integration of molecular subclasses. J. Pathol. 2018, 244, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Yang, W.; Lum, A.; Senz, J.; Boyd, N.; Pike, J.; Anglesio, M.; Kwon, J.S.; et al. Confirmation of ProMisE: A simple, genomics-based clinical classifier for endometrial cancer. Cancer 2017, 123, 802–813. [Google Scholar] [CrossRef]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Li-Chang, H.H.; Kwon, J.S.; Melnyk, N.; Yang, W.; Senz, J.; Boyd, N.; Karnezis, A.N.; et al. A clinically applicable molecular-based classification for endometrial cancers. Br. J. Cancer 2015, 113, 299–310. [Google Scholar] [CrossRef]
- Kommoss, S.; McConechy, M.K.; Kommoss, F.; Leung, S.; Bunz, A.; Magrill, J.; Britton, H.; Kommoss, F.; Grevenkamp, F.; Karnezis, A.; et al. Final validation of the ProMisE molecular classifier for endometrial carcinoma in a large population-based case series. Ann. Oncol. 2018, 29, 1180–1188. [Google Scholar] [CrossRef]
- Talhouk, A.; Hoang, L.N.; McConechy, M.K.; Nakonechny, Q.; Leo, J.; Cheng, A.; Leung, S.; Yang, W.; Lum, A.; Kobel, M.; et al. Molecular classification of endometrial carcinoma on diagnostic specimens is highly concordant with final hysterectomy: Earlier prognostic information to guide treatment. Gynecol. Oncol. 2016, 143, 46–53. [Google Scholar] [CrossRef]
- Church, D.N.; Briggs, S.E.; Palles, C.; Domingo, E.; Kearsey, S.J.; Grimes, J.M.; Gorman, M.; Martin, L.; Howarth, K.M.; Hodgson, S.V.; et al. DNA polymerase epsilon and delta exonuclease domain mutations in endometrial cancer. Hum. Mol. Genet. 2013, 22, 2820–2828. [Google Scholar] [CrossRef] [PubMed]
- Church, D.N.; Stelloo, E.; Nout, R.A.; Valtcheva, N.; Depreeuw, J.; der Haar, N.; Noske, A.; Amant, F.; Tomlinson, I.P.; Wild, P.J.; et al. Prognostic significance of POLE proofreading mutations in endometrial cancer. J. Natl. Cancer Inst. 2015, 107, 402. [Google Scholar] [CrossRef]
- McConechy, M.K.; Talhouk, A.; Leung, S.; Chiu, D.; Yang, W.; Senz, J.; Reha-Krantz, L.J.; Lee, C.H.; Huntsman, D.G.; Gilks, C.B.; et al. Endometrial Carcinomas with POLE Exonuclease Domain Mutations Have a Favorable Prognosis. Clin. Cancer Res. 2016, 22, 2865–2873. [Google Scholar] [CrossRef] [PubMed]
- Leon-Castillo, A.; Britton, H.; McConechy, M.K.; McAlpine, J.N.; Nout, R.; Kommoss, S.; Brucker, S.Y.; Carlson, J.W.; Epstein, E.; Rau, T.T.; et al. Interpretation of somatic POLE mutations in endometrial carcinoma. J. Pathol. 2020, 250, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Hohn, A.K.; Brambs, C.E.; Hiller, G.G.R.; May, D.; Schmoeckel, E.; Horn, L.C. 2020 WHO Classification of Female Genital Tumors. Geburtshilfe Frauenheilkd 2021, 81, 1145–1153. [Google Scholar] [CrossRef]
- Oaknin, A.; Bosse, T.J.; Creutzberg, C.L.; Giornelli, G.; Harter, P.; Joly, F.; Lorusso, D.; Marth, C.; Makker, V.; Mirza, M.R.; et al. Endometrial cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2022, 33, 860–877. [Google Scholar] [CrossRef]
