Simple Summary
As recurrence in glioblastoma is locally generated around the resection cavity, surviving glioma stem-like cells may cause recurrence. Glioma stem-like cells expressing high levels of CD44 are highly invasive and are required to change to less invasive, more proliferative types to generate a recurrent tumor. CD44 promotes both the invasion and proliferation of tumor cells via various signaling pathways. Among these, paired pathways show reciprocal activation of invasion and proliferation under different conditions of oxygenation. Severe hypoxia activates genes related to cell invasion, whereas moderate hypoxia activates genes related to cell proliferation. CD44 is associated with both pathways and plays a critical role in regulating the balance between the promotion of cell invasion and cell proliferation. These results may indicate that CD44 is a key molecule for executing tumor recurrence in glioblastoma.
Abstract
High invasiveness is a characteristic of glioblastoma (GBM), making radical resection almost impossible, and thus, resulting in a tumor with inevitable recurrence. GBM recurrence may be caused by glioma stem-like cells (GSCs) that survive many kinds of therapy. GSCs with high expression levels of CD44 are highly invasive and resistant to radio-chemotherapy. CD44 is a multifunctional molecule that promotes the invasion and proliferation of tumor cells via various signaling pathways. Among these, paired pathways reciprocally activate invasion and proliferation under different hypoxic conditions. Severe hypoxia (0.5–2.5% O2) upregulates hypoxia-inducible factor (HIF)-1α, which then activates target genes, including CD44, TGF-β, and cMET, all of which are related to tumor migration and invasion. In contrast, moderate hypoxia (2.5–5% O2) upregulates HIF-2α, which activates target genes, such as vascular endothelial growth factor (VEGF)/VEGFR2, cMYC, and cyclin D1. All these genes are related to tumor proliferation. Oxygen environments around GBM can change before and after tumor resection. Before resection, the oxygen concentration at the tumor periphery is severely hypoxic. In the reparative stage after resection, the resection cavity shows moderate hypoxia. These observations suggest that upregulated CD44 under severe hypoxia may promote the migration and invasion of tumor cells. Conversely, when tumor resection leads to moderate hypoxia, upregulated HIF-2α activates HIF-2α target genes. The phenotypic transition regulated by CD44, leading to a dichotomy between invasion and proliferation according to hypoxic conditions, may play a crucial role in GBM recurrence.
Keywords:
glioblastoma; invasion; proliferation; CD44; tumor recurrence; hypoxia; phenotypic transition 1. Introduction
Glioblastoma (GBM) is the most common malignant brain tumor in adults and is characterized by a highly invasive nature and an extremely poor prognosis; the median survival is 15 months under the current standard treatment regimen based on the protocol outlined by Stupp et al. [1]. The poor prognosis may be largely attributable to early recurrence caused by tumor cells that escape tumor resection and develop resistance to radio-chemotherapy. Emerging evidence indicates that glioma stem-like cells (GSCs), which constitute a small subpopulation in GBM, may play a central role in GBM recurrence [2,3]. GSCs show stem cell properties of self-renewal and multi-lineage differentiation, and the ability to initiate a tumor on transplantation and recapitulation of the initial phenotype [4]. GSCs characteristically show highly invasive features and high resistance to radio-chemotherapy [5,6].
Solid tumors constitute tumor cells and a tumor microenvironment (TME). The TME in GBM includes astrocytes; neurons; endothelial cells; immune cells, such as non-tumor cells; extracellular matrix (ECM) components; and various growth factors and cytokines. In particular, the TME of cancer stem cells represents a niche in which cancer stem cells reside and mutually communicate with various components of the TME. Such bidirectional crosstalk between cancer stem cells and the TME can enable a wide variety of cellular events, such as invasion, proliferation, and angiogenesis [7,8].
In the TME, ECM molecules and their receptors play crucial roles in enacting this variety of cellular events. In the brain, the major component of the ECM is hyaluronic acid (HA) and its primary receptor is CD44. CD44 is a multi-functional transmembrane glycoprotein, and the binding of CD44 to HA can promote various tumor cell activities [9]. CD44 can mediate not only cell-to-cell interactions but also cell-to-ECM interactions, resulting in the transduction of microenvironmental signals to membrane-associated cytoskeletal proteins or the cell nucleus to activate the expression of various genes participating in cellular processes of migration, invasion, proliferation, and tumorigenesis in malignant tumors [10,11]. CD44 is also regarded as the most common stem cell surface marker in many cancers, such as those of the stomach, breast, lung, colon, and brain [12,13,14,15]. Thus, CD44 is not only an adhesion molecule but also a signal-transducing molecule that coordinately regulates numerous cellular behaviors through multiple signaling pathways.
GBM recurrences primarily occur at local sites of the marginal area around the resection cavity of the tumor [16,17]. Migrating GSCs with high invasive activity could be present in the peritumoral normal brain, escaping surgical resection and the following radio-chemotherapy and migrating further into the peritumoral area. Migrating GSCs that could cause tumor recurrence at the margins of the resection cavity would require at least two activities to accomplish recurrence. One is a homing ability to return to the original tumor site. The other is the ability to rapidly proliferate, i.e., to generate a tumor mass. According to the theoretical “Go or Grow” dichotomy of tumor cells [18,19], such behaviors of GSCs may require a phenotypic transition from invasive to proliferative tumor types. Herrlich et al. suggested that CD44 might display dual activities promoting the proliferation or invasiveness of tumor cells [20]. CD44 may thus play a crucial role in the regulation of invasion and proliferation as essential behaviors of tumor cells developing into GBM recurrence.
In this narrative review, we comprehensively review the literature that relates to the functional activities of CD44 and summarize recent advances in the activities of CD44. Using these data, we elucidate how CD44 can participate in the generation of tumor recurrence in GBM. A broad mapping of CD44 activities in the various cellular events, including invasion, proliferation, and epithelial-to-mesenchymal transition (EMT), and an understanding of how CD44-activated cellular events can be co-operatively regulated in GBM, particularly glioma stem-like cells, may bring new insight, not only clarifying the molecular mechanisms underlying GBM recurrence but also establishing a therapeutic strategy by effectively targeting CD44 to eradicate GBM.
2. Structure and Function of CD44
The functional variety of CD44 is attributed to the diversity of molecular structures of CD44, including isoforms generated by the alternative splicing of the CD44 gene. The CD44 gene encodes 20 exons, with exons 6–15 alternatively spliced and inserted into the stem region of CD44 as variant exons (v1–v10) [21,22]. The first five (1–5) and last five (16–20) exons are constant and encode the shortest isoform of CD44, which is the CD44 standard (CD44s) isoform. Splice variants with variant exons are designated CD44 variant (CD44v) isoforms (Figure 1).
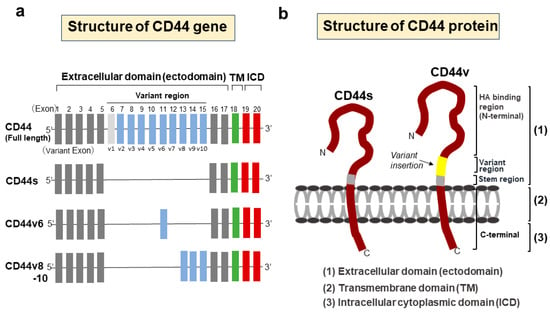
Figure 1.
Structures of CD44 gene and protein. (a) Schematic structures of the CD44 gene. The CD44 gene encodes 20 exons, with exons 6–15 alternatively spliced and inserted into the stem region of CD44 as variant exons. The first five (1–5) and last five (16–20) exons are constant and encode the CD44 standard (CD44s) isoform. Splice variants with variant exons are designated CD44 variant (CD44v) isoforms. The structure of CD44v6 is shown as a representative variant isoform. In humans, v1 exon is lacking due to an in-frame termination codon. (b) Structural domains of the CD44 protein. CD44 consists of three domain regions, including an extracellular domain (ectodomain), transmembrane domain, and cytoplasmic domain. In CD44v, a variant region containing splice variants is inserted in the ectodomain. All CD44 isoforms have a hyaluronic acid (HA)-binding site in the extracellular domain.
CD44 consists of three domains: an extracellular amino-terminal domain (ectodomain), a transmembrane domain, and an intracellular cytoplasmic domain (ICD) [23]. In CD44v isoforms, the stem region containing splice variants in the ectodomain provides a co-receptor for the binding of various growth factors and cytokines, thus transducing signals to promote invasion, progression, and tumorigenesis via specific signaling pathways [10]. Different isoforms of CD44 show different functions according to the variant exon inserted in the CD44 ectodomain. The amino-terminal globular region of the ectodomain is an HA-binding domain (HABD), where the HABD is common to all isoforms. HA binding to CD44-HABD causes conformational changes in CD44, resulting in the promotion of cell-signaling pathways to regulate a variety of cellular processes, such as migration and proliferation [24]. The transmembrane domain provides a pathway for interactions with co-factors, adaptor proteins, and receptor tyrosine kinases (RTKs) [25]. CD44-ICD is generated via intramembranous cleavage of CD44 via the proteolytic action of an enzyme complex formed by presenilin 1-presenilin 2-γ-secretase [26,27]. Proteolytic cleavage of CD44 is known to be sequentially performed in two steps [28]. Cleavage of the transmembrane domain of CD44 (second step) requires the preceding cleavage of the extracellular domain of CD44 (first step). The first step in the sequential cleavage of CD44 is achieved by two matrix metalloproteinases (MMPs), namely, a disintegrin and metalloproteinase (ADAM)10 and ADAM17, which are sequentially activated through the two separate pathways of elevated Ca2+ influx and activation of protein kinase C (PKC)-Rac, respectively [29]. The released CD44-ICD fragments then translocate to the nucleus and activate various genes as a co-transcription factor for Runt-related transcription factor 2 (RUNX2) [30]. Furthermore, CD44-ICD binds to the CCTGCG consensus sequence of CD44 (that is, the CD44-ICD response element (CIRE)) located near the RUNX2 site in the promoter of MMP-9, leading to upregulated MMP-9 expression. The CIRE sequence is also present in HIF-1α-related genes, the expression of which is regulated under normoxic conditions [31]. The activation of all these genes by CD44-ICD is achieved under normoxic conditions. In contrast, under hypoxic conditions, CD44-ICD binds to HIF-2α and activates its expression, resulting in the enhanced activation of HIF-2α target genes [32] (Figure 2).
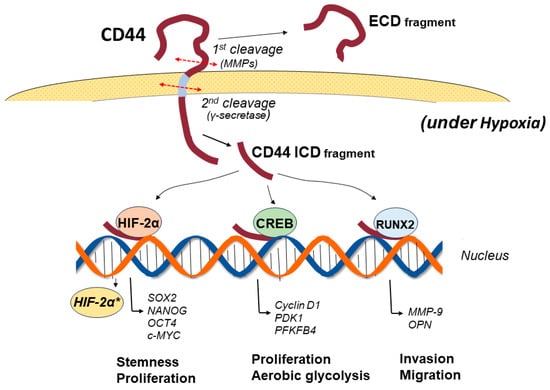
Figure 2.
Signaling pathways of the CD44 intracellular domain (CD44-ICD) under hypoxia. CD44 is cleaved in two steps. The first cleavage is promoted via proteolysis of the ectodomain by matrix metalloproteinases, among others, followed by a second cleavage of the transmembrane domain by presenilin-dependent γ-secretase, resulting in the release of CD44-ICD. The released CD44-ICD fragments translocate to the nucleus and activate various genes as a co-transcription factor for Runt-related transcription factor 2 (RUNX2). CD44-ICD also binds to cAMP response element-binding protein (CREB) to regulate the gene expression of cyclin D1 and oxidative glycolysis-related genes 3-phosphoinositide-dependent kinase 1 (PDK1) and 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (PFKFB4). Under hypoxic conditions, CD44-ICD binds to hypoxia-inducible factor (HIF)-2α and activates its expression, resulting in enhanced activation of HIF-2α target genes, including Sox2, NANOG, and OCT4, the genes of which are required for maintaining the stemness of GSCs. *: activated gene.
The interactions of CD44 and HA can enhance, inhibit, or sometimes not show any effect on various cellular behaviors. CD44 was demonstrated to exist in three different activation states: inactive, inducibly active, and constitutively active [33]. Yang et al. reported that CD44 is inactive on normal cells, but active on cancerous cells [34,35]. The expression of the functional activities of CD44 and HA requires the activation of CD44 [36]. This activation can be induced by various factors, including growth factors, cytokines, and the TME [37]. In addition, although all CD44 molecules possess the HA-binding domain, affinities for HA binding differ. This suggests that variant isoforms of CD44 generated by alternative splicing in GSCs may participate in CD44 activation [38]. A more detailed understanding of the molecular mechanism underlying CD44 activation will no doubt provide new therapeutic strategies.
