Homologous Recombination Deficiency and Cyclin E1 Amplification Are Correlated with Immune Cell Infiltration and Survival in High-Grade Serous Ovarian Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient and Tumor Selection
2.2. Tissue Samples and Tissue Microarrays (TMA)
2.3. Immunohistochemical (IHC) Staining
2.4. Molecular Analyses
2.5. Somatic BRCA Analysis
2.6. BRCA Promotor Methylation
2.7. Low-Coverage Whole-Genome Sequencing (WGS)
2.8. Non-BRCAmut HRD Classification
2.9. CCNE1 Classification
2.10. Statistical Analyses
3. Results
3.1. Patient Characteristics
3.2. Molecular Profiles
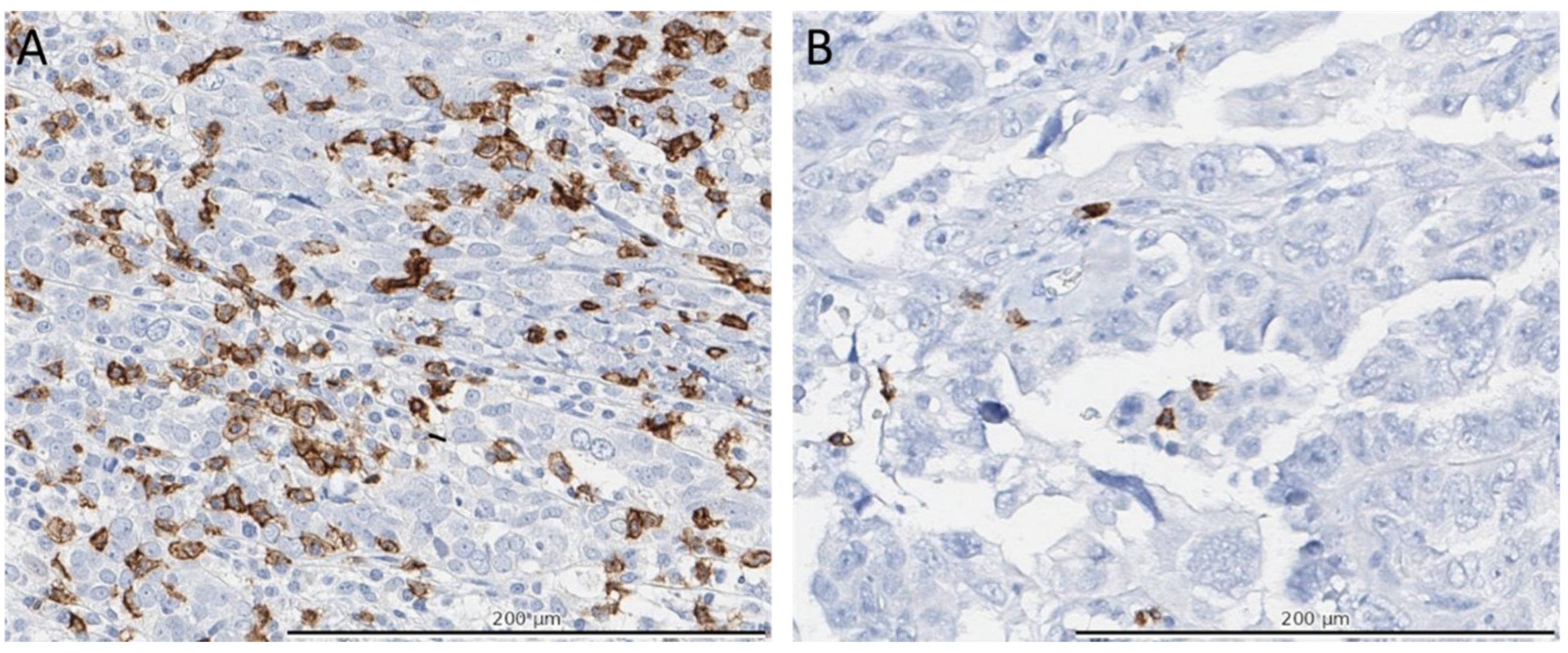
3.3. Immune Cell Infiltration in Molecular Profiles
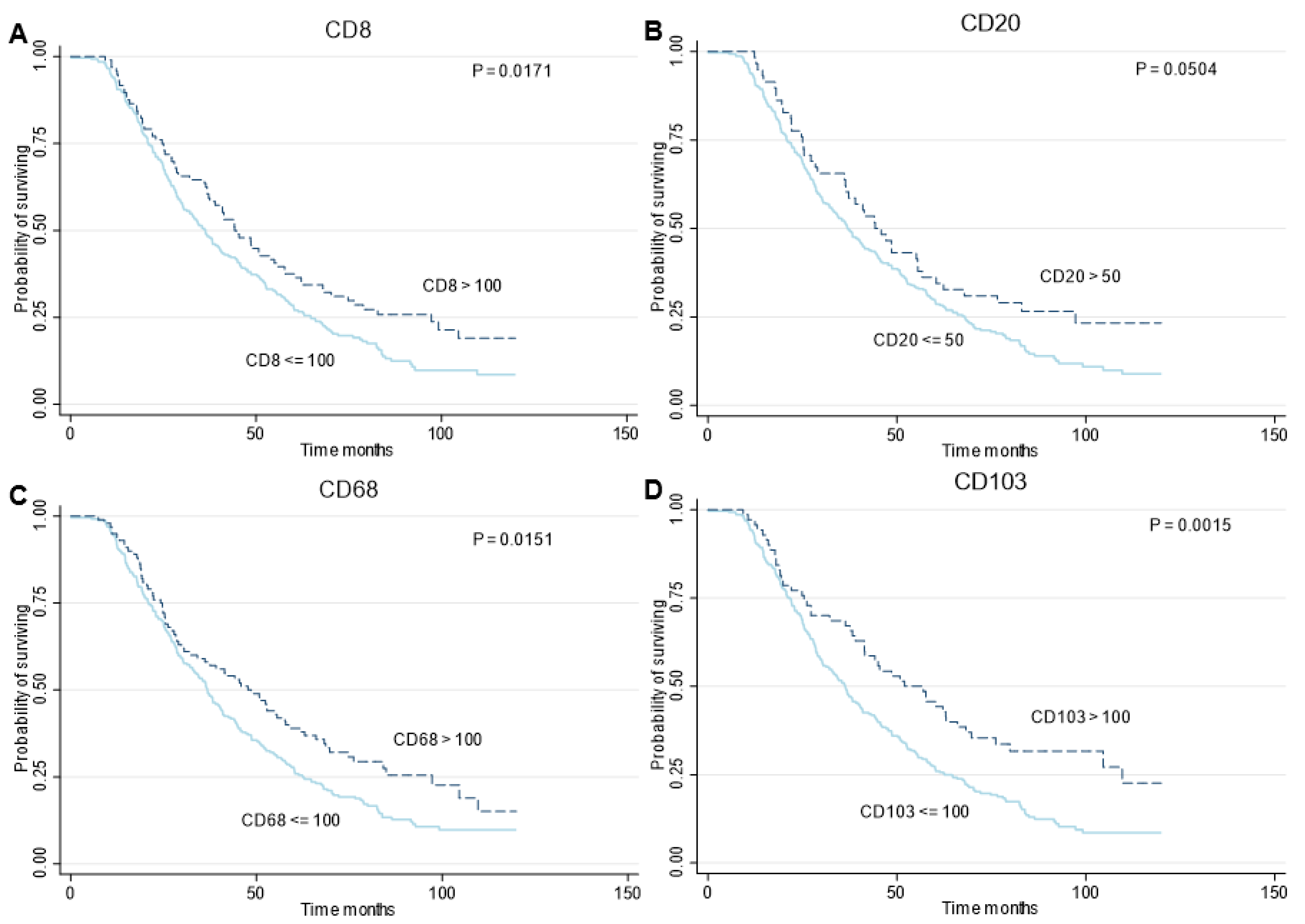
3.4. PFS and OS of Molecular Profiles and Immune Cell Infiltration
3.5. Multivariable Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Bowtell, D.D.; Bohm, S.; Ahmed, A.A.; Aspuria, P.J.; Bast, R.C., Jr.; Beral, V.; Berek, J.S.; Birrer, M.J.; Blagden, S.; Bookman, M.A.; et al. Rethinking ovarian cancer II: Reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer 2015, 15, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Gorringe, K.L.; Jacobs, S.; Thompson, E.R.; Sridhar, A.; Qiu, W.; Choong, D.Y.; Campbell, I.G. High-resolution single nucleotide polymorphism array analysis of epithelial ovarian cancer reveals numerous microdeletions and amplifications. Clin. Cancer Res. 2007, 13, 4731–4739. [Google Scholar] [CrossRef]
- Macintyre, G.; Goranova, T.E.; de Silva, D.; Ennis, D.; Piskorz, A.M.; Eldridge, M.; Sie, D.; Lewsley, L.A.; Hanif, A.; Wilson, C.; et al. Copy number signatures and mutational processes in ovarian carcinoma. Nat. Genet. 2018, 50, 1262–1270. [Google Scholar] [CrossRef]
- Patch, A.M.; Christie, E.L.; Etemadmoghadam, D.; Garsed, D.W.; George, J.; Fereday, S.; Nones, K.; Cowin, P.; Alsop, K.; Bailey, P.J.; et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 2015, 521, 489–494. [Google Scholar] [CrossRef]