- Kim, G.; Lee, S.K.; Suh, D.H.; Kim, K.; No, J.H.; Kim, Y.B.; Kim, H. Clinical evaluation of a droplet digital PCR assay for detecting POLE mutations and molecular classification of endometrial cancer. J. Gynecol. Oncol. 2022, 33, e15. [Google Scholar] [CrossRef]
- Berek, J.S.; Matias-Guiu, X.; Creutzberg, C.; Fotopoulou, C.; Gaffney, D.; Kehoe, S.; Lindemann, K.; Mutch, D.; Concin, N.; Endometrial Cancer Staging Subcommittee, F.W.s.C.C. FIGO staging of endometrial cancer: 2023. Int. J. Gynaecol. Obstet. 2023, 162, 383–394. [Google Scholar] [CrossRef]
- Concin, N.; Matias-Guiu, X.; Vergote, I.; Cibula, D.; Mirza, M.R.; Marnitz, S.; Ledermann, J.; Bosse, T.; Chargari, C.; Fagotti, A.; et al. ESGO/ESTRO/ESP guidelines for the management of patients with endometrial carcinoma. Int. J. Gynecol. Cancer 2021, 31, 12–39. [Google Scholar] [CrossRef]
- Abu-Rustum, N.; Yashar, C.; Arend, R.; Barber, E.; Bradley, K.; Brooks, R.; Campos, S.M.; Chino, J.; Chon, H.S.; Chu, C.; et al. Uterine Neoplasms, Version 1.2023, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2023, 21, 181–209. [Google Scholar] [CrossRef]
- Zeimet, A.G.; Reimer, D.; Huszar, M.; Winterhoff, B.; Puistola, U.; Azim, S.A.; Muller-Holzner, E.; Ben-Arie, A.; van Kempen, L.C.; Petru, E.; et al. L1CAM in early-stage type I endometrial cancer: Results of a large multicenter evaluation. J. Natl. Cancer Inst. 2013, 105, 1142–1150. [Google Scholar] [CrossRef]
- Asano, H.; Hatanaka, K.C.; Matsuoka, R.; Dong, P.; Mitamura, T.; Konno, Y.; Kato, T.; Kobayashi, N.; Ihira, K.; Nozaki, A.; et al. L1CAM Predicts Adverse Outcomes in Patients with Endometrial Cancer Undergoing Full Lymphadenectomy and Adjuvant Chemotherapy. Ann. Surg. Oncol. 2020, 27, 2159–2168. [Google Scholar] [CrossRef]
- Van Gool, I.C.; Stelloo, E.; Nout, R.A.; Nijman, H.W.; Edmondson, R.J.; Church, D.N.; MacKay, H.J.; Leary, A.; Powell, M.E.; Mileshkin, L.; et al. Prognostic significance of L1CAM expression and its association with mutant p53 expression in high-risk endometrial cancer. Mod. Pathol. 2016, 29, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, S.I.; Kim, N.R.; Kim, H.; Kim, H.S.; Chung, H.H.; Kim, J.W.; Lee, C.; Lee, M. Prognostic significance of L1CAM expression in addition to ProMisE in endometrial cancer. Gynecol. Oncol. 2023, 174, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Kajiwara, Y.; Ueno, H.; Hashiguchi, Y.; Shinto, E.; Shimazaki, H.; Mochizuki, H.; Hase, K. Expression of l1 cell adhesion molecule and morphologic features at the invasive front of colorectal cancer. Am. J. Clin. Pathol. 2011, 136, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Li, L.; Zhao, Z.S.; Wang, Y.X.; Ye, Z.Y.; Tao, H.Q. L1 and epithelial cell adhesion molecules associated with gastric cancer progression and prognosis in examination of specimens from 601 patients. J. Exp. Clin. Cancer Res. 2013, 32, 66. [Google Scholar] [CrossRef]