3. Expression of CD44 in GBM
Immunohistochemical investigations of CD44 and nestin in GBM demonstrated that CD44 is expressed on the cell membrane of GBM stem cells [39]. Si et al. recently reported that CD44 in GBM tissues is mainly expressed on the cell membrane, but some tumor cells express high levels of CD44 in the cytoplasm, with such high expression predicting a poor prognosis due to GBM [40]. We also reported that GBM expressing CD44 at much higher levels in the tumor periphery than in the tumor core shows much higher invasive features on magnetic resonance imaging (MRI), and patients presenting with high CD44 expression experience earlier recurrence and worse prognosis than those presenting with low CD44 expression [41]. Many studies reported that high expression of CD44 predicts poor prognosis in various cancers [42,43,44]. Immunohistochemical analysis of 125 breast cancer patients disclosed that the amount of CD44 protein correlates positively with poor disease-free survival and overall survival (OS) [45]. A meta-analysis of 583 patients with pancreatic cancer revealed that high levels of CD44 correlate adversely with the 5-year OS [46]. However, some studies reported no association between CD44 expression levels and survival. A meta-analysis of 1747 breast cancer patients showed no relationship between CD44 expression and OS [47]. Further, in GBM, high expression of CD44 is not significantly associated with low OS [48]. Although CD44 expression is recognized as a definite independent prognostic factor in various cancers, CD44v isoforms have increasingly attracted attention due to their much more specific expression in cancer stem cells and their various functions via signaling transduction compared with CD44. In esophageal squamous cell carcinoma, higher levels of CD44v9 protein at the invasive front of tumors correlate with worse progression-free survival (PFS) and OS [49], and CD44v9 independently predicts adverse prognosis in gastric cancer [50]. CD44v6 is reportedly predictive of metastasis for colon cancer [51] and has been associated with poor prognosis in several cancers, including prostate cancer and pancreatic cancer [52,53].
For GBM, few reports examined CD44v isoforms. To date, the expression of CD44v3, CD44v5, and CD44v6 isoforms have been identified in GBM, but their functions in terms of cellular processes remain unclarified, except for a role of CD44v6 in the regulation of GSC growth [54]. We investigated gene expressions of CD44s and CD44v in GSCs but found no significant difference in expression. GSCs obtained from the primary culture of tumor tissues in GBM expressing high CD44 showed that the expression of CD44v5 was the highest, followed by CD44v6, whereas differentiated GBM cells showed higher expression of CD44v6, followed by CD44v5. Stem-like cells obtained by introducing the Oct3/4 gene into differentiated glioma cells revealed the same expression pattern as GSCs (unpublished data). Wu et al. recently reported the results of a meta-analysis that evaluated the predictive value of higher CD44 expression for prognosis in patients with glioma [55]. That study disclosed that higher CD44 expression predicted poorer survival in patients with WHO grade II–III glioma but did not show any significant positive correlation with worse prognosis in patients with GBM. As potential reasons for this discrepancy, the authors suggested the molecular heterogeneity of GBM and the fact that the optimal cut-off values for CD44 expression had not been determined for each grade of tumor. We recently reported that patients with GBM showing high CD44 expression in the tumor periphery show significantly shorter survival in terms of both PFS and OS than those showing low CD44 expression [56]. In the future, a novel CD44v isoform that can more definitively predict prognosis in patients with GBM may be identified.
4. Signaling Pathways via CD44-HA Interactions
CD44 binds to various ligands and the interactions allow cancer cells to execute many cellular functions via the various signal transductions specific to the ligands. Among these, common ligands for CD44 include HA, fibronectin [57], osteopontin (OPN) [58], collagen [59], and serglycin/sulfated proteoglycan [60]. The effects of CD44 on tumor cell invasion, proliferation, and progression depend on the specificity of the interactions between CD44 and each ligand. HA, which is a major constituent of the ECM, is the most common ligand of CD44 and its interactions with CD44 can activate multiple pathways, leading to cell survival, proliferation, and invasion via signaling networks, including pathways through adaptor molecules; RTKs, such as EGFR and ErB2; and non-receptor kinases of the Src family [10,61] (Figure 3a,b).
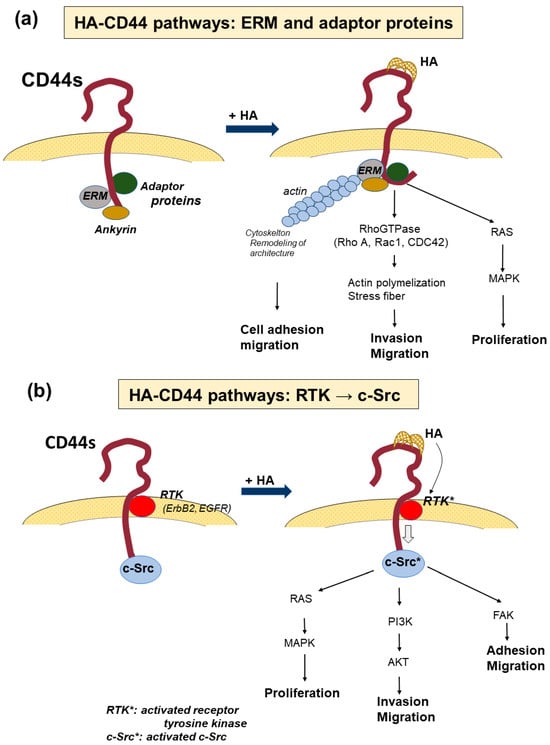
Figure 3.
The signaling pathway activated by HA-CD44 interaction. (a) The signaling pathway from the promotion of the binding of ezerin-radexin-moesin (ERM), ankyrin, and adaptor proteins to the cytoplasmic tail. Binding activates the interaction of CD44 and RAS and RhoGTPase, leading to the promotion of actin polymerization, remodeling of the cytoskeletal architectures, and facilitating cell migration and invasion. In contrast, the oncogenic gene RAS activates mitogen-activated protein kinase (MAPK) and promotes cell proliferation. (b) Signaling pathway activated by receptor tyrosine kinases (RTKs). HA-CD44 interaction activates RTKs, such as ErbB2 and EGFR, which elevate the activities of the non-receptor kinases of the Src family or Ras family GTPase. Such intracellular signaling enhances the activity of downstream signaling pathways, such as MAPK, phosphoinositide 3-kinase (PI3K)/AKT, and focal adhesion kinase (FAK). The former promotes cell proliferation, while the latter two enhance cell migration and invasion. *: activated gene.
As CD44 lacks kinase activity, several signaling systems are utilized to activate protein kinases. The binding of HA to CD44 causes a conformational change in CD44 to facilitate binding to the ezrin-radoxin-moesin (ERM) protein and other adaptor molecules via the cytoplasmic tail of CD44, which mediates CD44 interactions with Ras and RhoGTPase, including RhoA, Rac1, and CDC42 as effectors of upstream pathways [62]. Activation of these molecules initiates changes in the cytoskeletal architecture to facilitate tumor cell migration and invasion into the extracellular space of the ECM [61]. HA-CD44 interactions also activate receptor tyrosine kinases, such as ErbB2 and EGFR, in the signaling domain of CD44 and elevate the activities of non-receptor kinases of the Src family or Ras family GTPases [63]. These intracellular signaling pathways enhance the activity of downstream signaling pathways, such as mitogen-activated protein kinases (MAPK), phosphoinositide 3-kinase (PI3K)/Akt, and focal adhesion kinase [63]. Through the Ras/MAPK pathway, Src signaling promotes tumor cell division and proliferation. In contrast, the PI3K/AKT pathway contributes to tumor cell invasion and proliferation [61,64]. In addition, HA binding to CD44 activates multidrug transporters that participate in chemoresistance [65].
Specific exons at some sites in the ectodomain of CD44 are inserted via alternative splicing in CD44v isoforms, as mentioned above. These peptide sites in CD44v isoforms can act as co-receptors for various growth factors and receptors, resulting in participation in specific signaling pathways (Figure 4). In the CD44v3 isoform containing a heparin sulfate site, heparin sulfate-binding growth factors, such as fibroblast growth factor (FGF) and epidermal growth factor (EGF), bind to CD44 [24]. CD44v6 has a binding site for hepatocyte growth factor (HGF) and VEGF [25]. As HGF is a ligand for cMET, the interactions of these molecules activate cMET, leading to stimulation of the Ras signaling pathway and a subsequent increase in motility [66]. In the treatment of both primary and recurrent GBM, antiangiogenic therapy with bevacizumab, which is a monoclonal antibody that targets VEGF, is often performed. However, in patients that show resistance to bevacizumab, the tumor often presents much more invasive features than seen before treatment. McCarty reported that tumor cell proliferation and invasion are regulated via crosstalk between the VEGF and HGF signaling pathways [67]. When GBM proliferates and shows progression, VEGFR-2 and cMET form heterodimeric complexes that inhibit cell invasion via the tyrosine dephosphorylation of cMET. When VEGF is neutralized with bevacizumab, cMET is upregulated, forms homodimers that allow for the binding of HGF, and promotes mesenchymal-like phenotypes and tumor cell invasion. In our study, the inhibition of VEGF with siRNA or bevacizumab in GSCs also markedly increased the expressions of both CD44 and cMET and caused much higher tumor invasion than the control [68].
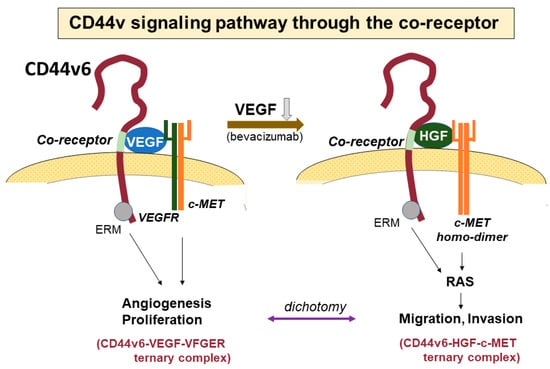
Figure 4.
The signaling pathway through co-receptors of CD44. The CD44v6 isoform has a co-receptor in the variant region of the CD44 ectodomain. Vascular endothelial growth factor (VEGF) and hepatocyte growth factor (HGF) can bind to the co-receptor. VEGF bound to the co-receptor further binds to the VEGF receptor (VEGFR), forming a ternary complex of CD44v6-VEGF-VEGFR to promote angiogenesis. VEGFR and cMET form a heterodimer and suppress migratory activity via the interaction of HGF and cMET. In contrast, the binding of VEGF to CD44 is inhibited by the anti-VEGF antibody bevacizumab, and thus, CD44v6 does not form the heterodimer of VEGFR and cMET and a homodimer with cMET is instead generated. HGF then binds to the co-receptor of CD44 instead of VEGF, creating a CD44v6-HGF-cMET ternary complex. This stimulates the Ras signaling pathway, resulting in increased motility.
CD44-intracellular cytoplasmic domain (ICD) plays crucial roles in the intracellular signaling pathways for executing special cellular events, such as tumor progression, the maintenance of the stemness of cancer stem cells, aerobic glycolysis, and the production of CD44. As the structure and generation of CD44-ICD were described in the previous section, the major pathways of CD44-ICD are detailed here (Figure 2, Table 1). The expression of CD44-ICD is specific to cancer stem cells, and the hypoxic environment greatly affects CD44-ICD pathways. Under normoxic conditions, CD44-ICD activates HIF-1α-related genes without the binding of HIF-1α to CD44-ICD [32]. In contrast, under hypoxic conditions, CD44-ICD can bind to HIF-2α and activate the expression of HIF-2α. The enhanced activity of HIF-2α upregulates HIF-2α-target genes, such as SOX2, OCT4, NANOG, and c-MYC [32,69]. Hypoxia-induced HIF-2α is preferentially expressed in cancer stem cells compared with differentiated tumor cells [70] and plays an important role as a transcription factor to activate genes related to the maintenance of stem cell features. In addition, CD44-ICD activates the transcription of genes containing 12-O-tetradecanoyl-phorbol-13-acetate response elements, such as CD44 [71]. The cleavage of CD44 facilitates detachment from HA in the ECM. CD44-ICD liberated by the second cleavage step can elevate the expression of newly generated CD44 molecules on the cell membrane, facilitating binding to other sites and resulting in the promotion of cell migration and invasion. CD44-ICD also binds to the cyclic adenosine monophosphate (cAMP) response element-binding protein (CREB) to regulate the expression of genes, such as cyclin D1 [72] and the oxidative glycolysis-related genes 3-phsphoinositide-dependent kinase 1 (PDK1) and 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (PFKFB4) [73]. Activation of these genes can promote the proliferation, aerobic glycolysis, and creation of new CD44 molecules in GSCs.

Table 1.
Summary of genes and transcription factors that are activated by CD44-ICD.
As mentioned above, HA-binding CD44 activates numerous signaling pathways that participate in various cellular events of cancers, including migration, invasion, proliferation, survival, stemness, drug resistance, and many other functions involved in malignant properties. However, the question remains as to how cancer cells can regulate the numerous signaling pathways in a coordinated manner, leading to the tumor-specific phenotypes and behaviors seen during cancer development and progression. Bourguignon et al. demonstrated that HA-CD44 interaction also activates microRNA (miRNA) signaling to regulate tumor-specific functions, such as tumor cell growth, migration, invasion, and chemoresistance, thus promoting tumor progression [74] (Figure 5, Table 2). As small, non-coding RNAs of approximately 22 nucleotides in length, miRNAs mediate the posttranscriptional silencing of specific target mRNAs and are recognized as important conductors that regulate tumor development and progression [75]. Many miRNAs were reported to show aberrant expression in various cancers. In this review, we describe primary miRNAs expressed in GBM and their pivotal roles in tumor-specific malignant behaviors. Oncogenic miRNA-21 is known to regulate tumor cell proliferation, invasion, survival, chemoresistance, and progression in various cancers [76,77,78]. HA-binding CD44 interactions were demonstrated to activate miRNA-21 via the phosphorylation of the JNK/c-Jun transcription factor [79]. Chen et al. found that the inhibition of JNK/c-Jun or miRNA-21 downregulated the expressions of survival proteins, such as Bcl2 and inhibitors of apoptosis protein (IAPs), resulting in increased apoptotic cell death and the decreased chemoresistance of tumor cells [80]. These findings suggest that miRNA-21 upregulated by JNK/c-Jun, which is activated by HA-CD44 interactions, may play a crucial role in tumorigenesis and chemoresistance. Several studies reported that miRNA-21 is significantly elevated in GBM. Gabriely et al. reported that the inhibition of miRNA-21 by antisense oligonucleotides leads to elevated levels of RECK and tissue inhibitor of metalloproteinase (TIMP) 3, thus reducing MMP activities, both in vitro and in vivo. In addition, the downregulation of miRNA-21 in glioma cells decreased their migratory and invasive abilities [81]. Luo et al. found that miRNA-21 promotes the migration and invasion of glioma cells by activating Sox2 and β-catenin signaling [82]. Further, miRNA-21 was reported to participate in the drug resistance of GBM [83].