- Zhong, Q.; Peng, H.L.; Zhao, X.; Zhang, L.; Hwang, W.T. Effects of BRCA1- and BRCA2-related mutations on ovarian and breast cancer survival: A meta-analysis. Clin. Cancer Res. 2015, 21, 211–220. [Google Scholar] [CrossRef]
- Ledermann, J.A.; Drew, Y.; Kristeleit, R.S. Homologous recombination deficiency and ovarian cancer. Eur. J. Cancer 2016, 60, 49–58. [Google Scholar] [CrossRef]
- Strickland, K.C.; Howitt, B.E.; Shukla, S.A.; Rodig, S.; Ritterhouse, L.L.; Liu, J.F.; Garber, J.E.; Chowdhury, D.; Wu, C.J.; D’Andrea, A.D.; et al. Association and prognostic significance of BRCA1/2-mutation status with neoantigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget 2016, 7, 13587–13598. [Google Scholar] [CrossRef]
- Turner, N.; Tutt, A.; Ashworth, A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat. Rev. Cancer 2004, 4, 814–819. [Google Scholar] [CrossRef]
- Lord, C.J.; Ashworth, A. BRCAness revisited. Nat. Rev. Cancer 2016, 16, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Ruscito, I.; Dimitrova, D.; Vasconcelos, I.; Gellhaus, K.; Schwachula, T.; Bellati, F.; Zeillinger, R.; Benedetti-Panici, P.; Vergote, I.; Mahner, S.; et al. BRCA1 gene promoter methylation status in high-grade serous ovarian cancer patients—A study of the tumour Bank ovarian cancer (TOC) and ovarian cancer diagnosis consortium (OVCAD). Eur. J. Cancer 2014, 50, 2090–2098. [Google Scholar] [CrossRef] [PubMed]
- Konstantinopoulos, P.A.; Ceccaldi, R.; Shapiro, G.I.; D’Andrea, A.D. Homologous Recombination Deficiency: Exploiting the Fundamental Vulnerability of Ovarian Cancer. Cancer Discov. 2015, 5, 1137–1154. [Google Scholar] [CrossRef] [PubMed]
- Sahnane, N.; Carnevali, I.; Formenti, G.; Casarin, J.; Facchi, S.; Bombelli, R.; Di Lauro, E.; Memoli, D.; Salvati, A.; Rizzo, F.; et al. BRCA Methylation Testing Identifies a Subset of Ovarian Carcinomas without Germline Variants That Can Benefit from PARP Inhibitor. Int. J. Mol. Sci. 2020, 21, 9708. [Google Scholar] [CrossRef]
