Impact of CRISPR/Cas9-Mediated CD73 Knockout in Pancreatic Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Flow Cytometry
2.3. FACS Sorting
2.4. Lentivirus Production
2.5. Lentivirus Transduction
2.6. Deep Sequencing and Data Analysis
2.7. CRISPR/Cas9 Gene Editing
2.8. Confirmation of CRISPR/Cas9-Mediated Knockout
2.9. Quantitative PCR
2.10. Western Blot Analysis
2.11. Cell Cycle Analysis and Apoptosis Assay
2.12. Proliferation Assay
2.13. Colony Formation Assay
2.14. Wound Healing Assay
2.15. Immunofluorescence
2.16. Statistical Analysis
3. Results
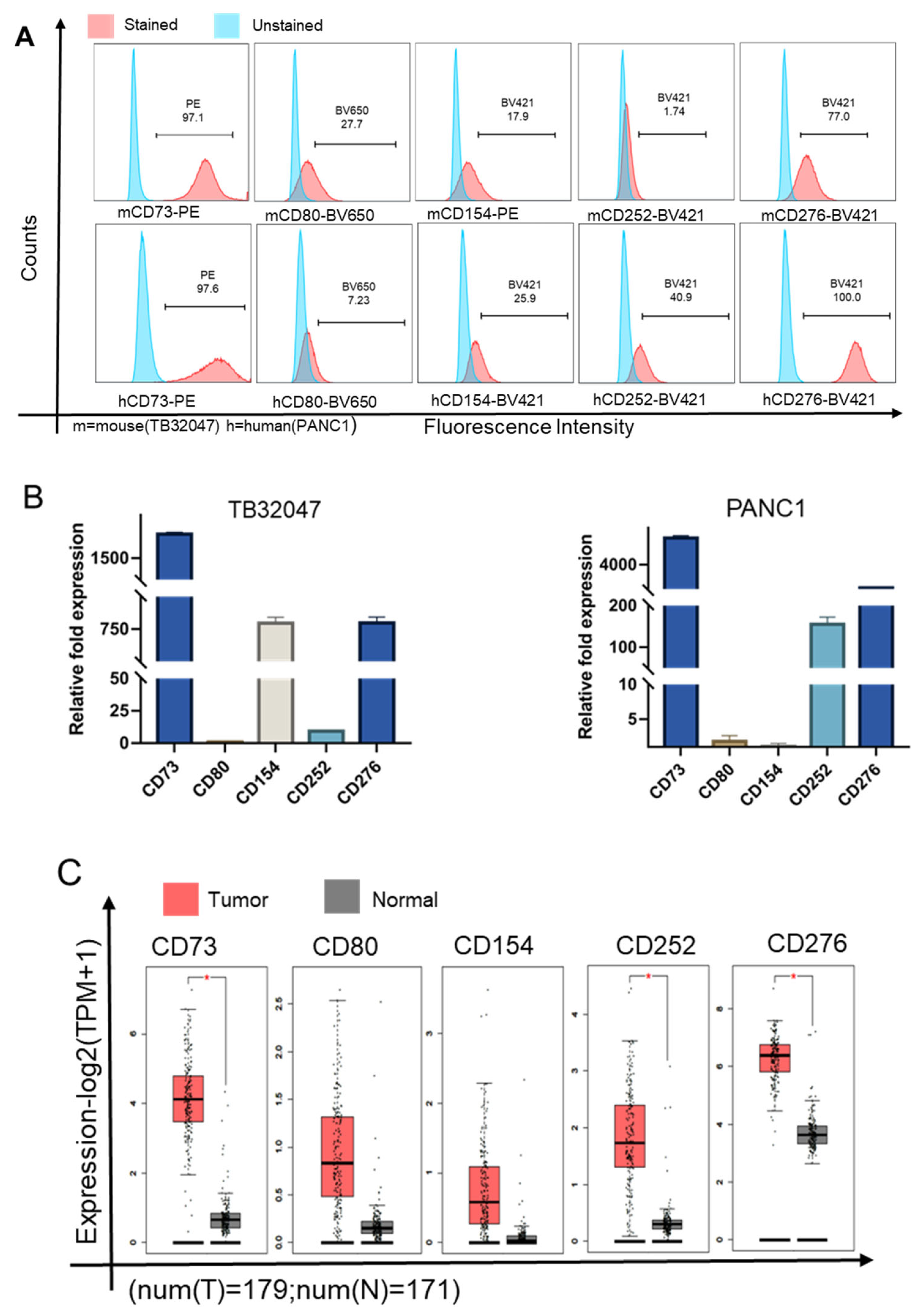
3.1. CD73 Is Highly Expressed in Pancreatic Cancer and Reduces Patient Survival
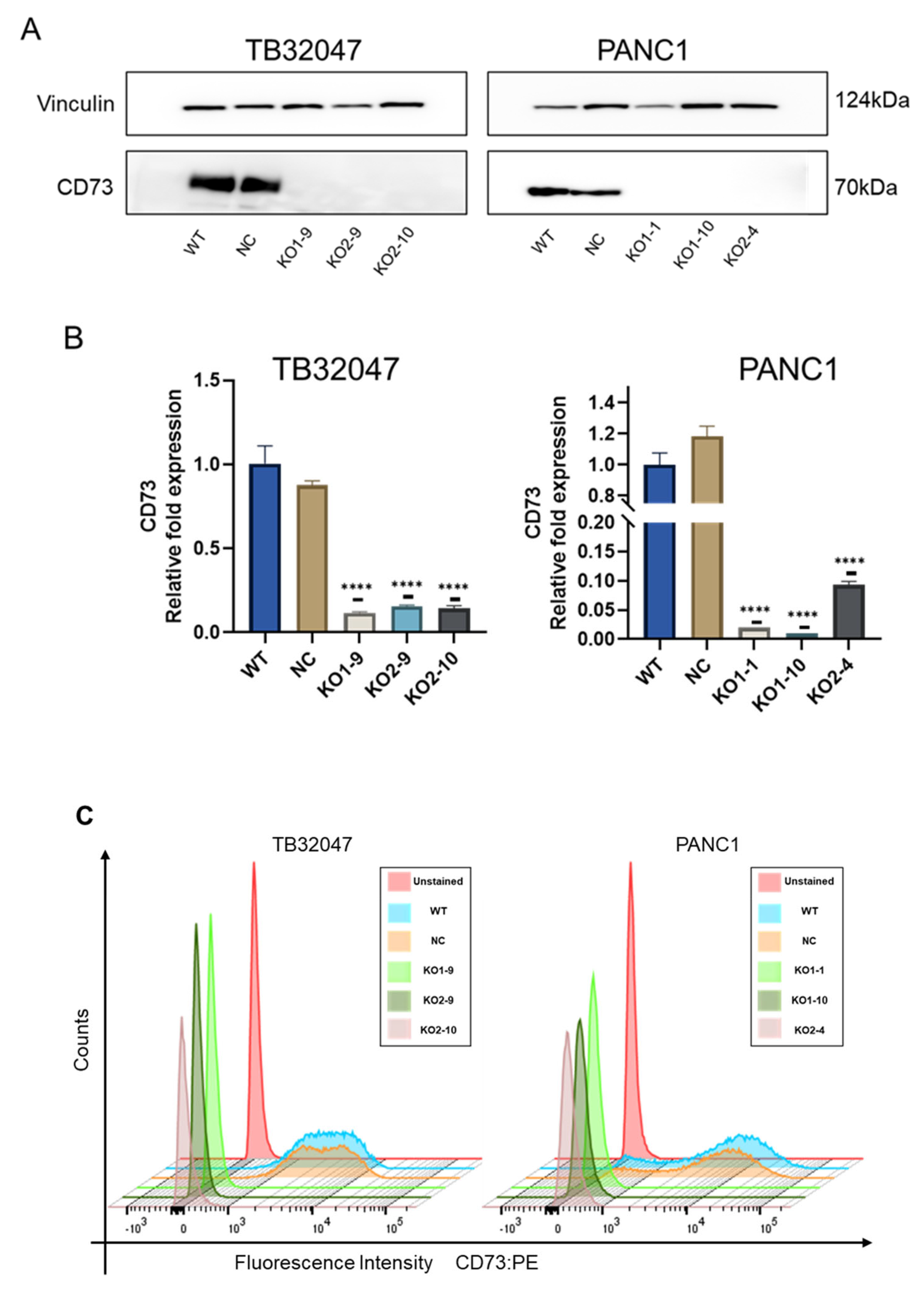
3.2. Expression of CD73 after CRISPR/Cas9-Mediated Knockout in TB32047 Cells and PANC1 Cells
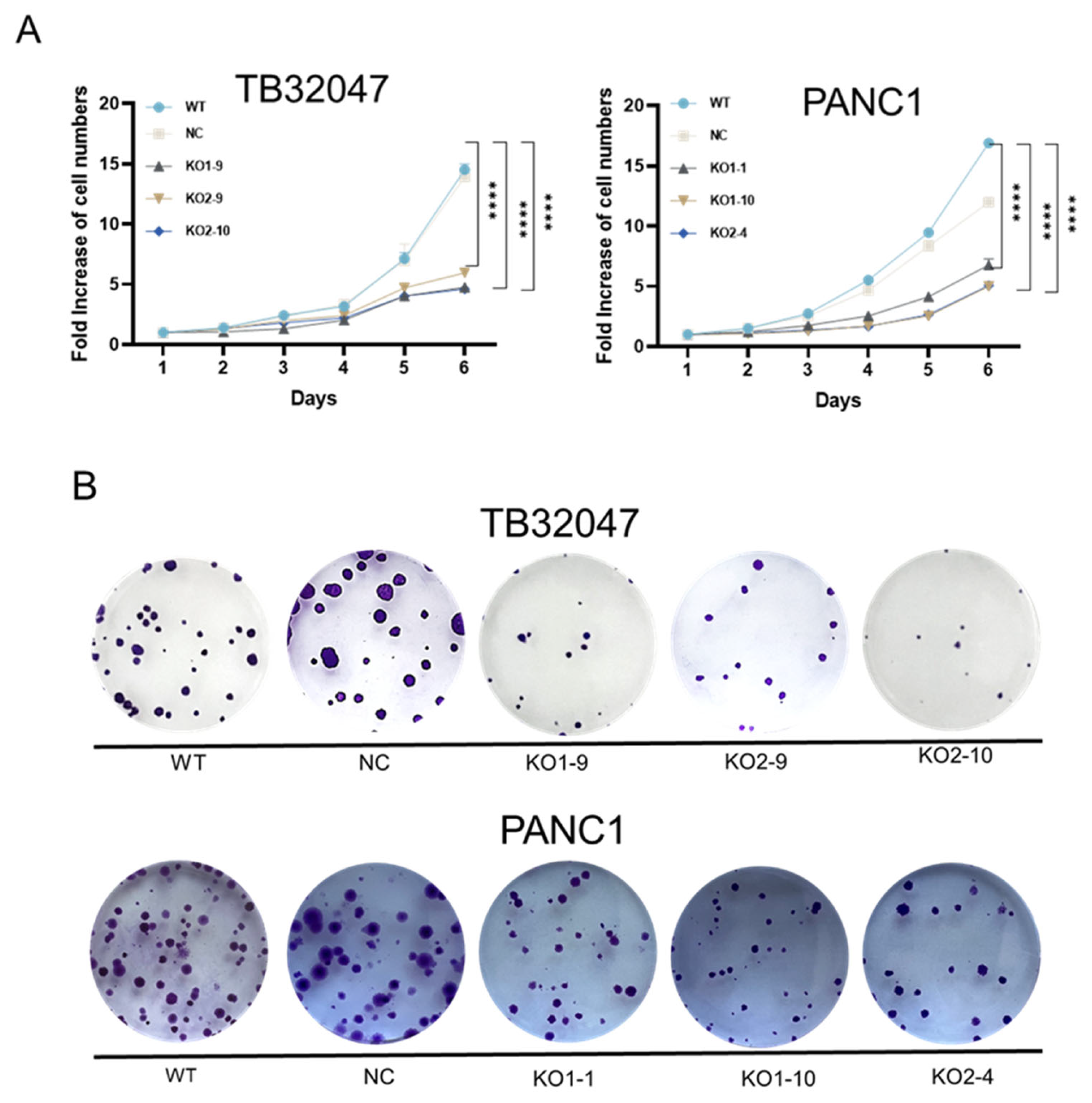
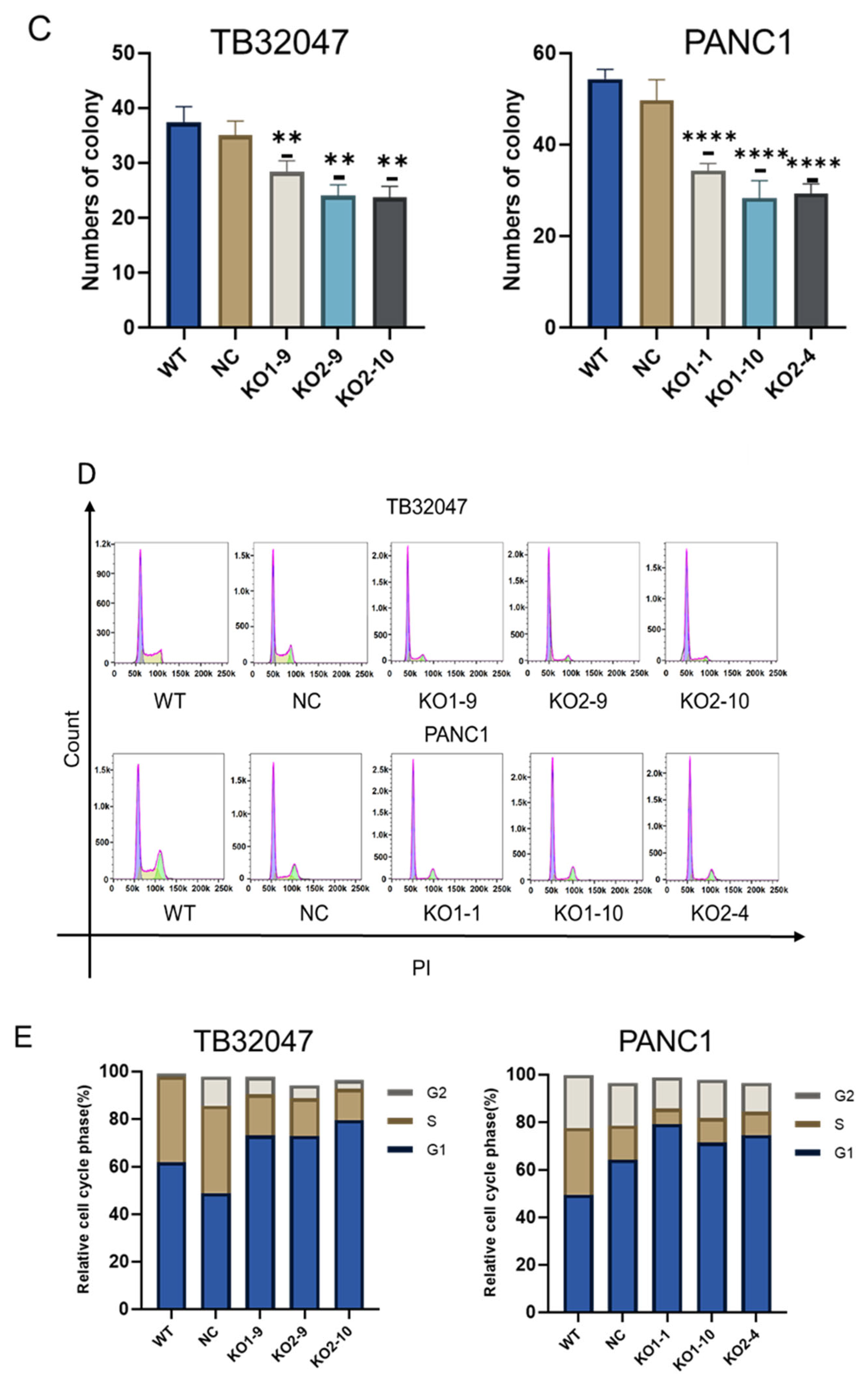
3.3. CD73 Knockout Inhibits Cell Proliferation and Induces G1 Phase Arrest with No Effect on Apoptosis
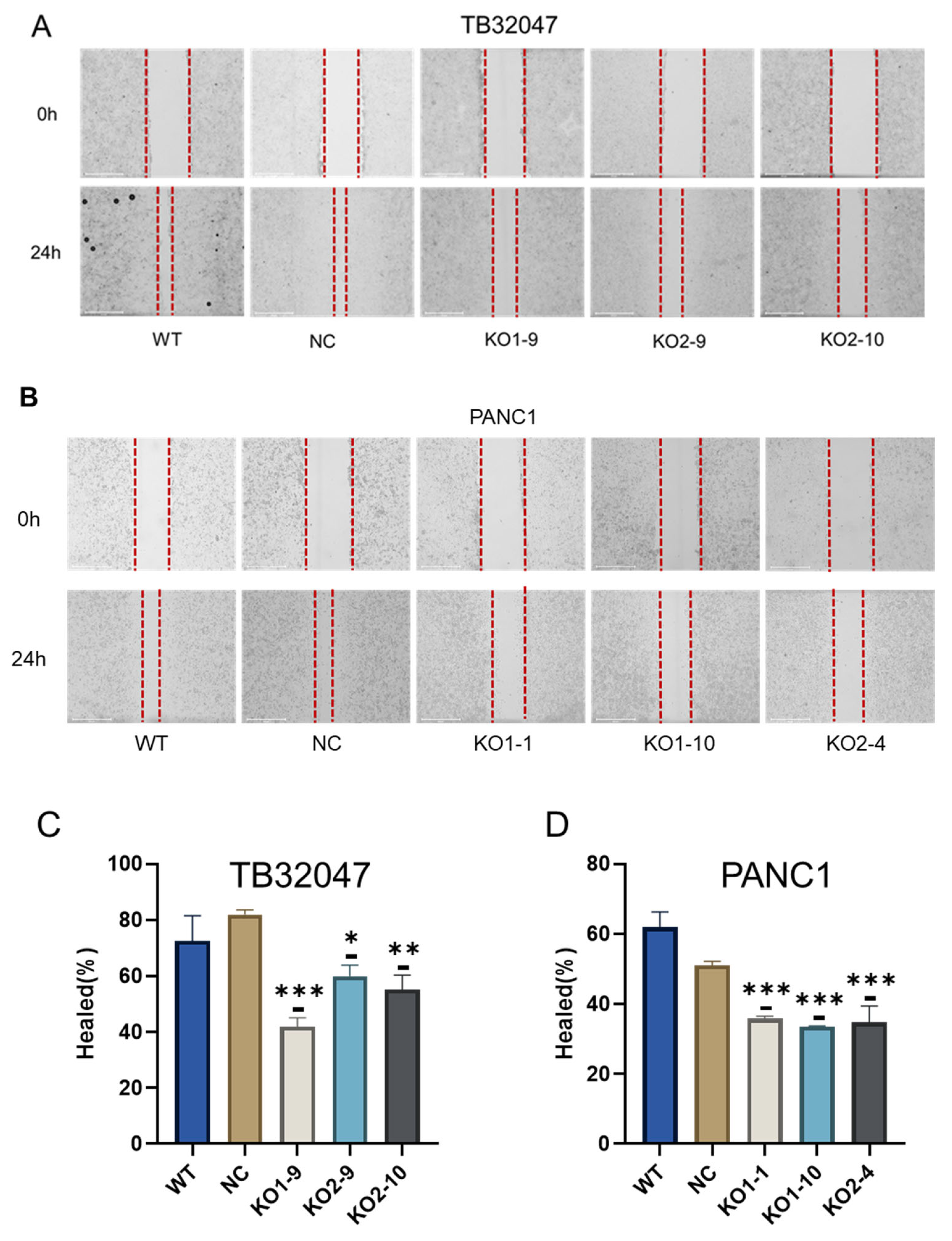
3.4. CD73 Knockout Inhibits Cell Migration In Vitro
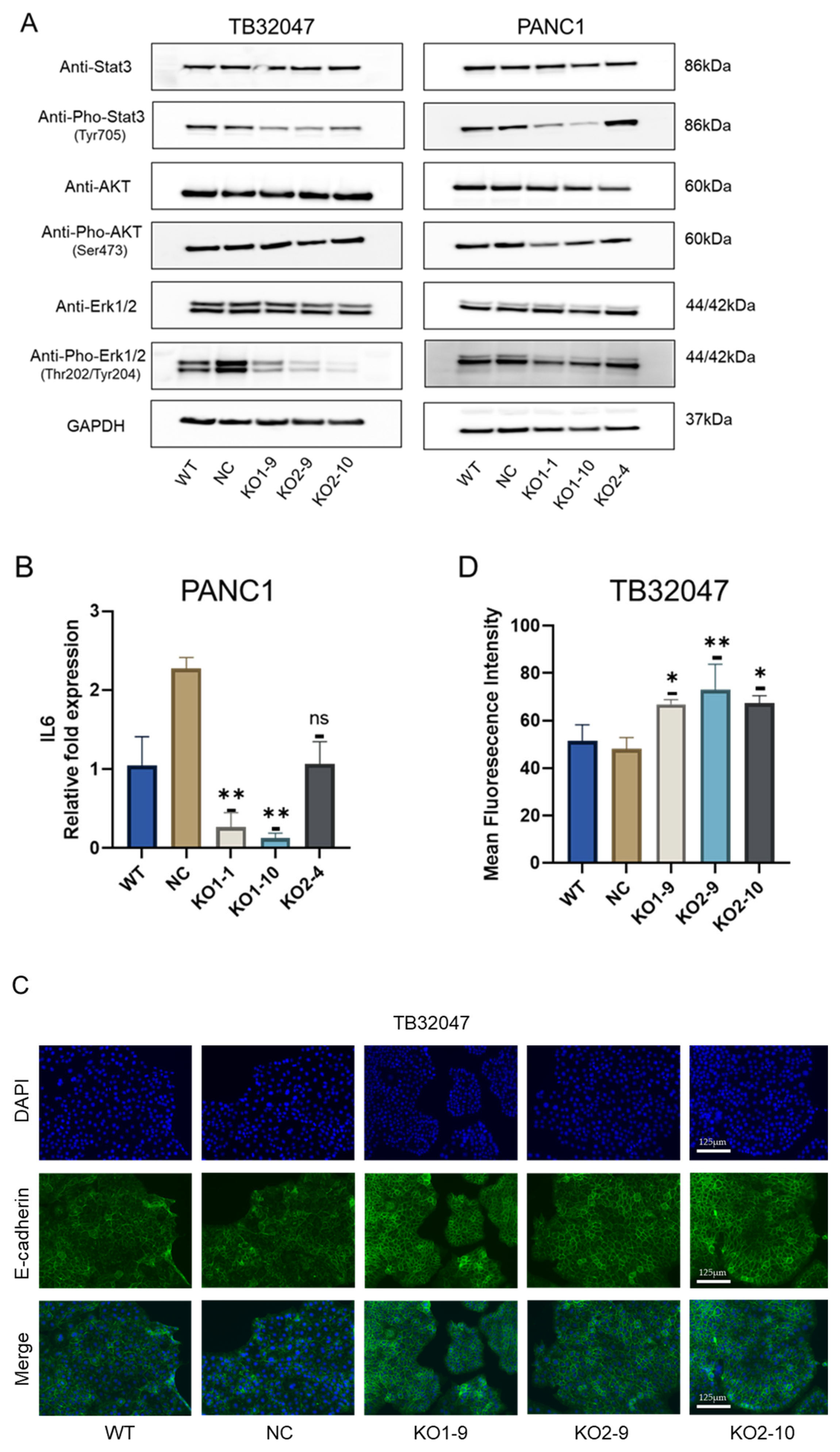