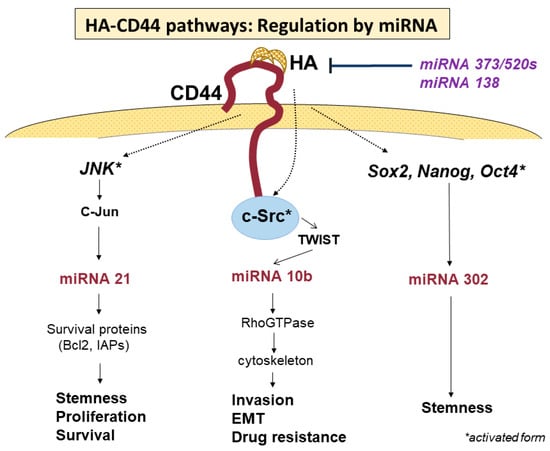
Figure 5.
Regulation of HA-CD44 binding-activated signaling pathways. Activity of the signaling pathways promoted by HA-CD44 binding is regulated by miRNAs from both upstream and downstream. Each miRNA regulates the signaling pathway to promote special cellular behaviors.
In addition to miRNA-21, the overexpression of miRNA-10b was found to be necessary to promote the migration and invasion of tumor cells in the metastatic process of breast cancer cells [84]. Sasayama et al. reported that miRNA-10b is overexpressed in malignant gliomas, being the highest in GBM, and that the overexpression of miRNA-10b is associated with the upregulation of Rho C, which stimulates glioma invasion and migration [61,85]. On the other hand, miRNA-10b is reportedly activated by phosphorylated TWIST via the activation of c-Src under the stimulation of HA-CD44 interaction, promoting tumor-cell-specific activities, such as EMT, tumor invasion, and drug resistance, in cancer cells [86]. Furthermore, miRNA-302 was demonstrated to promote the stemness of cancer stem cells [87,88,89], which is upregulated by transcription factors such as Nanog, Oct4, and Sox2 [89,90,91], the expressions of which are activated by HA-CD44 interaction [92]. The treatment of cancer stem cells with anti-miRNA-302 inhibitors upregulates lysine-specific histone demethylases and decreases DNA global demethylation, impairing HA-CD44-activated functions in cancer stem cells [93]. These results indicate that the signaling pathway of miRNA-302 may be regulated by stem cell markers, such as Nanog/Oct4/Sox2, for which the expressions are activated by HA-CD44 interactions [93].

Table 2.
microRNAs participating in regulation of the activites of CD44 in glioblastoma.
Table 2.
microRNAs participating in regulation of the activites of CD44 in glioblastoma.
| microRNA | Regulation | Activated or Inhibitted Cellular Function of CD44 | References |
|---|---|---|---|
| miR-21 | positive | Activation: Proliferation, stemness, invasion, chemotherapy resistant | [76,77,78,80,81,82] |
| Inhibition: senescence, apoptosis | [83] | ||
| miR-10b | positive | Activation: Invasion, migration, EMT, drug resistance | [61,84,85,86,93] |
| miR-302 | positive | Activation: stemness | [87,88,89,90,92] |
| miR-373 | negative | Inhibition: proliferation, invasion | [94] |
| miR-520s | |||
| miR-138 | negative | Inhibition: proliferation, cell cycle, migration, wound-healing ability | [95] |
EMT; epithelial-to-mesenchymal transition.
On the other hand, some miRNAs are known to regulate the expression and functional activities of CD44. Feng et al. demonstrated that CD44 is a direct target of miRNA-373 and miRNA-520s, and the expressions of miRNA-373 and miRNA-520s are negatively associated with CD44 expression in GBM [94]. Suppressing CD44 expression via the increased expressions of miRNA-373 and miRNA-520s leads to the inhibition of the proliferation and invasiveness of GBM cells [94]. Yeh et al. reported that whole-transcriptome analysis and miRNA expression profiling identified miRNA-138 as one of the most significantly downregulated miRNAs [95]. Downregulated miRNA-138 shows an inverse correlation with CD44 expression. An overexpression of miRNA-138 in GBM cells reduces CD44 expression and inhibits cell proliferation, cell cycle, migration, and wound-healing capabilities. They revealed that miRNA-138 negatively regulates the expression of CD44 by directly binding to the 3′-untranslated region (UTR) of CD44. Thus, CD44 inhibition by miRNA-138 results in an inhibition of GBM cell proliferation via cell cycle arrest by inducing p27 and subsequent translocation of this molecule into the nucleus. Such suppressive activities of miRNAs may have much more beneficial effects as therapeutic targets than anti-CD44 therapy targeting each signaling pathway. As mentioned above, CD44 signaling pathways are regulated in a very complicated manner by transcriptional factors, protein kinases, miRNAs, and so on. Among these, miRNAs may play primary roles in regulating the activities of CD44 related to cellular events. However, the detailed mechanisms remain unclear. The elucidation of these mechanisms underlying the regulation of CD44 signaling pathways will be required to develop novel therapeutic strategies.
5. CD44-Promoted Tumor Invasion in GSCs
CD44 is well known to promote tumor migration and invasion in GBM [96]. The mechanisms of glioma invasion are increasingly being understood via in vitro and in vivo models, but the results have yet to be translated into clinical applications. GBM cells, particularly GSCs, were shown to be more migratory and invasive than their non-GSC counterparts [97]. GSCs are highly migratory and invade normal brain parenchyma around the tumor mass in a coordinated manner, allowing for escape from surgical resection and adjuvant therapies and providing the major source of tumor recurrences. The invasive ability of tumor cells is determined by interactions between tumor cells and the ECM, as well as non-tumor cells, such as endothelial cells, astrocytes, and neurons. The ECM plays important roles, not only in providing a scaffold for tumor cells to adhere to and move along but also in intra- and inter-cellularly transducing signals. These interactions in GSCs are generally regulated by hypoxic conditions as special microenvironments called niches. Hypoxia enhances the invasiveness of GBMs with links to the increased production of HA [98]. In addition, higher levels of HA are expressed at the invasive edge of a GBM [99]. We found that severe hypoxia (1% O2) increases the expression of CD44 via the activation of HIF-1α expression, resulting in enhanced GSC migration and invasion. Enhancements in both CD44 expression and GSC invasion were significantly decreased by silencing the HIF-1α gene with its siRNA [100].
Cell migration and invasion are highly integrated, multistep processes that can be accomplished via the interactions of tumor cells and various components in the TME, particularly the ECM. These processes include the adhesion of tumor cells to adjacent cells, such as endothelial cells, neurons, astrocytes, and the ECM; changes in tumor cell morphology via reorganization of the intracellular cytoskeleton so that the tumor cell can more readily pass through the narrow spaces of the ECM; and diffuse migration into the normal brain tissues around the tumor mass with remodeling of the ECM via proteolytic degradation using matrix metalloproteinases, such as MMP-2 and MMP-9, to promote detachment from the adhered ECM and movement to a new site of adhesion [101,102]. The interaction of CD44 and HA activates PI3K, by which phosphatidylinositol diphosphate (PIP2) is converted to PIP3. This PIP3 then activates Rho GTPases, including Rho A, Rac 1, and Cdc42. Rho GTPase signaling plays critical roles in cell motility and the invasion of GBM cells via the polymerization of actin to produce stress fibers, filopodia, and lamellipodia [62].
GSCs expressing high levels of CD44 thus bind to HA, and the amounts of HA are increased via excretion from GBM cells, promoting cell migration and invasion via the signaling pathways of Rho GTPases and PI3K. On the other hand, integrins, which are another type of major cell adhesion receptor comprising α and β transmembrane glycoproteins [103], also stimulate tumor cell migration and invasion in GBMs via similar signaling pathways, including focal adhesion kinase, PIP3K, and AKT kinases [104]. An important question concerns how tumor cells use the two different signaling pathways of HA binding to CD44 or integrins binding to their ligands. HA is a major component of the ECM in the brain parenchyma, while the ligand molecules for integrins, such as collagen, fibronectin, and laminin, are generally lacking in the brain parenchyma. Accordingly, the major signaling pathway to elevate tumor cell motility and promote diffuse invasion into the brain parenchyma around the tumor mass in GBMs is thought to involve the interactions of HA and CD44 binding. In contrast, the interactions of integrins and their ligands are likely to have a critical function in the migration and invasion of tumor cells along the perivascular space and white matter tracts in GBM [105,106]. Binder et al. reported that the migration and invasion of GBM cells are unaffected by the knockdown of integrin α (ITGAV), but are decreased by the knockdown of CD44 using a mouse organotypic slice model [107]. Wolf et al. demonstrated a mechanism by which tumor cells adhere and migrate through the nano-porous, three-dimensional ECM of brain tissue [108]. As the mechanism, GBM cells are likely to adhere to and invade HA-rich ECM using microtentacles that extend tens of micrometers from the cell body via binding to the CD44 receptor. The study also provides insights into how CD44 supports cell motility. CD44 is sufficient to drive the formation of tension-bearing protrusions that can activate cell motility and invasion without the participation of integrins. They also previously reported that CD44-HA-promoted adhesion occurs more rapidly but is weaker than integrin-activated adhesion [109]. These data suggest that CD44-HA binding may be characterized by many rapid, weak interactions with the ECM, whereas integrin binding is characterized by fewer, slower interactions but with an ability to reinforce CD44-HA interactions.
Recently, the Ca2+ signaling pathway and ionotropic glutamate receptors were reported to affect the motility and invasiveness of GBM cells [110]. As described above, glioma cells shape the TME to promote their migration and invasion by modifying and degrading the ECM.
6. CD44-Promoted Tumor Proliferation in GBM (GSCs)
Although CD44 is a multi-functional receptor molecule that promotes invasion, proliferation, drug resistance, angiogenesis, tumorigenesis, and EMT in malignant tumor cells, the signaling pathways for these cellular events are thought to be activated according to the requirements of tumor cells to display each cellular event under pathological conditions. On the other hand, tumor cells were noted to show the dichotomy of either cell proliferation or migration, representing the “Go or Grow” theory [18]. That is, when tumor cells move, tumor cells do not grow, and vice versa [19,111]. How CD44 participates in the cellular processes of this dichotomy in the invasion and proliferation of GBM cells remains unclear. The possible mechanisms by which CD44 primarily promotes tumor cell proliferation with suppression of tumor cell invasion need to be determined. We describe the signaling pathways and their regulation in which CD44 transduces intracellular and extracellular molecules to promote tumor proliferation. Xu et al. demonstrated that CD44 promotes the proliferation of GBM cells, both in vitro and in vivo, by attenuating the activation of the Hippo signaling pathway as a single downstream pathway of CD44 [112]. This result supported a previous report that merlin, functioning upstream of the Hippo signaling pathway, binds to CD44 and inhibits GBM growth by interfering in the interaction of CD44 and HA [113]. Jijiwa et al. reported that the knockdown of the CD44v6 isoform with its siRNA inhibits in vitro growth in brain tumor (GBM) stem cells with high expression of CD44v6 cells, but not in GBM stem cells with low expression of CD44v6. Also, the overexpression of mouse CD44v6 or OPN enhances the growth of mouse stem-like tumor cells, and this growth may be dependent on the CD44/AKT pathway [54]. We also found that OPN upregulated by HIF-2α under 5% O2 (moderate hypoxia) enhances the proliferation of GSCs and that silencing the OPN gene with its siRNA inhibits the cell growth, but elevates the expression of CD44, leading to the promotion of tumor cell migration and invasion [100]. Pietras et al. reported that OPN-CD44 interactions not only enhance cancer stem cell phenotypes but also promote aggressive tumor growth in vivo in GSCs existing in the perivascular niche of proneural GBM [15]. They also demonstrated that these cellular activities are due to the activation of CD44-ICD that has been released into the cytoplasm via the cleavage of CD44 by γ-secretase, which, in turn, is activated by the interaction between OPN and CD44. As described above, CD44-ICD is an intracellular free fragment that translocates to the nucleus and activates the transcription factors of various genes, resulting in enhanced tumor proliferation, glycolytic metabolism, and increased stemness in tumors (particularly as cancer stem cells). Under moderately hypoxic conditions, CD44-ICD binds to HIF-2α and upregulates the expression of HIF target genes. De Falco et al. reported that CD44-ICD enhances the expression of cyclin D1 by promoting CREB recruitment to the cyclin D1 promoter and upregulating the transcription of cyclin D1, leading to the acceleration of cell proliferation in thyroid carcinoma [72,114].
Daniel et al. investigated the intratumoral activities of the MAPK and PI3K pathways in GBM, demonstrating a striking preponderance toward the mutual exclusivity of MAPK and PI3K activations in GBM tissue [115]. That is, MAPK activation correlates with proliferation and transcription factor CREB activation in the region of the tumor proliferation area, whereas PI3K activation correlates with CD44 expression, which strongly promotes tumor invasion in the region of the tumor border zone in GBM. Daniel et al. also showed that MAPK-active regions correlate with the expression of proliferating cell nuclear antigen (PCNA), MMP-9, and pCREB (a pro-oncogenic transcription factor that regulates genes related to proliferation) [114], but with little or no expression of CD44. In contrast, PI3K active regions exhibit high degrees of CD44 expression but little or no expressions of PCNA, MMP-9, or pCREB. These studies identified intratumoral heterogeneity in the activities between invasion and proliferation. The mutually exclusive expression of these activities may relate to a phenotypic transition that may be an essential behavior of tumor cells to develop local recurrence or distant metastasis in cancer cells. Consequently, a better understanding of the role of phenotype in tumor cells showing different activities of CD44 is required to create therapeutic strategies that target CD44 in GBM.