- Miller, R.E.; Leary, A.; Scott, C.L.; Serra, V.; Lord, C.J.; Bowtell, D.; Chang, D.K.; Garsed, D.W.; Jonkers, J.; Ledermann, J.A.; et al. ESMO recommendations on predictive biomarker testing for homologous recombination deficiency and PARP inhibitor benefit in ovarian cancer. Ann. Oncol. 2020, 31, 1606–1622. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.C.; Clurman, B.E. Cyclin E in normal and neoplastic cell cycles. Oncogene 2005, 24, 2776–2786. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, N.; Nakayama, K.; Shamima, Y.; Ishikawa, M.; Katagiri, A.; Iida, K.; Miyazaki, K. Gene amplification CCNE1 is related to poor survival and potential therapeutic target in ovarian cancer. Cancer 2010, 116, 2621–2634. [Google Scholar] [CrossRef]
- Etemadmoghadam, D.; deFazio, A.; Beroukhim, R.; Mermel, C.; George, J.; Getz, G.; Tothill, R.; Okamoto, A.; Raeder, M.B.; Harnett, P.; et al. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin. Cancer Res. 2009, 15, 1417–1427. [Google Scholar] [CrossRef]
- Webb, J.R.; Milne, K.; Watson, P.; Deleeuw, R.J.; Nelson, B.H. Tumor-infiltrating lymphocytes expressing the tissue resident memory marker CD103 are associated with increased survival in high-grade serous ovarian cancer. Clin. Cancer Res. 2014, 20, 434–444. [Google Scholar] [CrossRef]
- Clarke, B.; Tinker, A.V.; Lee, C.H.; Subramanian, S.; van de Rijn, M.; Turbin, D.; Kalloger, S.; Han, G.; Ceballos, K.; Cadungog, M.G.; et al. Intraepithelial T cells and prognosis in ovarian carcinoma: Novel associations with stage, tumor type, and BRCA1 loss. Mod. Pathol. 2009, 22, 393–402. [Google Scholar] [CrossRef]
- Milne, K.; Kobel, M.; Kalloger, S.E.; Barnes, R.O.; Gao, D.; Gilks, C.B.; Watson, P.H.; Nelson, B.H. Systematic analysis of immune infiltrates in high-grade serous ovarian cancer reveals CD20, FoxP3 and TIA-1 as positive prognostic factors. PLoS ONE 2009, 4, e6412. [Google Scholar] [CrossRef]
- Montfort, A.; Owen, S.; Piskorz, A.M.; Supernat, A.; Moore, L.; Al-Khalidi, S.; Bohm, S.; Pharoah, P.; McDermott, J.; Balkwill, F.R.; et al. Combining measures of immune infiltration shows additive effect on survival prediction in high-grade serous ovarian carcinoma. Br. J. Cancer 2020, 122, 1803–1810. [Google Scholar] [CrossRef]