3.5. CD73 Deficiency Inhibits the Phosphorylation of ERK and STAT3 in Pancreatic Cancer, and It Promotes E-Cadherin Expression in TB32047 Cells
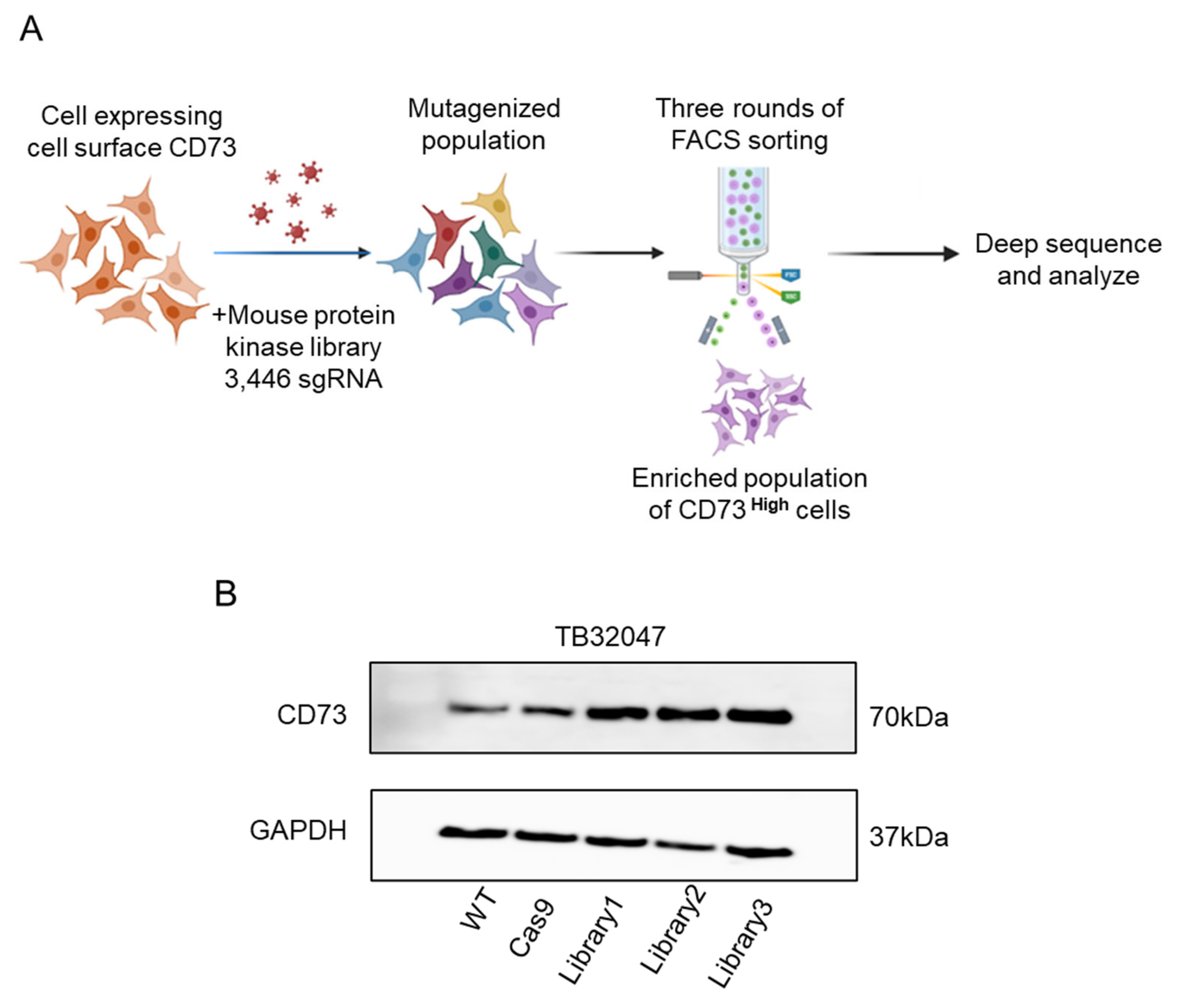
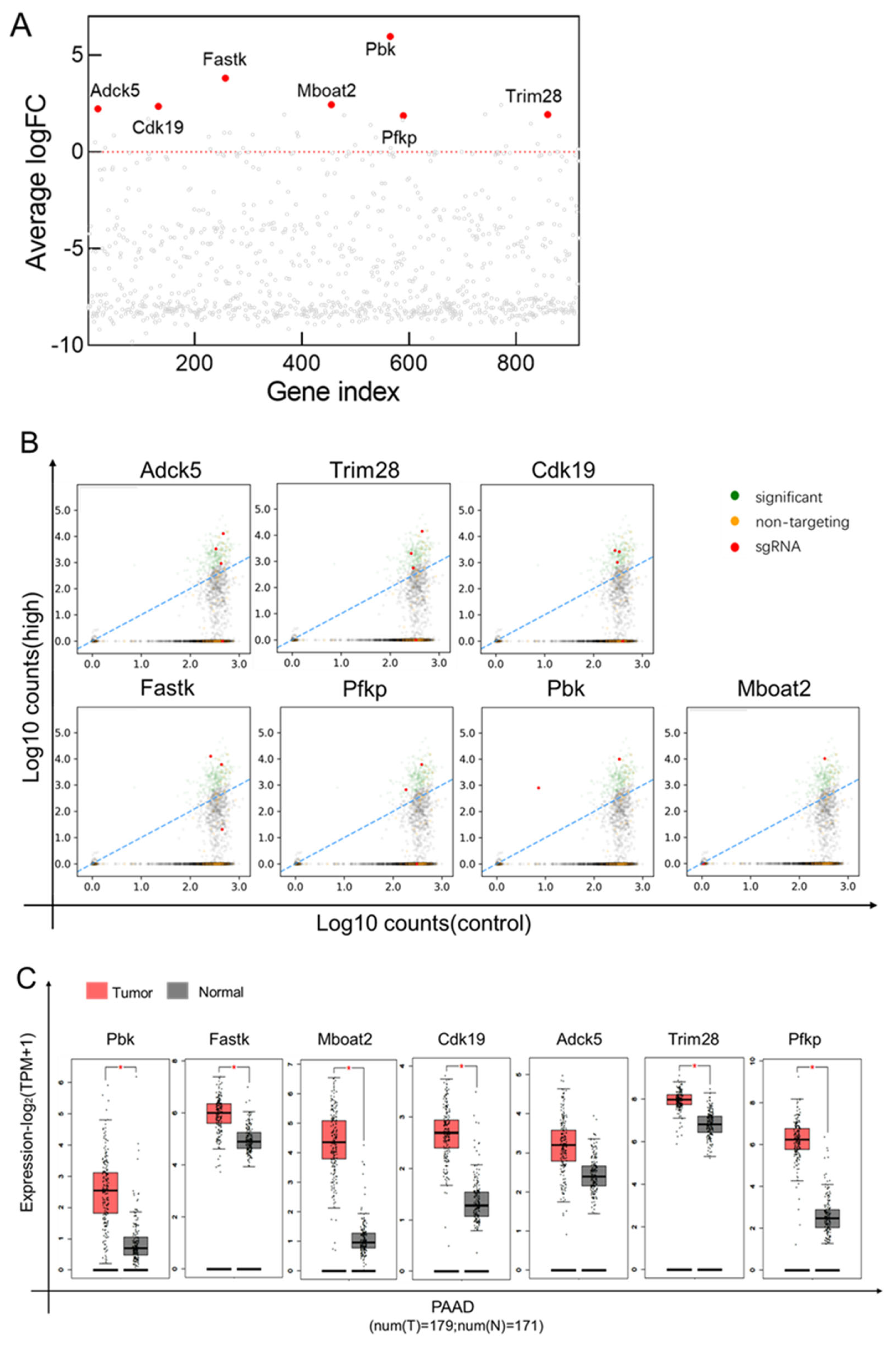
3.6. CRISPR/Cas9 Screen of Inducible Regulators of CD73 Expression in Pancreatic Cancer Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tonini, V.; Zanni, M. Pancreatic Cancer in 2021: What You Need to Know to Win. World J. Gastroenterol. 2021, 27, 5851–5889. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Pancreatic Cancer: An Increasing Global Public Health Concern|Gut. Available online: https://gut.bmj.com/content/71/8/1686.abstract (accessed on 16 January 2023).
- Carioli, G.; Malvezzi, M.; Bertuccio, P.; Boffetta, P.; Levi, F.; La Vecchia, C.; Negri, E. European Cancer Mortality Predictions for the Year 2021 with Focus on Pancreatic and Female Lung Cancer. Ann. Oncol. 2021, 32, 478–487. [Google Scholar] [CrossRef]
- Lianos, G.D.; Christodoulou, D.K.; Katsanos, K.H.; Katsios, C.; Glantzounis, G.K. Minimally Invasive Surgical Approaches for Pancreatic Adenocarcinoma: Recent Trends. J. Gastrointest. Cancer 2017, 48, 129–134. [Google Scholar] [CrossRef]
- Jain, A.; Bhardwaj, V. Therapeutic Resistance in Pancreatic Ductal Adenocarcinoma: Current Challenges and Future Opportunities. World J. Gastroenterol. 2021, 27, 6527–6550. [Google Scholar] [CrossRef]
- Mellman, I.; Coukos, G.; Dranoff, G. Cancer Immunotherapy Comes of Age. Nature 2011, 480, 480–489. [Google Scholar] [CrossRef] [PubMed]
- A Guide to Cancer Immunotherapy: From T Cell Basic Science to Clinical Practice|Nature Reviews Immunology. Available online: https://www.nature.com/articles/s41577-020-0306-5 (accessed on 16 January 2023).
- The Immune System in Cancer Pathogenesis: Potential Therapeutic Approaches. Available online: https://www.hindawi.com/journals/jir/2016/4273943/ (accessed on 16 January 2023).
- The Blockade of Immune Checkpoints in Cancer Immunotherapy|Nature Reviews Cancer. Available online: https://www.nature.com/articles/nrc3239 (accessed on 16 January 2023).
- IJMS|Free Full-Text|Immune Checkpoints as a Target for Colorectal Cancer Treatment. Available online: https://www.mdpi.com/1422-0067/18/6/1324 (accessed on 16 January 2023).