The activity of multiple functions promoted by CD44 is regulated by various factors, as described above. Among these, suppressive miRNAs, such as miRNA-138 (which can not only suppress the proliferation of GBM but also downregulate whole activities of CD44), may be potential targets for the establishment of effective therapies against GBM. While miRNAs may be effective at inhibiting tumor cell proliferation, examination is required to clarify whether such miRNA-regulated inhibition of proliferation affects the invasive activity in GBM cells.
7. Roles of CD44 in Phenotypic Transition in GBM Progression
GBM exhibits a high degree of inter- and intra-tumoral heterogeneity, reducing the anti-tumor effects of radiotherapy and chemotherapy in the treatment of both primary and recurrent GBM. Heterogeneity is confirmed to occur at various levels, including the histopathological, transcriptional, and genomic stages [116,117,118]. The heterogeneity, particularly intra-tumoral heterogeneity, is believed to be largely due to the phenotypic transition of GBM to a mesenchymal (MES) subtype [119]. The Cancer Genomic Atlas (TCGA) identified four molecular subtypes of GBM, which were associated with prognostic values via a large-scale genomic analysis of GBM, including proneural (PN), neural (NL), classical (CL), and MES types [120,121]. In 2017, among those four types, TCGA defined three subtypes, namely, PN, CL, and MES subtypes, in which the intrinsic genetic signature of each subtype correlated with the tumor immune microenvironment [122]. In the study, Wang et al. selected patients with GBM without the IDH mutation and established a relationship between treatment and phenotypic transition in recurrent tumors of GBM. By comparing 91 recurrent tumors, they found that the PN subtype showed the highest transition rate in which 59% of patients with the PN subtype shifted to MES or CL types, while 33% of patients with the MES subtype shifted to other subtypes [122]. These subtypes show different prognoses, with the worst in the MES subtype [122]. Over the course of tumor progression, GBM often shifts in subtype from the original to other types, and the most prominent shift was found in the PN-to-MES transition (PMT) [123]. The PMT is synonymous with the EMT in cancer cells of epithelial origin [124]. These MES-type tumors show increased motility, invasiveness, and metastatic potential, in addition to greater resistance to radio- and chemotherapies [125]. However, to develop a tumor mass at the metastatic site, the tumor is required to reproduce the original epithelial features via a reverse transition from an MES to an epithelial phenotype (MET) because the epithelial phenotype has features better suited to aggressively proliferating tumor cells compared with the MES phenotype [126]. Also, in the process of tumorigenesis on recurrence in GBM, highly migratory and invasive GSCs, both primarily and secondarily after radio-chemotherapy, are expected to show a change in phenotype to a less invasive, more proliferative phenotype (epithelial-like type) to reproduce the original tumor mass. Accordingly, an understanding of the mechanisms underlying the EMT in GBM cells may give clues to the elucidation of the molecular mechanisms underlying tumor generation on GBM recurrence.
Extensive studies revealed that high CD44 expression correlates with a mesenchymal phenotype of cancer stem cells and participates in the EMT, resulting in tumors with worse prognosis showing highly invasive, metastatic, and chemo-radiation-resistant features [125,127]. The EMT represents the adaptive plasticity of cancer cells [128], a cell behavior by which tumor cells change phenotype in response to a microenvironment where favorable conditions are provided to tumor cells to enhance invasiveness, proliferation, and survival from anti-tumor therapy. Accumulating evidence demonstrates that the EMT is closely related to the aggressive invasiveness and metastasis of epithelial cancer cells [129,130]. In epithelial cancer cells, the EMT process changes the tumor phenotype to a mesenchymal type. Expressions of adhesion molecules that are characteristic of epithelial-type tumors (such as E-cadherin, occludin, and a-catenin) are thus decreased, while expressions of molecules specific to mesenchymal-type tumors (including vimentin, fibronectin, and N-cadherin) are upregulated [131], thus promoting the motility and invasiveness of tumor cells and allowing for the development of tumor cells that are more resistant to apoptosis. As a result, the morphology of tumor cells is changed to that of invasive cells with reduced polarity and a more motile cyto-architecture [132]. The EMT process is activated via a network of EMT-activating transcription factors (EMT-TFs), including TWIST1 [128,133], SNAI1 [134], ZEB1 [135], and SLUG [136]. These transcription factors are regulated by microenvironmental conditions, including the ECM; immune cells, such as macrophages and hypoxia; and miRNAs, including miRNA-200 family members, to acquire the mesenchymal phenotype. Activation of these EMT-TFs is known to correlate positively with CD44 expression. In MCF-7 breast cancer cells, Twist markedly elevates the expression levels of CD44 via activation of the β-catenin and Akt pathways [131]. The overexpression of Slug in MCF-10A and MCF-7 breast cancer cells (CD44-/CD24+) generates a subpopulation of cancer stem cells with a CD44+/CD24+ phenotype showing an enhanced mammosphere-forming ability [136]. On the other hand, CD44 in SW480 colon cancer cells can promote EMT in various cancers by upregulating mesenchymal genes and downregulating epithelial phenotype-related genes [137,138,139]. The overexpression of CD44 downregulates E-cadherin expression; upregulates N-cadherin, α-actin, vimentin, and fibronectin; and disassociates membrane-associated E-cadherin-β-catenin complexes, promoting the translocation of β-catenin into the nucleus and transcriptional activation of genes related to cell invasion and migration [139].
The EMT is also affected by cytokines and hypoxia of the microenvironment surrounding tumor cells. Accumulating evidence suggests that TGF-β is an important cytokine for inducing the EMT and increasing the number of cancer stem cells [140]. The treatment of GBM cells with TGF-β upregulates the expressions of fibronectin and COL5A1 mesenchymal markers, resulting in aggressive tumors showing increased sphere formation and a spindle-shaped morphology, and the effect is reversed with the inhibition of TGF-β [141]. TGF-β receptor type I (RI) is known to contain a CD44-binding site. HA binding to CD44 induces a complex between CD44 and TGF-βRI and stimulates TGF-βRI serine/threonine kinase activity, leading to increased phosphorylation of Smad2/Smad3 and activation of ZEB1, thus causing increased invasiveness [142]. In addition, activated TGF-βRI kinase phosphorylates CD44, resulting in enhanced interactions of CD44 with the cytoskeletal protein ankyrin, in turn promoting HA-CD44 signaling. Luo et al. reported that TGF-β can induce MES transformation via the PDK1/c-Jun pathway by activating N-cadherin, ZEB1, SNAIL, and TWIST1 [143]. TNF-α, which is a common cytokine in the TME, induces mesenchymal transition through the activation of nuclear factor-kappa B (NF-κB) in various types of cancer, including GBM. The treatment of PN-type GSCs with TNF-α causes an MES phenotypic shift with a high expression of CD44 [144]. Iwata et al. reported that the treatment of PN GSCs with TNF-α dramatically increases NF-κB activity, whereas the silencing of NF-κB decreases inducible T-cell costimulator ligand (ICOSLG) expression, which is typically associated with the MES phenotype after TNF-α treatment [145].
Isoforms of CD44 were reported to play a crucial role in causing the EMT in cancer cells [146]. Generally, a switch in CD44 isoform expression from CD44v to CD44s is required to execute the EMT [147]. CD44 alternative splicing is differentially regulated during the EMT [147] so that CD44v can change to CD44s in human immortalized epithelial cells. The CD44s isoform can promote the EMT and its knockdown inhibits the EMT. The re-expression of CD44s fully recovers the phenotype that cannot be obtained from the EMT due to inhibition via CD44 knockdown. Although CD44v is reportedly predominantly expressed in cancer stem-like cells, not only expression patterns and frequency but also the functional significance of CD44s and CD44v are not fully understood. Jijiwa et al. reported that CD44v6 is expressed on GBM stem-like cells and enhances in vitro and in vivo tumor growth, with a link to the upregulated expression of OPN (a ligand of CD44v6) [54]. This result suggests that CD44v6 may promote the generation of GSCs with the epithelial phenotype. Many studies reported CD44v6 as an isoform that promotes the EMT, where its expression is largely restricted to the advanced stages of tumor progression and is more prevalent in metastatic tumors than in non-metastatic ones [51].
How the phenotypic transition of GBM or GBM stem cells plays a role in GBM recurrence has yet to be clarified. A better understanding of the regulation of the functional activities of CD44 in invasion, proliferation, and phenotypic transitions between the invasive type and proliferative type may present a clue to elucidating the molecular mechanisms underlining recurrence in GBM and identifying a therapeutic target for effective treatment against GBM.
8. Molecular Mechanism Underlying GBM Recurrence: From the Perspective of the Dual Activity of CD44: Invasion and Proliferation
CD44 participates in multiple cellular processes in cancers, including GBM, as not only an adhesion molecule but also a signal-transducing molecule. In the various cellular processes, invasion and proliferation are two major events in the tumor progression and recurrence of GBM. These two events cannot be executed simultaneously in the same cells, as mentioned above. When tumor recurrence occurs locally at the margin of the resection cavity in GBM, migrated GSCs too far from the original tumor site appear to move to the tumor resection site as GSCs home in on their tumor niche [148,149,150]. Then, to allow for highly migratory GSCs to colonize marginal sites around the resection cavity, a shift in phenotype from highly migratory to less migratory, more proliferative types is necessary. These behaviors of tumor cells are likely to represent the two-way shifts of the PMT and its reversion in the mesenchymal-to-proneural transition (MET). In CD44-regulated signaling pathways, the MAPK and PI3K downstream pathways show mutually exclusive expressions; that is, the former activates the proliferation of tumor cells (corresponding to the epithelial type) and the latter promotes invasion by tumor cells (corresponding to the mesenchymal type), as mentioned earlier [115]. How these signaling pathways are regulated to achieve specific behaviors in tumor cells remains elusive, but a phenotypic transition is likely to play a critical role in displaying the features of each phenotype. A recently proposed definition is that the EMT is a process showing a shift toward the mesenchymal state and includes multiple and dynamic transitional states between the epithelial and mesenchymal phenotypes, as opposed to a process involving a single binary switch [151]. Studies in mouse tumor models and human circulating tumor cells revealed that the EMT is not a binary process during cancer metastasis. In human breast cancer patients, circulating tumor cells show diverse EMT statuses with many varieties of expressions of epithelial and mesenchymal markers [152]. These not-fully EMT cells represent intermediate EMT cells, called partial EMT cells, in which the tumor cells exhibit both mesenchymal and epithelial properties [152]. As described in the section on phenotypic transition, a reversion of the EMT is thought to be essential for the regrowth of invading tumor cells at distant sites. Such EMT/MET switches require the status of partial EMT cells. A full EMT is difficult to reverse in terms of phenotypic features at distant sites in various organs [153]. Further, CD44 isoform switching from the expression of CD44v to CD44s is essential for the EMT [147]. CD44 alternative splicing was differentially regulated during the EMT, resulting in a switch in expression from CD44v to CD44s among human immortalized epithelial mammary cells, as well as breast cancer progression [147]. Zhao et al. reported that the CD44s isoform may play a role in the EMT and CD44v may play a role in tumor engraftment and growth [146].
Various factors including cytokines, non-tumor cells, and oxygen concentration in the TME can affect the activity of CD44 signaling pathways [154,155]. Among them, hypoxia is a potential factor for executing the phenotypic transition, as the core-to-edge cellular transition of progressive tumor cells is achieved with hypoxia [156]. Consequently, an understanding of the molecular mechanisms underlying GBM recurrence requires clarification of the expression and pathogenesis of various genes and molecules that could be altered under differing levels of hypoxia before and after tumor resection (Figure 6).

Figure 6.
Illustration of hypothetical differential expression of genes related to cell invasion/migration and cell proliferation in glioblastoma according to hypoxic conditions before tumor resection and 1–3 months (in the period of tissue repair) after tumor resection. HIF-1α is upregulated by severe hypoxia of 0.5–2.5% O2, corresponding to the tumor rim (tumor border area) before resection. In this area, HIF-1α target genes (including CD44, TGF-β, TWIST, SNAIL, cMET, and NF-kB) are activated, resulting in promotion of cell migration and invasion and enhancement of stemness. HIF-2α is upregulated by moderate hypoxia of 2.5–5% O2, corresponding to the marginal area of the resection cavity once tissue injury is repaired and presenting with moderate hypoxia. These cellular processes represent the phenotypic transition of GBM.