- Yuan, X.; Zhang, J.; Li, D.; Mao, Y.; Mo, F.; Du, W.; Ma, X. Prognostic significance of tumor-associated macrophages in ovarian cancer: A meta-analysis. Gynecol. Oncol. 2017, 147, 181–187. [Google Scholar] [CrossRef]
- Casparie, M.; Tiebosch, A.T.; Burger, G.; Blauwgeers, H.; van de Pol, A.; van Krieken, J.H.; Meijer, G.A. Pathology databanking and biobanking in The Netherlands, a central role for PALGA, the nationwide histopathology and cytopathology data network and archive. Cell Oncol. 2007, 29, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Malpica, A.; Deavers, M.T.; Lu, K.; Bodurka, D.C.; Atkinson, E.N.; Gershenson, D.M.; Silva, E.G. Grading ovarian serous carcinoma using a two-tier system. Am. J. Surg. Pathol. 2004, 28, 496–504. [Google Scholar] [CrossRef]
- Komdeur, F.L.; Prins, T.M.; van de Wall, S.; Plat, A.; Wisman, G.B.A.; Hollema, H.; Daemen, T.; Church, D.N.; de Bruyn, M.; Nijman, H.W. CD103+ tumor-infiltrating lymphocytes are tumor-reactive intraepithelial CD8+ T cells associated with prognostic benefit and therapy response in cervical cancer. Oncoimmunology 2017, 6, e1338230. [Google Scholar] [CrossRef] [PubMed]
- Moelans, C.B.; Atanesyan, L.; Savola, S.P.; van Diest, P.J. Methylation-Specific Multiplex Ligation-Dependent Probe Amplification (MS-MLPA). Methods Mol. Biol. 2018, 1708, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Schouten, P.C.; Richters, L.; Vis, D.J.; Kommoss, S.; van Dijk, E.; Ernst, C.; Kluin, R.J.C.; Marme, F.; Lips, E.H.; Schmidt, S.; et al. Ovarian Cancer-Specific BRCA-like Copy-Number Aberration Classifiers Detect Mutations Associated with Homologous Recombination Deficiency in the AGO-TR1 Trial. Clin. Cancer. Res. 2021, 27, 6559–6569. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http.://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 17 May 2022).
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://broadinstitute.github.io/picard (accessed on 17 May 2022).