- Kodera, Y.; Nakanishi, H.; Ito, S.; Misawa, K.; Ito, Y.; Nakayama, G.; Koike, M.; Fujiwara, M.; Yamamura, Y.; Nakao, A. Expression of L1 cell adhesion molecule is a significant prognostic factor in pT3-stage gastric cancer. Anticancer Res. 2009, 29, 4033–4039. [Google Scholar] [PubMed]
- Fogel, M.; Gutwein, P.; Mechtersheimer, S.; Riedle, S.; Stoeck, A.; Smirnov, A.; Edler, L.; Ben-Arie, A.; Huszar, M.; Altevogt, P. L1 expression as a predictor of progression and survival in patients with uterine and ovarian carcinomas. Lancet 2003, 362, 869–875. [Google Scholar] [CrossRef]
- Schroder, C.; Schumacher, U.; Fogel, M.; Feuerhake, F.; Muller, V.; Wirtz, R.M.; Altevogt, P.; Krenkel, S.; Janicke, F.; Milde-Langosch, K. Expression and prognostic value of L1-CAM in breast cancer. Oncol. Rep. 2009, 22, 1109–1117. [Google Scholar]
- Pecorelli, S. Revised FIGO staging for carcinoma of the vulva, cervix, and endometrium. Int. J. Gynaecol. Obstet. 2009, 105, 103–104. [Google Scholar] [CrossRef] [PubMed]
- Stelloo, E.; Nout, R.A.; Osse, E.M.; Jurgenliemk-Schulz, I.J.; Jobsen, J.J.; Lutgens, L.C.; van der Steen-Banasik, E.M.; Nijman, H.W.; Putter, H.; Bosse, T.; et al. Improved Risk Assessment by Integrating Molecular and Clinicopathological Factors in Early-stage Endometrial Cancer-Combined Analysis of the PORTEC Cohorts. Clin. Cancer Res. 2016, 22, 4215–4224. [Google Scholar] [CrossRef] [PubMed]
- Kommoss, F.K.; Karnezis, A.N.; Kommoss, F.; Talhouk, A.; Taran, F.A.; Staebler, A.; Gilks, C.B.; Huntsman, D.G.; Kramer, B.; Brucker, S.Y.; et al. L1CAM further stratifies endometrial carcinoma patients with no specific molecular risk profile. Br. J. Cancer 2018, 119, 480–486. [Google Scholar] [CrossRef]
- Bosse, T.; Nout, R.A.; Stelloo, E.; Dreef, E.; Nijman, H.W.; Jurgenliemk-Schulz, I.M.; Jobsen, J.J.; Creutzberg, C.L.; Smit, V.T. L1 cell adhesion molecule is a strong predictor for distant recurrence and overall survival in early stage endometrial cancer: Pooled PORTEC trial results. Eur. J. Cancer 2014, 50, 2602–2610. [Google Scholar] [CrossRef]
- Jang, M.; Kwon, Y.; Kim, H.; Kim, H.; Min, B.S.; Park, Y.; Kim, T.I.; Hong, S.P.; Kim, W.K. Microsatellite instability test using peptide nucleic acid probe-mediated melting point analysis: A comparison study. BMC Cancer 2018, 18, 1218. [Google Scholar] [CrossRef]
- Colombo, N.; Creutzberg, C.; Amant, F.; Bosse, T.; Gonzalez-Martin, A.; Ledermann, J.; Marth, C.; Nout, R.; Querleu, D.; Mirza, M.R.; et al. ESMO-ESGO-ESTRO Consensus Conference on Endometrial Cancer: Diagnosis, treatment and follow-up. Ann. Oncol. 2016, 27, 16–41. [Google Scholar] [CrossRef]