Like other solid tumors, the center of the tumor tissue in GBM shows hypoxic features with an oxygen tension of 0.75–4 mmHg (0.1–0.5% O2) [157]. In the invasive area at the tumor periphery, oxygen tension may be a little higher than in tumor tissues while remaining hypoxic (0.5–2.5% O2) compared with the normal brain (2.5–12.5% O2) [157,158]. Under such a hypoxic microenvironment, the expression of various molecules in GSCs may be modified by hypoxia-responding signaling mediated through transcription factors called hypoxia-inducible factors (HIFs) [159]. Among the HIFs, the expression of HIF-1α is upregulated under severe hypoxia (≤1% O2), whereas the expression of HIF-2α is upregulated under moderate hypoxia (approximately 5% O2) and HIF-2α is expressed predominantly in GSCs [160]. In highly invasive GBM, GSCs with high expression levels of CD44 reside in the peritumoral invasion area when oxygen concentrations show severe-to-moderate hypoxia of 0.5–2.5% O2 [100,157]. Such a low oxygen concentration activates HIF-1α, and then the activated HIF-1α upregulates various genes and molecules, including CD44, TGF-β, TWIST, SNAIL, cMET, and NF-κB [161]. Signaling pathways from almost all these genes and molecules are related to the promotion of migration/invasion and the EMT of tumor cells, as shown in the section on signaling pathways. Upregulated CD44 under such severe hypoxia may primarily activate tumor cell migration and invasion rather than tumor cell proliferation [100]. In contrast, the area surrounding the tumor resection cavity in which brain tissues are injured by surgical resection of the tumor shows a severely hypoxic condition in the period immediately after tumor resection. The area including the cavity wall is repaired at least a month after tumor resection, resulting in conditions of moderate hypoxia (2.5–5% O2) [162]. In this area, HIF-2α can be predominantly activated, and the activated HIF-2α upregulates different genes and molecules from those activated by HIF-1α. These include VEGF/VEGFR2, cMYC, TGFα, OPN, and Oct4 [70]. Most of these genes and molecules are related to the promotion of proliferation in tumor cells. Under such moderate hypoxia, HIF-2α binds to CD44-ICD and activates various transcription factors, as mentioned above. Kolliopoulos et al. demonstrated that the knockout of CD44 in U251MG cells suppresses cell proliferation, diminishes phosphorylation of CREB, increases the levels of the cell cycle inhibitor p16, and decreases stemness. Similar results were obtained when cells were treated with the γ-secretase inhibitor N-[N-(3,5 difluorophenacetyl-L-alanyl)-(S)-phenylglycine t-butyl ester (DAPT), which inhibits CD44 cleavage, and thus, inhibits the release of ICD from CD44, resulting in the inhibited binding of activated HIF-2α to the released CD44-ICD. This result indicates that transcriptional factors for activating proliferation and stemness in GSCs are dependent on free CD44-ICD [163]. As described in the “CD44-promoted tumor proliferation in GBM (GSCs)” section, OPN can activate γ-secretase by binding to CD44 [15]. We demonstrated that silencing the OPN gene with siRNA increases CD44 expression in GSCs and enhances the CD44-promoted migration and invasion of GSCs [100]. OPN may be a key molecule to promote phenotypic transition in addition to the upregulation of certain transcription factors. The transcription factors NF-κB and c-MYC are reciprocally activated and show the dichotomy between invasion and proliferation according to differences in hypoxic conditions. Under severe hypoxia (<1% O2), NF-kB is activated by HIF-1α to enhance the invasive nature of tumor cells at the rim of GBM, whereas c-MYC is upregulated by HIF-2α under moderate hypoxia (2.5–5% O2) to promote the proliferation of tumor cells in GBM [164]. In addition, cMET and VEGFR2 are well known to show mutually exclusive expression, as described above. The former is activated by HIF-1α under severe hypoxia and the latter is activated by HIF-2α under moderate hypoxia. These differential expressions of cMET and VEGFR2 can regulate tumor cell growth and invasion according to the binding of ligands HCG and VEGF to the corresponding receptor [67]. These studies indicate that the level of hypoxia may play a critical role in determining the direction of the phenotypic shift induced by CD44. Musah-Eroje, A. et al. described how oxygen gradient regulates phenotypic and genetic changes in GBM and expression of the genes is reversible, showing the plasticity of GBM cells [165]. Understanding the degree of oxygen gradient in GBM will be crucial in clarifying the molecular mechanism underlying GBM recurrence and identifying a target molecule in the treatment for GBM patients.
Hypoxia thus has a strong potential for differentially activating the expression of various genes and molecules for achieving intended cellular processes in tumor cells according to the level of hypoxia. In the process of GBM recurrence, CD44 is expected to play a crucial role in shifting from the mesenchymal type to the epithelial type and vice versa, resulting in the local recurrence of GBM. To confirm whether these mechanisms are operating in the tumor recurrence of GBM, further studies are required. These include clarification of the expressions and functions of related genes and molecules for invasion and proliferation in different levels of hypoxia in much greater detail and whether the same molecular and cellular events can be reproduced using in vivo brain tumor models.
9. Conclusions
CD44 is a multi-functional molecule that participates in not only cell adhesion but also signal transduction, and plays a critical role in executing cellular events, such as invasion, migration, proliferation, and tumorigenesis in GBM. CD44 shows functional activity in transducing intra- and intercellular signals in GBM cells. These signaling pathways require CD44 activation to precede signal transduction. The binding of CD44 to HA activates various signaling pathways. Among the pathways used to activate the invasion and proliferation of tumor cells, some pathways show mutually exclusive expression under hypoxic conditions and in different locations in GBM tissues. The PI3K/AKT pathway, which promotes cell invasion and migration, is activated at the tumor rim, whereas the RAS-MARK pathway, which primarily enhances cell proliferation, is activated in the tumor core in the same tumor. NF-κB, which promotes the migration and invasion of tumor cells, is activated by HIF-1α under severe hypoxia. In contrast, cMYC, which enhances cell proliferation, is activated by HIF-2α under moderate hypoxia.
On the other hand, proteolytic cleavage of CD44 can provide specific signaling pathways. The proteolytic action of γ-secretase promotes the cleavage of the transmembrane domain of CD44 and can release CD44-ICD, which translocates to the nucleus and activates various genes as co-transcription factors. Under hypoxic conditions, CD44-ICD binds to HIF-2α and activates HIF-2α expression, which further activates HIF-2α target genes, including cyclin D1, Nanog, Oct4, Sox2, and OPN, resulting in the promotion of tumor proliferation and stemness of GSCs. These results indicate that the dichotomy theory of “Go or Grow” in tumor cells can be executed. At tumor recurrence, GSCs that survive any kind of therapy are required to undergo a phenotypic transition from an invasive type to a proliferative type. Differences in oxygen concentration can allow GSCs to regulate phenotypes to obtain the intended behaviors. These results suggest that CD44 presenting with dual functional activities of invasion and proliferation can play a central role in the generation of GBM recurrence. To elucidate the mechanism underlying the phenotypic transition of CD44 from an invasive to a proliferative type may present a clue to the development of a novel therapeutic method to prevent recurrence in GBM.
Author Contributions
Conceptualization, A.I. and T.O.; methodology, A.I., T.O. and M.N.; software, A.I. and T.O.; validation, A.I., T.O. and M.N.; formal analysis, A.I., T.O. and M.N.; investigation, A.I., T.O., M.N., Y.O., K.K. and H.Y.; resources, A.I.; data curation, A.I. and T.O.; writing—original draft preparation, A.I.; writing—review and editing, T.O.; visualization, A.I.; supervision, J.T. and T.K.; project administration, A.I. and T.O.; funding acquisition, A.I. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Number JP 22K09208.
Data Availability Statement
All data used for analysis are presented in the tables in this article.
Acknowledgments
The authors would like to express their gratitude to Taichi Furumochi, Yasuhiro Shiraishi, Yoshihiro Takimoto, and Mizusa Harada of the Department of Neurology, Ehime University Hospital, Japan, and Riko Kitazawa, Mashio Taniwaki, Eiji Katayama, Takuya Kondo and Satsuki Myoga of the Department of Pathology, Ehime University Hospital, Japan, for their help in obtaining pathological and radiological findings.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper. None of the authors have any financial interest related to this work.
References
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, Y.; Yu, T.S.; Mckay, R.M.; Burns, D.K.; Kernie, S.G.; Parada, L.F. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 2012, 488, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Lathia, J.D.; Gallagher, J.; Myers, J.T.; Li, M.; Vasanji, A.; McLendon, R.E.; Hjelmeland, A.B.; Huang, A.Y.; Rich, J.N. Direct in vivo evidence for tumor propagation by glioblastoma cancer stem cells. PLoS ONE 2011, 6, e24807. [Google Scholar] [CrossRef] [PubMed]
- Schonberg, D.L.; Lubelski, D.; Miller, T.E.; Rich, J.N. Brain tumor stem cells: Molecular characteristics and their impact on therapy. Mol. Aspects Med. 2014, 39, 82–101. [Google Scholar] [CrossRef] [PubMed]
- Galli, R.; Binda, E.; Orfanelli, U.; Cipelletti, B.; Gritti, A.; De Vitis, S.; Fiocco, R.; Foroni, C.; Dimeco, F.; Vescovi, A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004, 64, 7011–7021. [Google Scholar] [CrossRef] [PubMed]
- Beier, D.; Scriefer, B.; Brawanski, K.; Hau, P.; Weis, J.; Schulz, J.B.; Beier, C.P. Efficacy of clinically relevant temozolomide dosing schemes in glioblastoma cancer stem cell lines. J. Neuro Oncol. 2012, 109, 45–52. [Google Scholar] [CrossRef]
- Codrici, E.; Enciu, A.M.; Popescu, I.D.; Mihai, S.; Tanase, C. Glioma stem cells and their microenvironments: Providers of challenging therapeutic targets. Stem Cells Int. 2016, 2016, 5728438. [Google Scholar] [CrossRef]
- Schiffer, D.; Annovazzi, L.; Casalone, C.; Corona, C.; Mellai, M. Glioblastoma: Microenvironment and niche concept. Cancers 2019, 11, 5. [Google Scholar] [CrossRef]
- Chaudhry, G.E.; Akim, A.; Naveed Zafar, M.; Safdar, N.; Sung, Y.Y.; Muhammad, T.S.T. Understanding hyaluronan receptor (CD44) interaction, HA-CD44 activated potential targets in cancer therapeutics. Adv. Pharm. Bull. 2021, 11, 426–438. [Google Scholar] [CrossRef]
- Ponta, H.; Sherman, L.; Herrich, P.A. CD44 from adhesion molecules to signaling regulators. Nat. Rev. Mol. Cell Biol. 2003, 4, 33–45. [Google Scholar] [CrossRef]
- Naor, D.; Nedvetzkis, S.; Golan, I.; Melnik, L.; Faitelson, Y. CD44 in cancer. Crit. Rev. Clin. Lab. Sci. 2002, 39, 527–579. [Google Scholar] [CrossRef] [PubMed]
- Hirata, K.; Suzuki, H.; Imaeda, H.; Matsuzaki, J.; Tsugawa, H.; Nagano, O.; Asakura, K.; Saya, H.; Hibi, T. CD44 variant 9 expression in primary early gastric cancer as a predictive marker for recurrence. Br. J. Cancer 2013, 109, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Yae, T.; Tsuchihashi, K.; Ishimoto, T.; Motohara, T.; Yoshikawa, M.; Yoshida, G.J.; Wada, T.; Masuko, T.; Mogushi, K.; Tanaka, H.; et al. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat. Commun. 2012, 3, 883. [Google Scholar] [CrossRef]