- Scheinin, I.; Sie, D.; Bengtsson, H.; van de Wiel, M.A.; Olshen, A.B.; van Thuijl, H.F.; van Essen, H.F.; Eijk, P.P.; Rustenburg, F.; Meijer, G.A.; et al. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014, 24, 2022–2032. [Google Scholar] [CrossRef] [PubMed]
- Bedrosian, I.; Lee, C.; Tucker, S.L.; Palla, S.L.; Lu, K.; Keyomarsi, K. Cyclin E-associated kinase activity predicts response to platinum-based chemotherapy. Clin. Cancer Res. 2007, 13, 4800–4806. [Google Scholar] [CrossRef] [PubMed]
- Alsop, K.; Fereday, S.; Meldrum, C.; deFazio, A.; Emmanuel, C.; George, J.; Dobrovic, A.; Birrer, M.J.; Webb, P.M.; Stewart, C.; et al. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: A report from the Australian Ovarian Cancer Study Group. J. Clin. Oncol. 2012, 30, 2654–2663. [Google Scholar] [CrossRef]
- Moule, R.; Sohaib, A.; Eeles, R. Dramatic response to platinum in a patient with cancer with a germline BRCA2 mutation. Clin. Oncol. 2009, 21, 444–447. [Google Scholar] [CrossRef]
- Pinto, M.L.; Rios, E.; Duraes, C.; Ribeiro, R.; Machado, J.C.; Mantovani, A.; Barbosa, M.A.; Carneiro, F.; Oliveira, M.J. The Two Faces of Tumor-Associated Macrophages and Their Clinical Significance in Colorectal Cancer. Front. Immunol. 2019, 10, 1875. [Google Scholar] [CrossRef]
- Yang, M.; McKay, D.; Pollard, J.W.; Lewis, C.E. Diverse Functions of Macrophages in Different Tumor Microenvironments. Cancer Res. 2018, 78, 5492–5503. [Google Scholar] [CrossRef]
- Martin de la Fuente, L.; Westbom-Fremer, S.; Arildsen, N.S.; Hartman, L.; Malander, S.; Kannisto, P.; Masback, A.; Hedenfalk, I. PD-1/PD-L1 expression and tumor-infiltrating lymphocytes are prognostically favorable in advanced high-grade serous ovarian carcinoma. Virchows. Arch. 2020, 477, 83–91. [Google Scholar] [CrossRef]
- McAlpine, J.N.; Porter, H.; Kobel, M.; Nelson, B.H.; Prentice, L.M.; Kalloger, S.E.; Senz, J.; Milne, K.; Ding, J.; Shah, S.P.; et al. BRCA1 and BRCA2 mutations correlate with TP53 abnormalities and presence of immune cell infiltrates in ovarian high-grade serous carcinoma. Mod. Pathol. 2012, 25, 740–750. [Google Scholar] [CrossRef]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.; Robert, L.; Chmielowski, B.; Spasic, M.; Henry, G.; Ciobanu, V.; et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef]
- Konstantinopoulos, P.A.; Spentzos, D.; Karlan, B.Y.; Taniguchi, T.; Fountzilas, E.; Francoeur, N.; Levine, D.A.; Cannistra, S.A. Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J. Clin. Oncol. 2010, 28, 3555–3561. [Google Scholar] [CrossRef] [PubMed]