- Leon-Castillo, A.; Gilvazquez, E.; Nout, R.; Smit, V.T.; McAlpine, J.N.; McConechy, M.; Kommoss, S.; Brucker, S.Y.; Carlson, J.W.; Epstein, E.; et al. Clinicopathological and molecular characterisation of ‘multiple-classifier’ endometrial carcinomas. J. Pathol. 2020, 250, 312–322. [Google Scholar] [CrossRef]
- Betella, I.; Fumagalli, C.; Rafaniello Raviele, P.; Schivardi, G.; De Vitis, L.A.; Achilarre, M.T.; Aloisi, A.; Garbi, A.; Maruccio, M.; Zanagnolo, V.; et al. A novel algorithm to implement the molecular classification according to the new ESGO/ESTRO/ESP 2020 guidelines for endometrial cancer. Int. J. Gynecol. Cancer 2022, 32, 003480. [Google Scholar] [CrossRef]
- Imboden, S.; Nastic, D.; Ghaderi, M.; Rydberg, F.; Siegenthaler, F.; Mueller, M.D.; Rau, T.T.; Epstein, E.; Carlson, J.W. Implementation of the 2021 molecular ESGO/ESTRO/ESP risk groups in endometrial cancer. Gynecol. Oncol. 2021, 162, 394–400. [Google Scholar] [CrossRef]
- Matsumoto, S.; Uchiumi, T.; Noda, N.; Ueyanagi, Y.; Hotta, T.; Kang, D. Droplet digital polymerase chain reaction to measure heteroplasmic m.3243A>G mitochondrial mutations. Lab. Med. 2023; online ahead of print. [Google Scholar] [CrossRef]
- Boldrin, E.; Mazza, M.; Piano, M.A.; Alfieri, R.; Montagner, I.M.; Magni, G.; Scaini, M.C.; Vassallo, L.; Rosato, A.; Pilati, P.; et al. Putative Clinical Potential of ERBB2 Amplification Assessment by ddPCR in FFPE-DNA and cfDNA of Gastroesophageal Adenocarcinoma Patients. Cancers 2022, 14, 2180. [Google Scholar] [CrossRef] [PubMed]
- Boldrin, E.; Piano, M.A.; Bernaudo, F.; Alfieri, R.; Biasin, M.R.; Montagner, I.M.; Volpato, A.; Mattara, G.; Lamacchia, F.; Magni, G.; et al. p53/TP53 Status Assessment in Gastroesophageal Adenocarcinoma. Cancers 2023, 15, 2783. [Google Scholar] [CrossRef] [PubMed]
- Colozza-Gama, G.A.; Callegari, F.; Besic, N.; Paniza, A.C.J.; Cerutti, J.M. Machine learning algorithm improved automated droplet classification of ddPCR for detection of BRAF V600E in paraffin-embedded samples. Sci. Rep. 2021, 11, 12648. [Google Scholar] [CrossRef] [PubMed]
- Schöniger, S.; Rüschoff, J. Mismatch Repair Deficiency and Microsatellite Instability. Encyclopedia 2022, 2, 1559–1576. [Google Scholar] [CrossRef]


| Characteristics | Total |
|---|---|
| Age | 55.93 (30–83) |
| OP | |
| Hysterectomy | 7 (3.8%) |
| Hys+BSO | 19 (10.4%) |
| Hys+BSO+LD | 157 (85.8%) |
| Histologic type | |
| Endometrioid | 166 (90.7%) |
| Non-endometrioid | 17 (9.3%) |
| Histologic grade | |
| Low | 146 (79.8%) |
| High | 37 (20.2%) |
| LVSI | |
| Absent | 152 (83.1%) |
| Present | 31 (16.9%) |
| Myometrial invasion | |
| <50% | 145 (79.2%) |
| >50% | 38 (20.8%) |
| FIGO stage 2009 | |
| IA | 133 (72.7%) |
| IB | 33 (18.0%) |
| II | 15 (8.2%) |
| III | 2 (1.1%) |
| FIGO stage updated 2023 | |
| IA | 109 (59.6%) |
| IB | 18 (9.8%) |
| IC | 8 (4.4%) |
| IIA | 9 (4.9%) |
| IIB | 10 (5.5%) |
| IIC | 29 (15.8%) |
| Prognostic risk group * | |
| Low | 100 (54.6%) |
| Intermediate | 20 (10.9%) |
| High intermediate | 47 (25.7%) |