- Ozawa, M.; Ichikawa, Y.; Zheng, Y.W.; Oshima, T.; Miyata, H.; Nakazawa, K.; Guan, H.B.; Shiozawa, M.; Akaike, M.; Watanabe, K.; et al. Prognostic significance of CD44 variant 2 upregulation in colorectal cancer. Br. J. Cancer 2014, 111, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Pietras, A.; Katz, A.M.; Ekström, E.J.; Wee, B.; Halliday, J.J.; Pitter, K.L.; Werbeck, J.L.; Amankulor, N.M.; Huse, J.T.; Holland, E.C. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell 2014, 14, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Sahgal, A.; Sanghera, P.; Tsao, M.N.; Davey, P.; Lam, K.; Symons, S.; Aviv, R.; Perry, J.R. Glioblastoma: Patterns of recurrence and efficacy of salvage treatments. Can. J. Neurol. Sci. 2011, 38, 621–625. [Google Scholar] [CrossRef]
- Giese, A.; Kucinski, T.; Knopp, U.; Goldbrunner, R.; Hamel, W.; Mehdorn, H.M.; Tonn, J.C.; Hilt, D.; Westphal, M. Patterns of recurrence following local chemotherapy with biodegradable carmustine (BCNU) implants in patients with glioblastoma. J. Neuro Oncol. 2004, 66, 351–360. [Google Scholar] [CrossRef]
- Hatzikirou, H.; Basanta, D.; Simon, M.; Schaller, K.; Deutsch, A. ‘Go or grow’: The key to the emergence of invasion in tumour progression? Math. Med. Biol. 2012, 29, 49–65. [Google Scholar] [CrossRef]
- Giese, A.; Loo, M.A.; Tran, N.; Haskett, D.; Coons, S.W.; Berens, M.E. Dichotomy of astrocytoma migration and proliferation. Int. J. Cancer 1996, 67, 275–282. [Google Scholar] [CrossRef]
- Herrich, P.; Morrison, H.; Sleeman, J.; Orian-Rousseau, V.; Konig, H.; Weg-Remers, S.; Ponta, H. CD44 acts both as a growth- and invasiveness-promoting molecule and as a tumor-suppressing cofactor. Ann. N. Y. Acad. Sci. 2000, 910, 106–118. [Google Scholar] [CrossRef]
- Lesley, J.; Hyman, R. CD44 structure and function. Front. Biosci. 1998, 3, d616–d630. [Google Scholar] [CrossRef] [PubMed]
- Prochazka, L.; Tesarik, R.; Turanek, J. Regulation of alternative splicing of CD44 in cancer. Cell Signal. 2014, 26, 2234–2239. [Google Scholar] [CrossRef]
- Iczkowski, K.A. Cell adhesion molecule CD44: Its functional roles in prostate cancer. Am. J. Transl. Res. 2010, 3, 1–7. [Google Scholar] [PubMed]
- Bennett, K.L.; Jackson, D.G.; Simon, J.C.; Tanczos, E.; Peach, R.; Modrell, B.; Stamenkovic, I.; Plowman, G.; Aruffo, A. CD44 isoforms containing exon V3 are responsible for the presentation of heparin-binding growth factor. J. Cell Biol. 1995, 128, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Tremmel, M.; Matzke, A.; Albrecht, I.; Laib, A.M.; Olaku, V.; Ballmer-Hofer, K.; Christofori, G.; Héroult, M.; Augustin, H.G.; Ponta, H.; et al. A CD44v6 peptide reveals a role of CD44 in VEGFR-2 signaling and angiogenesis. Blood 2009, 114, 5236–5244. [Google Scholar] [CrossRef]
- Medina, M.; Dotti, C.G. RIPped out by presenilin-dependent γ-secretase. Cell Signal. 2003, 15, 829–841. [Google Scholar] [CrossRef]
- Murakami, D.; Okamoto, I.; Nagano, O.; Kawano, Y.; Tomita, T.; Iwatsubo, T.; De Strooper, B.; Yumoto, E.; Saya, H. Presenilin-dependent gamma-secretase activity mediates the intramembranous cleavage of CD44. Oncogene 2003, 22, 1511–1516. [Google Scholar] [CrossRef]
- Nagano, O.; Saya, H. Mechanism and biological significance of CD44 cleavage. Cancer Sci. 2004, 95, 830–835. [Google Scholar] [CrossRef]
- Nagano, O.; Murakami, D.; Hartmann, D.; De Strooper, B.; Saftig, P.; Iwatsubo, T.; Nakajima, M.; Shinohara, M.; Saya, H. Cell-matrix interaction via CD44 is independently regulated by different metalloproteinases activated in response to extracellular Ca2+ influx and PKC activation. J. Cell Biol. 2004, 165, 893–902. [Google Scholar] [CrossRef]
- Senbanjo, L.T.; Aljohani, H.; Majumdar, S.; Chellaiah, M.A. Characterization of CD44 intracellular domain interaction with RUNX2 in PC3 human prostate cancer cells. Cell. Commun. Signal. 2019, 17, 80. [Google Scholar] [CrossRef]
- Miletti-Gonzaalez, K.E.; Murphy, K.; Kumaran, M.N.; Ravindranath, A.K.; Wernyj, R.P.; Kaur, S.; Miles, G.D.; Lim, E.; Chan, R.; Chekmareva, M.; et al. Identification of function for CD44 intracytoplasmic domain (CD44-ICD): Modulation of matrix metalloproteinase 9 (MMP-9) transcription via novel promoter response element. J. Biol. Chem. 2012, 287, 18995–19007. [Google Scholar] [CrossRef] [PubMed]
- Yohansson, E.; Grassi, E.S.; Pantazopoulou, V.; Tong, B.; Lindgren, D.; Berg, T.J.; Pietras, E.J.; Axelson, H.; Pietras, A. CD44 Interacts with HIF-2α to modulate the hypoxic phenotype of perinecrotic and perivascular glioma cells. Cell Rep. 2017, 20, 1641–1653. [Google Scholar] [CrossRef] [PubMed]
- Ogino, S.; Nishida, N.; Umemoto, R.; Suzuki, M.; Takeda, M.; Terasawa, H.; Kitayama, J.; Matsumoto, M.; Hayasaka, H.; Miyasaka, M.; et al. Two-state conformations in the hyaluronan-binding domain regulate CD44 adhesiveness under flow condition. Structure 2010, 18, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; He, Y.; Zang, H.; Liu, Y.; Wang, W.; Du, Y.; Gao, F. Selective killing of breast cancer cells expressing activated CD44 using CD44 ligand-coated nanoparticles in vitro and in vivo. Oncotarget 2015, 6, 15283–15296. [Google Scholar] [CrossRef]
- Lesley, J.; Hyman, R. CD44 can be activated to function as an hyaluronic acid receptor in normal murine T cells. Eur. J. Immunol. 1992, 22, 2719–2723. [Google Scholar] [CrossRef] [PubMed]
- Moll, J.; Khaldoyanidi, S.; Sleeman, J.P.; Achtnich, M.; Preus, I.; Ponta, H.; Herrlich, P. Two different functions for CD44 proteins in human myelopoiesis. J. Clin. Investig. 1998, 102, 1024–1034. [Google Scholar] [CrossRef]
- Guo, Q.; Yang, C.; Gao, F. The state of CD44 activation in cancer progression and therapeutic targeting. FEBS J. 2022, 289, 7970–7986. [Google Scholar] [CrossRef]
- Spadea, A.; Rios de la Rosa, J.M.; Tirella, A.; Ashford, M.B.; Williams, K.J.; Stratford, I.J.; Tirelli, N.; Mehibel, M. Evaluating the efficiency of hyaluronic acid for tumor targeting via CD44. Mol. Pharm. 2019, 16, 2481–2493. [Google Scholar] [CrossRef]
- Guadagno, E.; Borrelli, G.; Califano, M.; Cali, G.; Solari, D.; Del Basso De Caro, M. Immunohistochemical expression of stem cell markers CD44 and nestin in glioblastoma: Evaluation of their prognostic significance. Pathol. Res. Pract. 2016, 212, 825–832. [Google Scholar] [CrossRef]
- Si, D.; Yin, F.; Peng, J.; Zhang, G. High expression of CD44 predicts a poor prognosis in glioblastomas. Cancer Manag. Res. 2020, 12, 769–775. [Google Scholar] [CrossRef]
- Nishikawa, M.; Inoue, A.; Ohnishi, T.; Kohno, S.; Ohue, S.; Matsumoto, S.; Suehiro, S.; Yamashita, D.; Ozaki, S.; Watanabe, H.; et al. Significance of glioma stem-like cells in the tumor periphery that express high levels of CD44 in tumor invasion, early progression, and poor prognosis in glioblastoma. Stem Cells Int. 2018, 23, 5387041. [Google Scholar] [CrossRef]
- Sawant, S.; Ahire, C.; Dongre, H.; Joshi, S.; Jamghare, S.; Rane, P.; Kane, S.; Chaukar, D. Prognostic significance of elevated serum CD44 levels in patients with oral squamous cell carcinoma. J. Oral Pathol. Med. 2018, 47, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Karan Križanac, D.; Krasić Arapović, A.; Skočibušić, S.; Pintarić, I.; Trgo, G.; Tomić, S. CD44 Immunoexpression is Unfavorable Predictor in Ovarian Serous Cancer. Appl. Immunohistochem. Mol. Morphol. 2018, 26, 398–402. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.H.; Wang, Y.C.; Chou, Y.C.; Yu, M.H.; Chao, T.K. The combination of aldehyde dehydrogenase 1 (ALDH1) and CD44 is associated with poor outcomes in endometrial cancer. PLoS ONE 2018, 13, e0206685. [Google Scholar] [CrossRef]
- Xie, X.; Huang, X.; Tang, H.; Ye, F.; Yang, L.; Guo, X.; Tian, Z.; Xie, X.; Peng, C.; Xie, X. Diallyl Disulfide inhibits breast cancer stem cell progression and glucose metabolism by targeting CD44/PKM2/AMPK signaling. Curr. Cancer Drug Targets 2018, 18, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, T.; Lu, D.; Zhen, J.; Zhang, L. CD44 overexpression related to lymph node metastasis and poor prognosis of pancreatic cancer. Int. J. Biol. Markers 2018, 33, 308–313. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Q.; Wang, Q.; Wang, Y.; Chen, J. Prognostic significance of CD24 and CD44 in breast cancer: A meta-analysis. Int. J. Biol. Markers 2017, 32, e75–e82. [Google Scholar] [CrossRef]
- Tsidulko, A.Y.; Kazanskaya, G.M.; Kostromskaya, D.V.; Aidagulova, S.V.; Kiselev, R.S.; Volkov, A.M.; Kobozev, V.V.; Gaitan, A.S.; Krivoshapkin, A.L.; Grigorieva, E.V. Prognostic relevance of NG2/CSPG4, CD44 and Ki-67 in patients with glioblastoma. Tumour Biol. 2017, 39, 1010428317724282. [Google Scholar] [CrossRef]
- Taniguchi, D.; Saeki, H.; Nakashima, Y.; Kudou, K.; Nakanishi, R.; Kubo, N.; Ando, K.; Oki, E.; Oda, Y.; Maehara, Y. CD44v9 is associated with epithelial-mesenchymal transition and poor outcomes in esophageal squamous cell carcinoma. Cancer Med. 2018, 7, 6258–6268. [Google Scholar] [CrossRef]
- Yamakawa, Y.; Kusuhara, M.; Terashima, M.; Kinugasa, Y.; Sugino, T.; Abe, M.; Mochizuki, T.; Hatakeyama, K.; Kami, K.; Yamaguchi, K. CD44 variant 9 expression as a predictor for gastric cancer recurrence: Immunohistochemical and metabolomic analysis of surgically resected tissues. Biomed Res. 2017, 38, 41–52. [Google Scholar] [CrossRef]
- Todaro, M.; Gaggianesi, M.; Catalano, V.; Benfante, A.; Iovino, F.; Biffoni, M.; Apuzzo, T.; Sperduti, I.; Volpe, S.; Cocorullo, G.; et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell 2014, 14, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Cozzi, P.J.; Hao, J.L.; Beretov, J.; Chang, L.; Duan, W.; Shigdar, S.; Delprado, W.J.; Graham, P.H.; Bucci, J.; et al. CD44 variant 6 associated with prostate cancer metastasis and chemo-/radioresistance. Prostate 2014, 74, 602–617. [Google Scholar] [CrossRef] [PubMed]
- Gotoda, T.; Matsumura, Y.; Kondo, H.; Saitoh, D.; Shimada, Y.; Kosuge, T.; Kanai, Y.; Kakizoe, T. Expression of CD44 variants and its association with survival in pancreatic cancer. Jpn. J. Cancer Res. 1998, 89, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Jijiwa, M.; Demir, H.; Gupta, S.; Leung, C.; Joshi, K.; Orozco, N.; Huang, T.; Yildiz, V.O.; Shibahara, I.; de Jesus, J.A.; et al. CD44v6 regulates growth of brain tumor stem cells partially through the AKT-mediated pathway. PLoS ONE 2011, 6, e24217. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Song, X.; Liu, J.; Li, S.; Gao, W.; Qiu, M.; Yang, C.; Ma, Y.; Chen, Y. Expression of CD44 and the survival in glioma: A meta-analysis. Biosci. Rep. 2020, 40, BSR20200520. [Google Scholar] [CrossRef]
- Inoue, A.; Ohnishi, T.; Nishikawa, M.; Watanabe, H.; Kusakabe, K.; Taniwaki, M.; Yano, H.; Ohtsuka, Y.; Matsumoto, S.; Suehiro, S.; et al. Identification of CD44 as a reliable biomarker for glioblastoma invasion: Based on magnetic resonance imaging and spectroscopic analysis of 5-aminolevulinic acid fluorescence. Biomedicines 2023, 11, 2369. [Google Scholar] [CrossRef]
- Jalkanen, M.; Elenius, K.; Salmivirta, M. Syndecan—A cell surface proteoglycan that selectively binds extracellular effector molecules. Adv. Exp. Med. Biol. 1992, 313, 79–85. [Google Scholar]
- Weber, G.F.; Ashkar, S.; Glimcher, M.J.; Cantor, H. Receptor-ligand interaction between CD44 and osteopontin (Eta-1). Science 1996, 271, 509–512. [Google Scholar] [CrossRef]
- Knutson, J.R.; Iida, J.; Fields, G.B.; McCarthy, J.B. CD44/chondroitin sulfate proteoglycan and alpha 2 beta 1 integrin mediate human melanoma cell migration on type IV collagen and invasion of basement membranes. Mol. Biol. Cell 1996, 7, 383–396. [Google Scholar] [CrossRef]
- Toyama-Sorimachi, N.; Sorimachi, H.; Tobita, Y.; Kitamura, F.; Yagita, H.; Suzuki, K.; Miyasaka, M. A novel ligand for CD44 is serglycin, a hematopoietic cell adherence and activation. J. Biol. Chem. 1995, 270, 7437–7444. [Google Scholar] [CrossRef]