- Vollebergh, M.A.; Lips, E.H.; Nederlof, P.M.; Wessels, L.F.; Schmidt, M.K.; van Beers, E.H.; Cornelissen, S.; Holtkamp, M.; Froklage, F.E.; de Vries, E.G.; et al. An aCGH classifier derived from BRCA1-mutated breast cancer and benefit of high-dose platinum-based chemotherapy in HER2-negative breast cancer patients. Ann. Oncol. 2011, 22, 1561–1570. [Google Scholar] [CrossRef] [PubMed]
- Schouten, P.C.; van Dyk, E.; Braaf, L.M.; Mulder, L.; Lips, E.H.; de Ronde, J.J.; Holtman, L.; Wesseling, J.; Hauptmann, M.; Wessels, L.F.; et al. Platform comparisons for identification of breast cancers with a BRCA-like copy number profile. Breast Cancer Res. Treat. 2013, 139, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Schouten, P.C.; Marme, F.; Aulmann, S.; Sinn, H.P.; van Essen, H.F.; Ylstra, B.; Hauptmann, M.; Schneeweiss, A.; Linn, S.C. Breast cancers with a BRCA1-like DNA copy number profile recur less often than expected after high-dose alkylating chemotherapy. Clin. Cancer Res. 2015, 21, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, G.; Xue, F.; Edwards, R.; Sood, A.K.; Zhang, W.; Yang, D. Copy number deletion of RAD50 as predictive marker of BRCAness and PARP inhibitor response in BRCA wild type ovarian cancer. Gynecol. Oncol. 2016, 141, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Etemadmoghadam, D.; Weir, B.A.; Au-Yeung, G.; Alsop, K.; Mitchell, G.; George, J.; Australian Ovarian Cancer Study, G.; Davis, S.; D’Andrea, A.D.; Simpson, K.; et al. Synthetic lethality between CCNE1 amplification and loss of BRCA1. Proc. Natl. Acad. Sci. USA 2013, 110, 19489–19494. [Google Scholar] [CrossRef]
- Kanska, J.; Zakhour, M.; Taylor-Harding, B.; Karlan, B.Y.; Wiedemeyer, W.R. Cyclin E as a potential therapeutic target in high grade serous ovarian cancer. Gynecol. Oncol. 2016, 143, 152–158. [Google Scholar] [CrossRef]
- Au-Yeung, G.; Lang, F.; Azar, W.J.; Mitchell, C.; Jarman, K.E.; Lackovic, K.; Aziz, D.; Cullinane, C.; Pearson, R.B.; Mileshkin, L.; et al. Selective Targeting of Cyclin E1-Amplified High-Grade Serous Ovarian Cancer by Cyclin-Dependent Kinase 2 and AKT Inhibition. Clin. Cancer Res. 2017, 23, 1862–1874. [Google Scholar] [CrossRef]
- Polcher, M.; Braun, M.; Friedrichs, N.; Rudlowski, C.; Bercht, E.; Fimmers, R.; Sauerwald, A.; Keyver-Paik, M.D.; Kubler, K.; Buttner, R.; et al. Foxp3(+) cell infiltration and granzyme B(+)/Foxp3(+) cell ratio are associated with outcome in neoadjuvant chemotherapy-treated ovarian carcinoma. Cancer Immunol. Immunother. 2010, 59, 909–919. [Google Scholar] [CrossRef]
- Mesnage, S.J.L.; Auguste, A.; Genestie, C.; Dunant, A.; Pain, E.; Drusch, F.; Gouy, S.; Morice, P.; Bentivegna, E.; Lhomme, C.; et al. Neoadjuvant chemotherapy (NACT) increases immune infiltration and programmed death-ligand 1 (PD-L1) expression in epithelial ovarian cancer (EOC). Ann. Oncol. 2017, 28, 651–657. [Google Scholar] [CrossRef]