| High | 16 (8.8%) |
| Advanced | 0 (0.0%) |
| Adjuvant treatment | |
| None | 147 (80.3%) |
| Radiotherapy | 25 (13.7%) |
| Chemotherapy | 7 (3.8%) |
| Chemoradiotherapy | 4 (2.2%) |
| Recur/Distant meta | |
| Absent | 160 (87.4%) |
| Present | 20 (10.9%) |
| NA | 3 (1.6%) |
| Characteristics | n = 183 | POLEmut n = 29 | MMR-D n = 53 | p53abn n = 16 | NSMP-L1CAM Neg n = 76 | NSMP-L1CAM Pos n = 9 | p-Value |
|---|---|---|---|---|---|---|---|
| Age | 0.038 | ||||||
| <60 | 134 | 22 (16.4%) | 43 (32.1%) | 8 (6.0%) | 57 (42.5%) | 4 (3.0%) | |
| ≥60 | 49 | 7 (14.3%) | 10 (20.4%) | 8 (16.3%) | 19 (38.8%) | 5 (10.2%) | |
| OP | 0.521 | ||||||
| Hysterectomy | 7 | 0 (0.0%) | 2 (28.6%) | 1 (14.3%) | 3 (42.9%) | 1 (14.3%) | |
| Hys+BSO | 19 | 1 (5.3%) | 4 (21.1%) | 2 (10.5%) | 10 (52.6%) | 2 (10.5%) | |
| Hys+BSO+LD | 157 | 28 (17.8%) | 47 (29.9%) | 13 (8.3%) | 63 (40.1%) | 6 (3.8%) | |
| Histologic type | <0.001 | ||||||
| Endometrioid | 166 | 29 (17.5%) | 51 (30.7%) | 4 (2.4%) | 75 (45.2%) | 7 (4.2%) | |
| Non-endometrioid | 17 | 0 (0.0%) | 2 (11.8%) | 12 (70.6%) | 1 (5.9%) | 2 (11.8%) | |
| Histologic grade | <0.001 | ||||||
| Low | 146 | 24 (16.4%) | 43 (29.5%) | 4 (2.7%) | 73 (50.0%) | 2 (1.4%) | |
| High | 37 | 5 (13.5%) | 10 (27.0%) | 12 (32.4%) | 3 (8.1%) | 7 (18.9%) | |
| LVSI | 0.329 | ||||||
| Absent | 152 | 26 (17.1%) | 40 (26.3%) | 15 (9.9%) | 64 (42.1%) | 7 (4.6%) | |
| Present | 31 | 3 (9.7%) | 13 (41.9%) | 1 (3.2%) | 12 (38.7%) | 2 (6.5%) | |
| Myometrial invasion | 0.324 | ||||||
| <50% | 145 | 24 (16.6%) | 40 (27.6%) | 14 (9.7%) | 62 (42.8%) | 5 (3.4%) | |
| >50% | 38 | 5 (13.2%) | 13 (34.2%) | 2 (5.3%) | 14 (36.8%) | 4 (10.5%) | |
| FIGO stage 2009 | 0.551 | ||||||
| IA | 133 | 23 (17.3%) | 36 (27.1%) | 14 (10.5%) | 56 (42.1%) | 4 (3.0%) | |
| IB | 33 | 5 (15.2%) | 11 (33.3%) | 0 (0.0%) | 13 (39.4%) | 4 (12.1%) | |
| II | 15 | 1 (6.7%) | 5 (33.3%) | 2 (13.3%) | 6 (40.0%) | 1 (6.7%) | |
| III | 2 | 0 (0.0%) | 1 (50.0%) | 0 (0.0%) | 1 (50.0%) | 0 (0.0%) | |
| FIGO stage updated 2023 | <0.001 | ||||||
| IA | 109 | 20 (18.3%) | 29 (26.6%) | 4 (3.7%) | 55 (50.5%) | 1 (0.9%) | |
| IB | 18 | 2 (11.1%) | 6 (33.3%) | 0 (0.0%) | 10 (55.6%) | 0 (0.0%) | |
| IC | 8 | 0 (0.0%) | 2 (25.0%) | 2 (25.0%) | 2 (25.0%) | 2 (25.0%) | |
| IIA | 9 | 1 (11.1%) | 2 (22.2%) | 0 (0.0%) | 6 (66.7%) | 0 (0.0%) | |
| IIB | 10 | 1 (10.0%) | 6 (60.0%) | 0 (0.0%) | 2 (20.0%) | 1 (10.0%) | |
| IIC | 29 | 5 (17.2%) | 8 (27.6%) | 10 (34.5%) | 1 (3.4%) | 5 (17.2%) | |
| Prognostic risk group * | <0.001 | ||||||
| Low | 100 | 20 (20.0%) | 28 (28.0%) | 3 (3.0%) | 48 (48.0%) | 1 (1.0%) | |
| Intermediate | 20 | 3 (15.0%) | 6 (30.0%) | 2 (10.0%) | 7 (35.0%) | 2 (10.0%) | |
| High intermediate | 47 | 6 (12.8%) | 17 (36.2%) | 1 (2.1%) | 20 (42.6%) | 3 (6.4%) | |
| High | 16 | 0 (0.0%) | 2 (12.5%) | 10 (62.5%) | 1 (6.3%) | 3 (18.8%) | |