- Toole, B.P.; Slomiany, M.G. Hyaluronan, CD44 and Emmprin: Patners in cancer cell chemoresistance. Drug Resist. Updates 2008, 11, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, L.Y. Hyaluronan-mediated CD44 activation of RhoGTPase signaling and cytoskeleton function promotes tumor progression. Semin. Cancer Biol. 2008, 18, 251–259. [Google Scholar] [CrossRef]
- Bourguignon, L.Y. Hyaluronan-mediated CD44 interaction with receptor and non-receptor kinases promotes oncogenic signaling, cytoskeleton activation and tumor progression. In Hyaluronan in Cancer Biology; Stern, R., Ed.; Academic Press: San Diego, CA, USA, 2009; pp. 89–107. [Google Scholar]
- Turley, E.A.; Noble, P.W.; Bourguignon, L.Y. Signaling properties of hyaluronan receptors. J. Biol. Chem. 2002, 277, 4589–4592. [Google Scholar] [CrossRef] [PubMed]
- Slomiany, M.G.; Dai, L.; Bomar, P.A.; Knackstedt, T.J.; Kranc, D.A.; Tolliver, L.; Maria, B.L.; Toole, B.P. Abrogating drug resistance in malignant peripheral nerve sheath tumors by disrupting hyaluronan-CD44 interactions with small hyaluronan oligosaccharides. Cancer Res. 2009, 69, 4992–4998. [Google Scholar] [CrossRef]
- Orian-Rousseau, V.; Morrison, H.; Matzke, A.; Kastilan, T.; Pace, G.; Herrlich, P.; Ponta, H. Hepatocyte growth factor-induced Ras activation requires ERM proteins linked to both CD44v6 and F-actin. Mol. Biol. Cell 2007, 18, 76–83. [Google Scholar] [CrossRef]
- McCarty, J.H. Glioblastoma resistance to anti-VEGF therapy: Has the challenge been MET? Clin. Cancer Res. 2013, 19, 1631–1633. [Google Scholar] [CrossRef]
- Nishikawa, M.; Inoue, A.; Ohnishi, T.; Yano, H.; Kanemura, Y.; Kohno, S.; Ohue, S.; Ozaki, S.; Matsumoto, S.; Suehiro, S.; et al. CD44 expression in the tumor periphery predicts the responsiveness to bevacizumab in the treatment of recurrent glioblastoma. Cancer Med. 2021, 10, 2013–2025. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Lee, H.W.; Kang, H.G.; Kim, H.Y.; Kim, S.J.; Chun, K.H. Cleaved CD44 intracellular domain supports activation of stemness factors and promotes tumorigenesis of breast cancer. Oncotarget 2015, 6, 8709–8721. [Google Scholar] [CrossRef]
- Löfstedt, T.; Fredlund, E.; Holmquist-Mengelbier, L.; Pietras, A.; Ovenberger, M.; Poellinger, L.; Påhlman, S. Hypoxia inducible factor-2alpha in cancer. Cell Cycle 2007, 6, 919–926. [Google Scholar] [CrossRef]
- Okamoto, I.; Kawano, Y.; Murakami, D.; Sasayama, T.; Araki, N.; Miki, T.; Wong, A.J.; Saya, H. Protelytic release of CD44intracellular domain and its role in the CD44 signaling pathway. J. Cell. Biol. 2001, 155, 755–762. [Google Scholar] [CrossRef]
- De Falco, V.; Tamburrino, A.; Ventre, S.; Castellone, M.D.; Malek, M.; Manié, S.N.; Santoro, M. CD44 proteolysis increases CREB phosphorylation and sustains proliferation of thyroid cancer cells. Cancer Res. 2012, 72, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Li, D.; Xun, J.; Zhou, W.; Li, J.; Wang, J.; Liu, C.; Li, X.; Shen, W.; Qiao, H.; et al. CD44ICD promotes breast cancer stemness via PFKFB4-mediated glucose metabolism. Theranostics 2018, 8, 6248–6262. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, L.Y. Matrix hyaluronan promotes specific microRNA upregulation leading to drug resistance and tumor progression. Int. J. Mol. Sci. 2016, 17, 517. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Asangani, I.A.; Rasheed, S.A.; Nikolova, D.A.; Leupold, J.H.; Colburn, N.H.; Post, S.; Allgayer, H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008, 27, 2128–2136. [Google Scholar] [CrossRef]
- Xu, L.F.; Wu, Z.P.; Chen, Y.; Zhu, Q.S.; Hamidi, S.; Navab, R. MicroRNA-21 (miR-21) regulates cellular proliferation, invasion, migration, and apoptosis by targeting PTEN, RECK and Bcl-2 in lung squamous carcinoma. PLoS ONE 2014, 9, e103698. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.; Earle, C.; Wong, G.; Spevak, C.C.; Krueger, K. Stem cell marker (Nanog) and Stat-3 signaling promote microRNA-21 expression and chemoresistance in hyaluronan/CD44-activated head and neck squamous cell carcinoma cells. Oncogene 2012, 31, 149–160. [Google Scholar] [CrossRef]
- Fujita, S.; Ito, T.; Mizutani, T.; Minoguchi, S.; Yamamichi, N.; Sakurai, K.; Iba, H. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J. Mol. Biol. 2008, 378, 492–504. [Google Scholar] [CrossRef]
- Chen, L.; Bourguignon, L.Y. Hyaluronan-CD44 interaction promotes c-Jun signaling and miRNA21 expression leading to Bcl-2 expression and chemoresistance in breast cancer cells. Mol. Cancer 2014, 13, 52. [Google Scholar] [CrossRef]
- Gabriely, G.; Wurdinger, T.; Kesarri, S.; Esau, C.C.; Burchard, J.; Linsley, P.S.; Krichevsky, A.M. MicroRNA21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol. Cell. Biol. 2008, 28, 5369–5380. [Google Scholar] [CrossRef]
- Luo, G.; Luo, W.; Sun, X.; Lin, J.; Wang, M.; Zhang, Y.; Luo, W.; Zhang, Y. MicroRNA 21 promotes migration and invasion of glioma cells via activation of Sox2 and β catenin signaling. Mol. Med. Rep. 2017, 15, 187–195. [Google Scholar] [CrossRef]
- Shi, L.; Chen, J.; Yang, J.; Pan, T.; Zhang, S.; Wang, Z. MiR-21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity. Brain Res. 2010, 1352, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Reinhardt, F.; Pan, E.; Soutschek, J.; Bhat, B.; Marcusson, E.G.; Teruya-Feldstein, J.; Bell, G.W.; Weinberg, R.A. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol. 2010, 28, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Sasayama, T.; Nishihara, M.; Kondoh, T.; Hosoda, K.; Kohmura, E. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int. J. Cancer 2009, 125, 1407–1413. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, L.Y.; Wong, G.; Earle, C.; Krueger, K.; Spevak, C.C. Hyaluronan-CD44 interaction promotes c-Src-mediated Twist signaling, microRNA-10b expression, and RhoA/RhoC up-regulation, leading to Rho-kinase-associated cytoskeleton activation and breast tumor cell invasion. J. Biol. Chem. 2010, 285, 36721–36735. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Deng, S.; Zhao, Z.; Zhang, H.; Xiao, J.; Song, W.; Gao, F.; Guan, Y. Oct4 regulates the miR-302 cluster in P19 mouse embryonic carcinoma cells. Mol. Biol. Rep. 2011, 38, 2155–2160. [Google Scholar] [CrossRef]
- Lin, S.L.; Chang, D.C.; Chang-Lin, S.; Lin, C.H.; Wu, D.T.; Chen, D.T.; Ying, S.Y. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA 2008, 14, 2115–2124. [Google Scholar] [CrossRef] [PubMed]
- Clay, M.R.; Tabor, M.; Owen, J.H.; Carey, T.E.; Bradford, C.R.; Wolf, G.T.; Wicha, M.S.; Prince, M.E. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck 2010, 32, 1195–1201. [Google Scholar] [CrossRef]
- Lin, S.L.; Chang, D.C.; Lin, C.H.; Ying, S.Y.; Leu, D.; Wu, D.T. Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res. 2011, 39, 1054–1065. [Google Scholar] [CrossRef]
- Chen, Y.C.; Chen, Y.W.; Hsu, H.S.; Tseng, L.M.; Huang, P.I.; Lu, K.H.; Chen, D.T.; Tai, L.K.; Yung, M.C.; Chang, S.C.; et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem. Biophys. Res. Commun. 2009, 385, 307–313. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.; Wong, G.; Earle, C.; Chen, L. Hyaluronan-CD44v3 interaction with Oct4-Sox2-Nanog promotes miR-302 expression leading to self-renewal, clonal formation, and cisplatin resistance in cancer stem cells from head and neck squamous cell carcinoma. J. Biol. Chem. 2012, 287, 32800–32824. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Wang, K.; Shao, Z.; Lin, Q.; Li, B.; Liu, P. Mir-373/miR-520s-CD44 axis significantly inhibits the growth and invasion of human glioblastoma cells. Arch. Med Res. 2022, 53, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Yeh, M.; Wang, Y.Y.; Yoo, J.; Oh, C.; Otani, Y.; Kang, J.M.; Park, E.S.; Kim, E.; Chung, S.; Jeon, Y.J.; et al. MicroRNA-138 suppresses glioblastoma proliferation through downregulation of CD44. Sci. Rep. 2021, 11, 9219. [Google Scholar] [CrossRef]
- Mooney, K.L.; Choy, W.; Sidhu, S.; Pelargos, P.; Bui, T.T.; Voth, B.; Barnette, N.; Yang, I. The role of CD44 in glioblastoma multiforme. J. Clin. Neurosci. 2016, 34, 1–5. [Google Scholar] [CrossRef]
- Cheng, L.; Wu, Q.; Guryanova, O.A.; Huang, Z.; Huang, Q.; Rich, J.N.; Bao, S. Elevated invasive potential of glioblastoma stem cells. Biochem. Biophys. Res. Commun. 2011, 406, 643–648. [Google Scholar] [CrossRef]
- Chen, J.E.; Lumibao, J.; Blazek, A.; Gaskins, H.R.; Harley, B. Hypoxia activates enhanced invasive potential and endogenous hyaluronic acid production by glioblastoma cells. Biomater. Sci. 2018, 6, 854–862. [Google Scholar] [CrossRef]
- Delpech, B.; Maingonnat, C.; Girard, N.; Chauzy, C.; Maunoury, R.; Olivier, A.; Tayot, J.; Creissard, P. Hyaluronan and hyaluronectin in the extracellular matrix of human brain tumor stroma. Eur. J. Cancer 1993, 29, 1012–1017. [Google Scholar] [CrossRef]
- Nishikawa, M.; Inoue, A.; Ohnishi, T.; Yano, H.; Ozaki, S.; Kanemura, Y.; Suehiro, S.; Ohtsuka, Y.; Kohno, S.; Ohue, S.; et al. Hypoxia-induced phenotypic transition from highly invasive to less invasive tumors in glioma stem-like cells: Significance of CD44 and osteopontin as therapeutic targets in glioblastoma. Transl. Oncol. 2021, 14, 101137. [Google Scholar] [CrossRef]
- Ridley, A.J.; Schwartz, M.A.; Burridge, K.; Firtel, R.A.; Ginsberg, M.H.; Borisy, G.; Parsons, J.T.; Horwitz, A.R. Cell migration: Integrating signals from front to back. Science 2003, 302, 1704–1709. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, P.; Yang, L.; Yu, X.; Ye, X.; Yang, J.; Qian, C.; Zhang, X.; Cui, Y.H.; Bian, X.W. Activation of toll-like receptor 2 promotes invasion by upregulating MMPs in glioma stem cells. Am. J. Transl. Res. 2015, 7, 607–615. [Google Scholar] [PubMed]
- Meldolesi, J. Pharmacology of the cell/matrix form of adhesion. Pharm. Res. 2016, 107, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Ellert-Miklaszewska, A.; Poleszak, K.; Pasierbinska, M.; Kaminska, B. Integrin Signaling in Glioma Pathogenesis: From Biology to Therapy. Int. J. Mol. Sci. 2020, 21, 888. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, V.P.; Moura Neto, V.; Mentlein, R. Glioma infiltration and extracellular matrix: Key players and modulators. Glia 2018, 66, 1542–1565. [Google Scholar] [CrossRef] [PubMed]
- Onishi, M.; Ichikawa, T.; Kurozumi, K.; Date, I. Angiogenesis and invasion in glioma. Brain Tumor Pathol. 2011, 28, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Binder, Z.A.; Kim, S.H.J.; Wu, P.H.; Giri, A.; Gallia, G.L.; Pardo, A.; Wirtz, D. The role of integrin v and CD44 in GBM migration in human brain. bioRxiv, 2019; preprint. [Google Scholar] [CrossRef]
- Wolf, K.J.; Shukla, P.; Springer, K.; Lee, S.; Coombes, J.D.; Choy, C.J.; Kenny, S.J.; Xu, K.; Kumar, S. A mode of cell adhesion and migration facilitated by CD44-dependent microtentacles. Proc. Natl. Acad. Sci. USA 2020, 117, 11432–11443. [Google Scholar] [CrossRef]
- Kim, Y.; Kumar, S. CD44-mediated adhesion to hyaluronic acid contributes to mechanosensing and invasive motility. Mol. Cancer Res. 2014, 12, 1416–1429. [Google Scholar] [CrossRef]
- So, J.S.; Kim, H.; Han, K.S. Mechanisms of invasion in glioblastoma: Extracellular matrix, Ca2+ signaling, and glutamate. Front. Cell. Neurosci. 2021, 15, 663092. [Google Scholar] [CrossRef]
- Giese, A.; Bjerkvig, R.; Berens, M.E.; Westphal, M. Cost of migration: Invasion of malignant gliomas and implications for treatment. J. Clin. Oncol. 2003, 21, 1624–1636. [Google Scholar] [CrossRef]
- Xu, Y.; Stamenkovic, I.; Yu, Q. CD44 attenuates activation of the Hippo signaling pathway and is a prime therapeutic target for glioblastoma. Cancer Res. 2010, 70, 2455–2464. [Google Scholar] [CrossRef]
- Bai, Y.; Liu, Y.J.; Wang, H.; Xu, Y.; Stamenkovic, I.; Yu, Q. Inhibition of the hyaluronan-CD44 interaction by merlin contributes to the tumor suppressor activity of merlin. Oncogene 2007, 26, 836–850. [Google Scholar] [CrossRef]