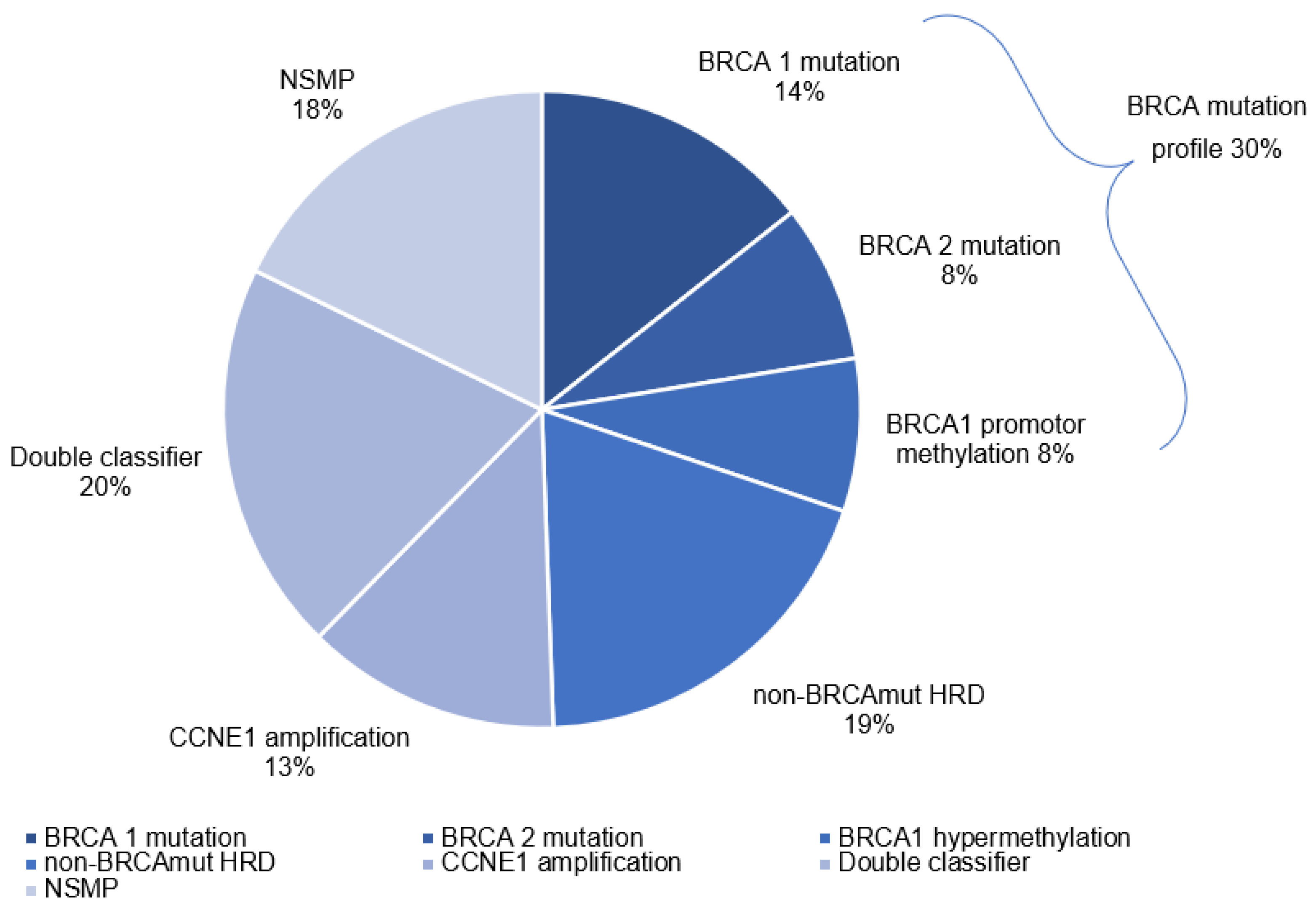

| BRCAm Profile | Non-BRCAmut HRD | CCNE1 Amplification | Double Classifier | NSMP | p-Value | |
|---|---|---|---|---|---|---|
| N = 105 (30.2%) | N = 67 (19.3%) | N = 45 (12.9%) | N = 69 (19.8%) | N = 62 (17.8%) | ||
| Age | <0.001 | |||||
| <65 | 69 (65.7) | 29 (43.3) | 16 (35.6) | 23 (33.3) | 21 (33.9) | |
| 65–75 | 31 (29.5) | 30 (44.8) | 14 (31.1) | 33 (47.8) | 22 (35.5) | |
| >75 | 5 (4.8) | 8 (11.9) | 15 (33.3) | 13 (18.8) | 19 (30.7) | |
| Median (IQR) | 60 (53–66) | 66 (61–71) | 71 (62–77) | 68 (63–73) | 68 (61–76) | <0.001 |
| FIGO stage | ||||||
| II | 7 (6.7) | 5 (7.5) | 1 (2.2) | 5 (7.3) | 0 (0.0) | 0.598 |
| III | 68 (64.8) | 41 (61.2) | 31 (68.9) | 41 (59.4) | 37 (59.7) | |
| IV | 29 (27.6) | 19 (28.4) | 11 (24.4) | 21 (30.4) | 24 (38.7) | |
| Unknown | 1 (1.0) | 2 (3.0) | 2 (4.4) | 2 (2.9) | 1 (1.6) | |
| Treatment sequence | ||||||
| PDS | 49 (46.7) | 27 (40.3) | 19 (42.2) | 32 (46.4) | 13 (21.0) | 0.014 |
| NACT-IDS | 56 (53.3) | 40 (59.7) | 26 (57.8) | 37 (53.6) | 49 (79.0) | |
| Surgical outcome | ||||||
| Complete | 73 (69.5) | 37 (55.2) | 21 (46.7) | 34 (49.3) | 29 (46.8) | 0.141 |
| Optimal | 22 (21.0) | 25 (37.3) | 19 (42.2) | 26 (37.7) | 24 (38.7) | |
| Suboptimal | 6 (5.7) | 3 (4.5) | 5 (11.1) | 7 (10.1) | 8 (12.9) | |
| Unknown | 4 (3.8) | 2 (3.0) | 0 (0.0) | 2 (2.9) | 1 (1.6) |
| BRCAm Profile | Non-BRCAmut HRD | CCNE1 Amplification | Double Classifier | NSMP | p-Value | |
|---|---|---|---|---|---|---|
| N = 105 (30.2%) | N = 67 (19.3%) | N = 45 (12.9%) | N = 69 (19.8%) | N = 62 (17.8%) | ||
| CD8+ cells | 0.082 | |||||
| <20 | 13 (12.4) | 13 (19.4) | 13 (28.9) | 14 (20.3) | 14 (22.6) | |
| 20–100 | 58 (55.2) | 42 (62.7) | 23 (51.1) | 39 (56.5) | 26 (41.9) | |
| >100 | 34 (32.4) | 12 (17.9) | 9 (20.0) | 16 (23.2) | 22 (35.5) | |
| CD103+ cells | ||||||
| <20 | 28 (26.7) | 26 (38.8) | 19 (42.2) | 21 (30.4) | 19 (30.7) | 0.290 |
| 20–100 | 48 (45.7) | 28 (41.8) | 21 (46.7) | 36 (52.2) | 32 (51.6) | |
| >100 | 29 (27.6) | 13 (19.4) | 5 (11.1) | 12 (17.4) | 11 (17.7) | |
| CD20+ cells | ||||||
| <20 | 55 (52.4) | 38 (56.7) | 31 (68.9) | 40 (58.0) | 31 (50.0) | 0.232 |
| 20–50 | 28 (26.7) | 22 (32.8) | 12 (26.7) | 18 (26.1) | 18 (29.0) | |
| >50 | 22 (20.9) | 7 (10.5) | 2 (4.4) | 11 (15.9) | 13 (21.0) | |
| CD68+ cells | ||||||
| <50 | 42 (40) | 41 (61.2) | 24 (53.3) | 44 (63.8) | 40 (64.5) | <0.001 |
| 50–100 | 16 (15.2) | 11 (16.4) | 12 (26.7) | 9 (13.0) | 12 (19.4) | |
| >100 | 47 (44.8) | 15 (22.4) | 9 (20) | 16 (23.2) | 10 (16.1) |
| Median Progression Free Survival, Months (IQR) | Crude HR (95% CI) | Adjusted * HR (95% CI) | |
|---|---|---|---|
| Molecular Subtype | |||