| Adjuvant treatment | 0.019 | ||||||
| None | 147 | 25 (17.0%) | 41 (27.9%) | 12 (8.2%) | 64 (43.5%) | 5 (3.4%) | |
| Radiotherapy | 25 | 4 (16.0%) | 8 (32.0%) | 0 (0.0%) | 11 (14.3%) | 2 (8.0%) | |
| Chemotherapy | 7 | 0 (0.0%) | 2 (28.6%) | 3 (42.9%) | 1 (44.0%) | 1 (14.3%) | |
| Chemoradiotherapy | 4 | 0 (0.0%) | 2 (50.0%) | 1 (25.0%) | 0 (0.0%) | 1 (25.0%) |
| RFS | OS | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | Univariate Analysis | Multivariate Analysis | |||||
| Parameters | Hazard Ratio (95% CI) | p-Value | Hazard Ratio (95% CI) | p-Value | Hazard Ratio (95% CI) | p-Value | Hazard Ratio (95% CI) | p-Value |
| age (<60 vs >60 years) | 4.336 (1.772–10.611) | 0.001 | - | 6.353 (1.911–21.122) | 0.003 | - | ||
| Histologic type (endometrioid vs non-endometrioid) | 6.309 (2.410–16.511) | <0.001 | - | 13.836 (4.424–43.269) | <0.001 | - | ||
| Histologic grade (grade 1, 2 vs grade 3, high grade) | 4.978 (2.067–11.989) | <0.001 | - | 14.322 (3.865–53.078) | <0.001 | - | ||
| Myometrial invasion (<50% vs >50%) | 3.330 (1.375–8.063) | 0.008 | 3.845 (1.568–9.428) | <0.001 | 0.264 (0.0.85–0.821) | 0.021 | 4.535 (1.443–14.251) | 0.010 |
| Prognostic risk group | ||||||||
| intermediate | 5.604 (1.448–21.693) | 0.013 | - | 8.853 (0.989–79.213) | 0.051 | - | ||
| high | 10.645 (2.927–38.720) | <0.001 | - | 21.700 (2.668–176.469) | 0.004 | - | ||
| Updated 2023 FIGO stage (stage 1 vs stage 2) | 5.882 (2.344–14.756) | <0.001 | - | 4.248 (1.348–13.389) | 0.014 | - | ||
| Molecular L1CAM classification (POLEmut, MMR-D, NSMP-L1CAMneg vs p53abn, NSMP-L1CAMpos) | 13.537 (5.401–33.932) | <0.001 | 15.005 (5.883–38.269) | <0.001 | 22.585 (6.104–83.560) | <0.001 | 24.807 (6.669–92.277) | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joe, S.; Lee, M.; Kang, J.; Kim, J.; Hong, S.-H.; Lee, S.J.; Lee, K.H.; Lee, A. Enhanced Risk Stratification in Early-Stage Endometrial Cancer: Integrating POLE through Droplet Digital PCR and L1CAM. Cancers 2023, 15, 4899. https://doi.org/10.3390/cancers15194899
Joe S, Lee M, Kang J, Kim J, Hong S-H, Lee SJ, Lee KH, Lee A. Enhanced Risk Stratification in Early-Stage Endometrial Cancer: Integrating POLE through Droplet Digital PCR and L1CAM. Cancers. 2023; 15(19):4899. https://doi.org/10.3390/cancers15194899
Chicago/Turabian StyleJoe, Seungyeon, Miseon Lee, Jun Kang, Joori Kim, Sook-Hee Hong, Sung Jong Lee, Keun Ho Lee, and Ahwon Lee. 2023. "Enhanced Risk Stratification in Early-Stage Endometrial Cancer: Integrating POLE through Droplet Digital PCR and L1CAM" Cancers 15, no. 19: 4899. https://doi.org/10.3390/cancers15194899
APA StyleJoe, S., Lee, M., Kang, J., Kim, J., Hong, S.-H., Lee, S. J., Lee, K. H., & Lee, A. (2023). Enhanced Risk Stratification in Early-Stage Endometrial Cancer: Integrating POLE through Droplet Digital PCR and L1CAM. Cancers, 15(19), 4899. https://doi.org/10.3390/cancers15194899