- Ibrahim, H.M.; AbdElbary, A.M.; Mohamed, S.Y.; Elwan, A.; Abdelhamid, M.I.; Ibrahim, A. Prognostic value of cyclin D1 and CD44 expression in gastric adenocarcinoma. J. Gastrointest. Cancer 2019, 50, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Daniel, P.M.; Filiz, G.; Tymmms, M.J.; Ramsay, R.G.; Kaye, A.H.; Stylli, S.S.; Mantamadiotis, T. Intratumor MAPK and PI3K signaling pathway heterogeneity in glioblastoma tissue correlates with CREB signaling and distinct target gene signatures. Exp. Mol. Pathol. 2018, 105, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Qazi, M.A.; Vora, P.; Venugopal, C.; Sidhu, S.S.; Moffat, J.; Swanton, C.; Singh, S.K. Intratumoral heterogeneity: Pathways to treatment resistance and relapse in human glioblastoma. Ann. Oncol. 2017, 28, 1448–1456. [Google Scholar] [CrossRef] [PubMed]
- Parker, N.R.; Khong, P.; Parkinson, J.F.; Howell, V.M.; Wheeler, H.R. Molecular heterogeneity in glioblastoma: Potential clinical implications. Front. Oncol. 2015, 5, 55. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef] [PubMed]
- Segerman, A.; Niklasson, M.; Haglund, C.; Bergström, T.; Jarvius, M.; Xie, Y.; Westermark, A.; Sönmez, D.; Hermansson, A.; Kastemar, M.; et al. Clonal variation in drug and radiation response among glioma-initiating cells is linked to proneural-mesenchymal transition. Cell Rep. 2016, 11, 2994–3009. [Google Scholar] [CrossRef]
- Phillips, H.S.; Kharbanda, S.; Chen, R.; Forrest, W.F.; Soriano, R.H.; Wu, T.D.; Misra, A.; Nigro, J.M.; Colman, H.; Soroceanu, L.; et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006, 9, 157–173. [Google Scholar] [CrossRef]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef]
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; Decarvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 32, 42–56. [Google Scholar] [CrossRef]
- Takashima, Y.; Kawaguchi, A.; Yamanaka, R. Promising prognosis marker candidates on the status of epithelial-mesenchymal transition and glioma stem cells in glioblastoma. Cells 2019, 8, 1312. [Google Scholar] [CrossRef] [PubMed]
- Fedele, M.; Cerchia, L.; Pegoraro, S.; Sgarra, R.; Manfioletti, G. Proneural-Mesenchymal Transition: Phenotypic Plasticity to Acquire Multitherapy Resistance in Glioblastoma. Int. J. Mol. Sci. 2019, 20, 2746. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Slomiany, M.G.; Grass, G.D.; Robertson, A.D.; Yang, X.Y.; Maria, B.L.; Beeson, C.; Toole, B.P. Hyaluronan, CD44, and emmprin regulate lactate efflux and membrane localization of monocarboxylate transporters in human breast carcinoma cells. Cancer Res. 2009, 69, 1293–1301. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhou, B.P. Activation of beta-catenin and Akt pathways by Twist are critical for the maintenance of EMT associated cancer stem cell-like characters. BMC Cancer 2011, 11, 49. [Google Scholar] [CrossRef]
- Jang, B.I.; Li, Y.; Graham, D.Y.; Cen, P. The role of CD44 in the pathogenesis, diagnosis, and therapy of gastric cancer. Gut Liver 2011, 5, 397–405. [Google Scholar] [CrossRef]
- Li, L.; Liu, C.; Amato, R.J.; Chang, J.T.; Du, G.; Li, W. CDKL2 promotes epithelial-mesenchymal transition and breast cancer progression. Oncotarget 2014, 5, 10840–10853. [Google Scholar] [CrossRef]
- Lee, J.K.; Joo, K.M.; Lee, J.; Yoon, Y.; Nam, D.H. Targeting the epithelial to mesenchymal transition in glioblastoma: The emerging role of MET signaling. OncoTargets Ther. 2014, 7, 1933–1944. [Google Scholar]
- Godar, S.; Ince, T.A.; Bell, G.W.; Feldser, D.; Donaher, J.L.; Bergh, J.; Liu, A.; Miu, K.; Watnick, R.S.; Reinhardt, F.; et al. Growth-inhibitory and tumor- suppressive functions of p53 depend on its repression of CD44 expression. Cell 2008, 134, 62–73. [Google Scholar] [CrossRef]
- Park, J.; Schwarzbauer, J.E. Mammary epithelial cell interactions with fibronectin stimulate epithelial-mesenchymal transition. Oncogene 2014, 33, 1649–1657. [Google Scholar] [CrossRef]
- Way, T.D.; Huang, J.T.; Chou, C.H.; Huang, C.H.; Yang, M.H.; Ho, C.T. Emodin represses TWIST1-induced epithelial-mesenchymal transitions in head and neck squamous cell carcinoma cells by inhibiting the beta-catenin and Akt pathways. Eur. J. Cancer 2014, 50, 366–378. [Google Scholar] [CrossRef] [PubMed]
- Masui, T.; Ota, I.; Yook, J.I.; Mikami, S.; Yane, K.; Yamanaka, T.; Hosoi, H. Snail-induced epithelial-mesenchymal transition promotes cancer stem cell-like phenotype in head and neck cancer cells. Int. J. Oncol. 2014, 44, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Ning, Z.; Zeng, J.; Fan, J.; Zhou, J.; Zhang, T.; Zhang, L.; Chen, Y.; Gao, Y.; Wang, B.; et al. Silibinin inhibits beta-catenin/ZEB1 signaling and suppresses bladder cancer metastasis via dual-blocking epithelial-mesenchymal transition and stemness. Cell Signal. 2013, 25, 2625–2633. [Google Scholar] [CrossRef]
- Bhat-Nakshatri, P.; Appaiah, H.; Ballas, C.; Pick-Franke, P.; Goulet, R., Jr.; Badve, S.; Srour, E.F.; Nakshatri, H. SLUG/SNAI2 and tumor necrosis factor generate breast cells with CD44+/CD24− phenotype. BMC Cancer 2010, 10, 411. [Google Scholar] [CrossRef]
- Ju, S.Y.; Chiou, S.H.; Su, Y. Maintenance of the stemness in CD44(+) HCT-15 and HCT-116 human colon cancer cells requires miR-203 suppression. Stem Cell Res. 2014, 12, 86–100. [Google Scholar] [CrossRef] [PubMed]
- Shang, Z.; Cai, Q.; Zhang, M.; Zhu, S.; Ma, Y.; Sun, L.; Jiang, N.; Tian, J.; Niu, X.; Chen, J.; et al. A switch from CD44(+) cell to EMT cell drives the metastasis of prostate cancer. Oncotarget 2015, 6, 1202–1216. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.H.; Park, Y.S.; Kim, H.J.; Kim, C.H.; Lim, S.W.; Huh, J.W.; Lee, J.H.; Kim, H.R. CD44 enhances the epithelial-mesenchymal transition in association with colon cancer invasion. Int. J. Oncol. 2012, 41, 211–218. [Google Scholar]
- Zubeldia, I.G.; Bleau, A.M.; Redrado, M.; Serrano, D.; Agliano, A.; Gil-Puig, C.; Vidal-Vanaclocha, F.; Lecanda, J.; Calvo, A. Epithelial to mesenchymal transition and cancer stem cell phenotypes leading to liver metastasis are abrogated by the novel TGFβ1-targeting peptides P17 and P144. Exp. Cell Res. 2013, 319, 12–22. [Google Scholar] [CrossRef]
- Joseph, J.V.; Conroy, S.; Tomar, T.; Eggens-Meijer, E.; Bhat, K.; Copray, S.; Walenlamp, A.M.; Boddeke, E.; Balasubramamyian, V.; Wagemakers, A.M.; et al. TGF-β is an inducer of ZEB1-dependent mesenchymal transdifferentiation in glioblastoma that is associated with tumor invasion. Cell Death Dis. 2014, 5, e1443. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.; Singleton, P.A.; Zhu, H.; Zhou, B. Hyaluronan promotes signaling interaction between CD44 and the transforming growth factor beta receptor I in metastatic breast tumor cells. J. Biol. Chem. 2002, 277, 39703–39712. [Google Scholar] [CrossRef]
- Luo, D.; Xu, X.; Li, J.; Chen, C.; Wang, F.; Xie, Y.; Li, F. The PDK1/c Jun pathway activated by TGF β induces EMT and promotes proliferation and invasion in human glioblastoma. Int. J. Oncol. 2018, 53, 2067–2080. [Google Scholar] [CrossRef] [PubMed]
- Bhat, K.P.L.; Balasubramaniyan, V.; Vaillant, B.; Ezhilarasan, R.; Hummelink, K.; Hollingsworth, F.; Wani, K.; Heathcock, L.; James, J.D.; Goodman, L.D.; et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell 2013, 24, 331–346. [Google Scholar] [CrossRef] [PubMed]
- Iwata, R.; Lee, J.H.; Hayashi, M.; Dianzani, U.; Ofune, K.; Maruyama, M.; Oe, S.; Ito, T.; Hashiba, T.; Yoshimura, K.; et al. ICOSLG-mediated regulatory T-cell expansion and IL-10 production promote progression of glioblastoma. Neuro Oncol. 2020, 22, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Chen, C.; Chang, K.; Karnad, A.; Jagirdar, J.; Kumar, A.P.; Freeman, J.W. CD44 expression level and isoform contributes to pancreatic cancer cell plasticity, invasiveness and response to therapy. Clin. Cancer Res. 2016, 22, 5592–5604. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.L.; Reinke, L.M.; Damerow, M.S.; Perez, D.; Chodosh, L.A.; Yang, J.; Cheng, C. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J. Clin. Investig. 2011, 121, 1064–1074. [Google Scholar] [CrossRef]
- Dhruv, H.D.; McDonough Winslow, W.S.; Armstrong, B.; Tuncali, S.; Eschbacher, J.; Kislin, K.; Loftus, J.C.; Tran, N.L.; Berens, M.E. Reciprocal activation of transcription factors underlies the dichotomy between proliferation and invasion of glioma cells. PLoS ONE 2013, 8, e72134. [Google Scholar] [CrossRef]
- Jackson, J.S.; Golding, J.P.; Chapon, C.; Jones, W.A.; Bhakoo, K.K. Homing of stem cells to sites of inflammatory brain injury after intracerebral and intravenous administration: A longitudinal imaging study. Stem Cell Res. Ther. 2010, 1, 17. [Google Scholar] [CrossRef]
- Mehta, S. Editorial: The Role of Microenvironment in the Homing, Maintenance, and Release of Glioma Stem-Like Cells. Front Oncol. 2018, 8, 7. [Google Scholar] [CrossRef]
- Nieto, M.A.; Hung, R.Y.J.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef]
- Luond, F.; Sugiyama, N.; Bill, R.; Bomes, L.; Hager, C.; Tang, F.; Santacroce, N.; Beisel, C.; Ivanek, R.; Bürglin, T.; et al. Distinct contributions of partial and full EMT to breast cancer malignancy. Dev. Cell 2021, 56, 3203–3221. [Google Scholar] [CrossRef]
- Manfioletti, G.; Fedele, M. Epithelial-mesenchymal transition (EMT) 2021. Int. J. Mol. Sci. 2022, 23, 5848. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Bao, S.; Wu, Q.; Wang, H.; Eyler, C.; Sathornsumetee, S.; Shi, Q.; Cao, Y.; Lathia, J.; McLendon, R.E.; et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell 2009, 15, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hu, Z.; Horta, C.A.; Yang, J. Regulation of epithelial-mesenchymal transition by tumor microenvironmental signals and its implication in cancer therapeutics. Semin. Cancer Biol. 2023, 88, 46–66. [Google Scholar] [CrossRef] [PubMed]
- Mortezaee, K. Hypoxia induces core-to-edge transition of progressive tumoral cells: A critical review on differential yet corroborative roles for HIF-1α and HIF-2α. Life Sci. 2020, 242, 114145. [Google Scholar] [CrossRef]
- Bar, E.E. Glioblastoma, cancer stem cells and hypoxia. Brain Pathol. 2011, 21, 119–129. [Google Scholar] [CrossRef]
- Chakhoyan, A.; Guillamo, J.S.; Collet, S.; Kauffmann, F.; Delcroix, N.; Lechapt-Zalcman, E.; Constans, J.M.; Petit, E.; MacKenzie, E.T.; Barré, L.; et al. FMISO-PET-derived brain oxygen tension maps: Application to glioblastoma and less aggressive gliomas. Sci. Rep. 2017, 7, 10210. [Google Scholar] [CrossRef]
- Majmundar, A.J.; Wong, W.J.; Simon, M.C. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell 2010, 40, 294–309. [Google Scholar] [CrossRef]
- Boyd, N.H.; Tran, A.N.; Bernstock, J.D.; Etminan, T.; Jones, A.B.; Gillespie, G.Y.; Friedman, G.K.; Hjelmeland, A.B. Glioma stem cells and their roles within the hypoxic tumor microenvironment. Theranostics 2021, 11, 665–683. [Google Scholar] [CrossRef]
- Tam, S.Y.; Wu, V.W.C.; Law, H.K.W. Hypoxia-Induced Epithelial-Mesenchymal Transition in Cancers: HIF-1α and beyond. Front. Oncol. 2020, 10, 486. [Google Scholar] [CrossRef]
- Cruickshank, G.S.; Rampling, R. Peri-tumoral hypoxia in human brain:preoperative of the tissue oxygen tension around malignant brain tumours. Acta Neurochir. Suppl. 1994, 60, 375–377. [Google Scholar]
- Kolliopoulos, C.; Ali, M.M.; Castillejo-Lopez, C.; Heldin, C.H.; Heldin, P. CD44 depletion in glioblastoma cells suppresses growth and stemness and induces senescence. Cancers 2022, 14, 3747. [Google Scholar] [CrossRef] [PubMed]
- Heddleston, J.M.; Li, Z.; MeLendon, R.E.; Hjelmeland, A.B.; Rich, J.N. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle 2009, 8, 3274–3284. [Google Scholar] [CrossRef] [PubMed]
- Musah-Eroje, A.; Watson, S. Adaptive changes of glioblastoma cells following exposure to hypoxic (1% Oxygen) tumour microenvironment. Int. J. Mol. Sci. 2019, 20, 2091. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).