| BRCAm profile | 22.3 (15–60) | REF | REF |
| non-BRCAmut HRD | 16.9 (11–47) | 1.28 (0.89–1.86) | 1.22 (0.84–1.79) |
| CCNE1 amplification | 14.8 (12–21) | 1.94 (1.29–2.91) | 1.57 (1.02–2.43) |
| Double classifier | 14.7 (11–29) | 1.90 (1.33–2.72) | 1.77 (1.22–2.56) |
| NSMP | 16.7 (11–32) | 1.60 (1.12–2.30) | 1.05 (0.72–1.55) |
| Median Survival, Months (IQR) | Crude HR (95% CI) | Adjusted * HR (95% CI) | |
|---|---|---|---|
| CD8 | |||
| ≤100 | 36.3 (21–65) | REF | REF |
| >100 | 44.2 (25–97) | 0.73 (0.56–0.95) | 0.72 (0.55–0.94) |
| CD20 | |||
| ≤50 | 36.9 (21–68) | REF | REF |
| >50 | 44.2 (25–97) | 0.72 (0.52–1.00) | 0.71 (0.51–0.98) |
| CD68 | |||
| ≤100 | 36.8 (21–63) | REF | REF |
| >100 | 47.7 (24–97) | 0.72 (0.56–0.94) | 0.94 (0.71–1.23) |
| CD103 | |||
| ≤100 | 36.3 (22–63) | REF | REF |
| >100 | 52.1 (25–110) | 0.61 (0.45–0.83) | 0.68 (0.50–0.93) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Wagensveld, L.; van Baal, J.O.A.M.; Timmermans, M.; Gaillard, D.; Borghuis, L.; Coffelt, S.B.; Rosenberg, E.H.; Lok, C.A.R.; Nijman, H.W.; Kooreman, L.F.S.; et al. Homologous Recombination Deficiency and Cyclin E1 Amplification Are Correlated with Immune Cell Infiltration and Survival in High-Grade Serous Ovarian Cancer. Cancers 2022, 14, 5965. https://doi.org/10.3390/cancers14235965
van Wagensveld L, van Baal JOAM, Timmermans M, Gaillard D, Borghuis L, Coffelt SB, Rosenberg EH, Lok CAR, Nijman HW, Kooreman LFS, et al. Homologous Recombination Deficiency and Cyclin E1 Amplification Are Correlated with Immune Cell Infiltration and Survival in High-Grade Serous Ovarian Cancer. Cancers. 2022; 14(23):5965. https://doi.org/10.3390/cancers14235965
Chicago/Turabian Stylevan Wagensveld, Lilian, Juliette O. A. M. van Baal, Maite Timmermans, Duco Gaillard, Lauri Borghuis, Seth B. Coffelt, Efraim H. Rosenberg, Christianne A. R. Lok, Hans W. Nijman, Loes F. S. Kooreman, and et al. 2022. "Homologous Recombination Deficiency and Cyclin E1 Amplification Are Correlated with Immune Cell Infiltration and Survival in High-Grade Serous Ovarian Cancer" Cancers 14, no. 23: 5965. https://doi.org/10.3390/cancers14235965
APA Stylevan Wagensveld, L., van Baal, J. O. A. M., Timmermans, M., Gaillard, D., Borghuis, L., Coffelt, S. B., Rosenberg, E. H., Lok, C. A. R., Nijman, H. W., Kooreman, L. F. S., Sanders, J., de Bruijn, M., Wessels, L. F. A., van der Wiel, R., Rausch, C., Broeks, A., Kruitwagen, R. F. P. M., van der Aa, M. A., Sonke, G. S., ... Horlings, H. M. (2022). Homologous Recombination Deficiency and Cyclin E1 Amplification Are Correlated with Immune Cell Infiltration and Survival in High-Grade Serous Ovarian Cancer. Cancers, 14(23), 5965. https://doi.org/10.3390/cancers14235965