- Buisseret, L.; Pommey, S.; Allard, B.; Garaud, S.; Bergeron, M.; Cousineau, I.; Ameye, L.; Bareche, Y.; Paesmans, M.; Crown, J.P.A.; et al. Clinical Significance of CD73 in Triple-Negative Breast Cancer: Multiplex Analysis of a Phase III Clinical Trial. Ann. Oncol. 2018, 29, 1056–1062. [Google Scholar] [CrossRef] [PubMed]
- Shahbandi, A.; Chiu, F.-Y.; Ungerleider, N.A.; Kvadas, R.; Mheidly, Z.; Sun, M.J.S.; Tian, D.; Waizman, D.A.; Anderson, A.Y.; Machado, H.L.; et al. Breast Cancer Cells Survive Chemotherapy by Activating Targetable Immune-Modulatory Programs Characterized by PD-L1 or CD80. Nat. Cancer 2022, 3, 1513–1533. [Google Scholar] [CrossRef]
- Tong, A.W.; Papayoti, M.H.; Netto, G.; Armstrong, D.T.; Ordonez, G.; Lawson, J.M.; Stone, M.J. Growth-Inhibitory Effects of CD40 Ligand (CD154) and Its Endogenous Expression in Human Breast Cancer1. Clin. Cancer Res. 2001, 7, 691–703. [Google Scholar] [PubMed]
- Gao, Y.; Zhao, J.; Huang, Z.; Zhao, H.; Guo, Z.; Ma, S.; Tang, X.; Song, W.; Chen, X. In Situ Reprogramming of Tumors for Activating the OX40/OX40 Ligand Checkpoint Pathway and Boosting Antitumor Immunity. ACS Biomater. Sci. Eng. 2022, 9, 4108–4116. [Google Scholar] [CrossRef]
- Picarda, E.; Ohaegbulam, K.C.; Zang, X. Molecular Pathways: Targeting B7-H3 (CD276) for Human Cancer Immunotherapy. Clin. Cancer Res. 2016, 22, 3425–3431. [Google Scholar] [CrossRef] [PubMed]
- Kordaß, T.; Osen, W.; Eichmüller, S.B. Controlling the Immune Suppressor: Transcription Factors and MicroRNAs Regulating CD73/NT5E. Front. Immunol. 2018, 9, 813. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Gao, Z.; Zhang, H. The Role of Adenosinergic Pathway in Human Autoimmune Diseases. Immunol. Res. 2016, 64, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Turiello, R.; Pinto, A.; Morello, S. CD73: A Promising Biomarker in Cancer Patients. Front. Pharmacol. 2020, 11, 609931. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Dong, K.; Zhang, H. The Roles of CD73 in Cancer. BioMed Res. Int. 2014, 2014, e460654. [Google Scholar] [CrossRef]
- Zhang, B. CD73: A Novel Target for Cancer Immunotherapy. Cancer Res. 2010, 70, 6407–6411. [Google Scholar] [CrossRef]
- Chambers, A.M.; Wang, J.; Dao, T.N.; Lupo, K.B.; Veenhuis, P.; Ayers, M.G.; Slivova, V.; Cohen-Gadol, A.A.; Matosevic, S. Functional Expression of CD73 on Human Natural Killer Cells. Cancer Immunol. Immunother. 2022, 71, 3043–3056. [Google Scholar] [CrossRef]
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.; Orchard, R.; et al. Optimized SgRNA Design to Maximize Activity and Minimize Off-Target Effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Spahn, P.N.; Bath, T.; Weiss, R.J.; Kim, J.; Esko, J.D.; Lewis, N.E.; Harismendy, O. PinAPL-Py: A Comprehensive Web-Application for the Analysis of CRISPR/Cas9 Screens. Sci. Rep. 2017, 7, 15854. [Google Scholar] [CrossRef]
- Hu, G.; Cheng, P.; Pan, J.; Wang, S.; Ding, Q.; Jiang, Z.; Cheng, L.; Shao, X.; Huang, L.; Huang, J. An IL6–Adenosine Positive Feedback Loop between CD73+ ΓδTregs and CAFs Promotes Tumor Progression in Human Breast Cancer. Cancer Immunol. Res. 2020, 8, 1273–1286. [Google Scholar] [CrossRef]
- Ansari, D.; Gustafsson, A.; Andersson, R. Update on the Management of Pancreatic Cancer: Surgery Is Not Enough. World J. Gastroenterol. 2015, 21, 3157–3165. [Google Scholar] [CrossRef] [PubMed]
- Johansson, H.; Andersson, R.; Bauden, M.; Hammes, S.; Holdenrieder, S.; Ansari, D. Immune Checkpoint Therapy for Pancreatic Cancer. World J. Gastroenterol. 2016, 22, 9457–9476. [Google Scholar] [CrossRef] [PubMed]
- Pennock, G.K.; Chow, L.Q.M. The Evolving Role of Immune Checkpoint Inhibitors in Cancer Treatment. Oncologist 2015, 20, 812–822. [Google Scholar] [CrossRef] [PubMed]
- Bazhin, A.V.; Shevchenko, I.; Umansky, V.; Werner, J.; Karakhanova, S. Two Immune Faces of Pancreatic Adenocarcinoma: Possible Implication for Immunotherapy. Cancer Immunol. Immunother. 2014, 63, 59–65. [Google Scholar] [CrossRef]
- Zhou, L.; Jia, S.; Chen, Y.; Wang, W.; Wu, Z.; Yu, W.; Zhang, M.; Ding, G.; Cao, L. The Distinct Role of CD73 in the Progression of Pancreatic Cancer. J. Mol. Med. 2019, 97, 803–815. [Google Scholar] [CrossRef]
- Lavoie, H.; Gagnon, J.; Therrien, M. ERK Signalling: A Master Regulator of Cell Behaviour, Life and Fate. Nat. Rev. Mol. Cell Biol. 2020, 21, 607–632. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, H.T. MAPK Signal Pathways in the Regulation of Cell Proliferation in Mammalian Cells. Cell Res. 2002, 12, 9–18. [Google Scholar] [CrossRef]
- Shahmarvand, N.; Nagy, A.; Shahryari, J.; Ohgami, R.S. Mutations in the Signal Transducer and Activator of Transcription Family of Genes in Cancer. Cancer Sci. 2018, 109, 926–933. [Google Scholar] [CrossRef]
- Furtek, S.L.; Backos, D.S.; Matheson, C.J.; Reigan, P. Strategies and Approaches of Targeting STAT3 for Cancer Treatment. ACS Chem. Biol. 2016, 11, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Qin, L.; Li, X. Role of STAT3 Signaling Pathway in Breast Cancer. Cell Commun. Signal. 2020, 18, 33. [Google Scholar] [CrossRef]
- Corcoran, R.B.; Contino, G.; Deshpande, V.; Tzatsos, A.; Conrad, C.; Benes, C.H.; Levy, D.E.; Settleman, J.; Engelman, J.A.; Bardeesy, N. STAT3 Plays a Critical Role in KRAS-Induced Pancreatic Tumorigenesis. Cancer Res. 2011, 71, 5020–5029. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 Signalling Axis in Cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Wong, A.S.T.; Gumbiner, B.M. Adhesion-Independent Mechanism for Suppression of Tumor Cell Invasion by E-Cadherin. J. Cell Biol. 2003, 161, 1191–1203. [Google Scholar] [CrossRef] [PubMed]
- Rachagani, S.; Senapati, S.; Chakraborty, S.; Ponnusamy, M.P.; Kumar, S.; Smith, L.M.; Jain, M.; Batra, S.K. Activated KrasG12D Is Associated with Invasion and Metastasis of Pancreatic Cancer Cells through Inhibition of E-Cadherin. Br. J. Cancer 2011, 104, 1038–1048. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Song, Y.; Ye, M.; Zhou, J.; Wang, Z.; Zhu, X. Targeting E-Cadherin Expression with Small Molecules for Digestive Cancer Treatment. Am. J. Transl. Res. 2019, 11, 3932–3944. [Google Scholar]
- Abe, Y.; Matsumoto, S.; Kito, K.; Ueda, N. Cloning and expression of a novel MAPKK-like protein kinase, lymphokine-activated killer T-cell-originated protein kinase, specifically expressed in the testis and activated lymphoid cells. J. Biol. Chem. 2000, 275, 21525–21531. [Google Scholar] [CrossRef]
- Li, W.; Simarro, M.; Kedersha, N.; Anderson, P. FAST is a survival protein that senses mitochondrial stress and modulates TIA-1-regulated changes in protein expression. Mol. Cell. Biol. 2004, 24, 10718–10732. [Google Scholar] [CrossRef] [PubMed]
- Becker, F.; Joerg, V.; Hupe, M.C.; Roth, D.; Krupar, R.; Lubczyk, V.; Kuefer, R.; Sailer, V.; Duensing, S.; Kirfel, J.; et al. Increased mediator complex subunit CDK19 expression associates with aggressive prostate cancer. Int. J. Cancer 2020, 146, 577–588. [Google Scholar] [CrossRef]
- Qiu, M.; Li, G.; Wang, P.; Li, X.; Lai, F.; Luo, R.; Liu, B.; Lin, J. aarF domain containing kinase 5 gene promotes invasion and migration of lung cancer cells through ADCK5-SOX9-PTTG1 pathway. Exp. Cell Res. 2020, 392, 112002. [Google Scholar] [CrossRef]
- Reymond, A.; Meroni, G.; Fantozzi, A.; Merla, G.; Cairo, S.; Luzi, L.; Riganelli, D.; Zanaria, E.; Messali, S.; Cainarca, S.; et al. The tripartite motif family identifies cell compartments. EMBO J. 2001, 20, 2140–2151. [Google Scholar] [CrossRef]
- Lang, L.; Chemmalakuzhy, R.; Shay, C.; Teng, Y. PFKP Signaling at a Glance: An Emerging Mediator of Cancer Cell Metabolism. Adv. Exp. Med. Biol. 2019, 1134, 243–258. [Google Scholar] [CrossRef] [PubMed]
- Flowers, B.M.; Xu, H.; Mulligan, A.S.; Hanson, K.J.; Seoane, J.A.; Vogel, H.; Curtis, C.; Wood, L.D.; Attardi, L.D. Cell-of-Origin Influences Pancreatic Cancer Subtype. Cancer Discov. 2021, 11, 660–677. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Zhang, S.; Dörflein, I.; Ren, X.; Pfeffer, S.; Britzen-Laurent, N.; Grützmann, R.; Duan, X.; Pilarsky, C. Impact of CRISPR/Cas9-Mediated CD73 Knockout in Pancreatic Cancer. Cancers 2023, 15, 4842. https://doi.org/10.3390/cancers15194842
Zhang J, Zhang S, Dörflein I, Ren X, Pfeffer S, Britzen-Laurent N, Grützmann R, Duan X, Pilarsky C. Impact of CRISPR/Cas9-Mediated CD73 Knockout in Pancreatic Cancer. Cancers. 2023; 15(19):4842. https://doi.org/10.3390/cancers15194842
Chicago/Turabian StyleZhang, Jinping, Shuman Zhang, Isabella Dörflein, Xiaofan Ren, Susanne Pfeffer, Nathalie Britzen-Laurent, Robert Grützmann, Xianglong Duan, and Christian Pilarsky. 2023. "Impact of CRISPR/Cas9-Mediated CD73 Knockout in Pancreatic Cancer" Cancers 15, no. 19: 4842. https://doi.org/10.3390/cancers15194842
APA StyleZhang, J., Zhang, S., Dörflein, I., Ren, X., Pfeffer, S., Britzen-Laurent, N., Grützmann, R., Duan, X., & Pilarsky, C. (2023). Impact of CRISPR/Cas9-Mediated CD73 Knockout in Pancreatic Cancer. Cancers, 15(19), 4842. https://doi.org/10.3390/cancers15194842





