The Prognostic and Immune Significance of CILP2 in Pan-Cancer and Its Relationship with the Progression of Pancreatic Cancer
Abstract
:Simple Summary
Abstract
1. Background
2. Methods
2.1. Data Collection and Preparation
2.2. Demographic and Clinical Characteristics
2.3. Pan-Cancer Differential Expression, Genetic Abbreviations Landscape, and DNA Methylation Analysis
2.4. Survival Analysis
2.5. Functional Enrichment and Gene Set Variation Analyses
2.6. TME Analysis, Immune Cell Infiltration, and Immunological Characteristics
2.7. Patients and Specimens
2.8. Cell Lines
2.9. Cell Transfection
2.10. RNA Extraction and qRT-PCR Analysis
2.11. Western Blot
2.12. Cell Counting Kit (CCK- 8) Assay and Colony Assay
2.13. Wound Healing Assay and Transwell Assay
2.14. BALB/c Mice Orthotopic Pancreatic Cancer Model
2.15. C57BL/6 Orthotopic Pancreatic Cancer Mice Model and Reagents
2.16. BALB/c Metastatic Pancreatic Cancer Mice Model
2.17. Immunohistochemistry (IHC) Staining
2.18. Statistical Analysis
3. Results
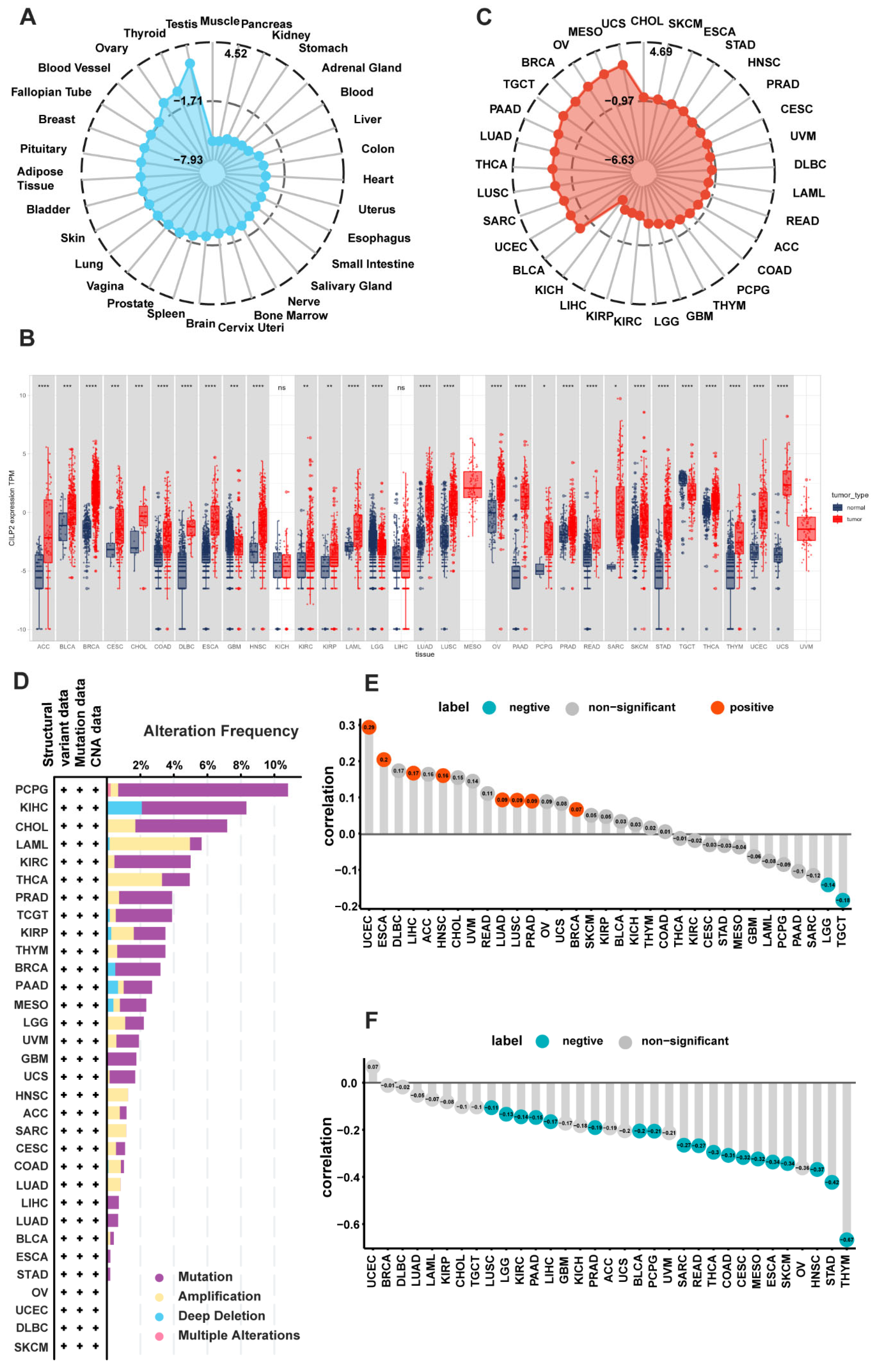
3.1. Tissue-Specific Expression Patterns of CILP2 in Pan-Cancer
3.2. Genetic Alterations and the Epigenetic Variation Landscape in CILP2
3.3. Clinical Prognostic Significance of CILP2 in Pan-Cancer
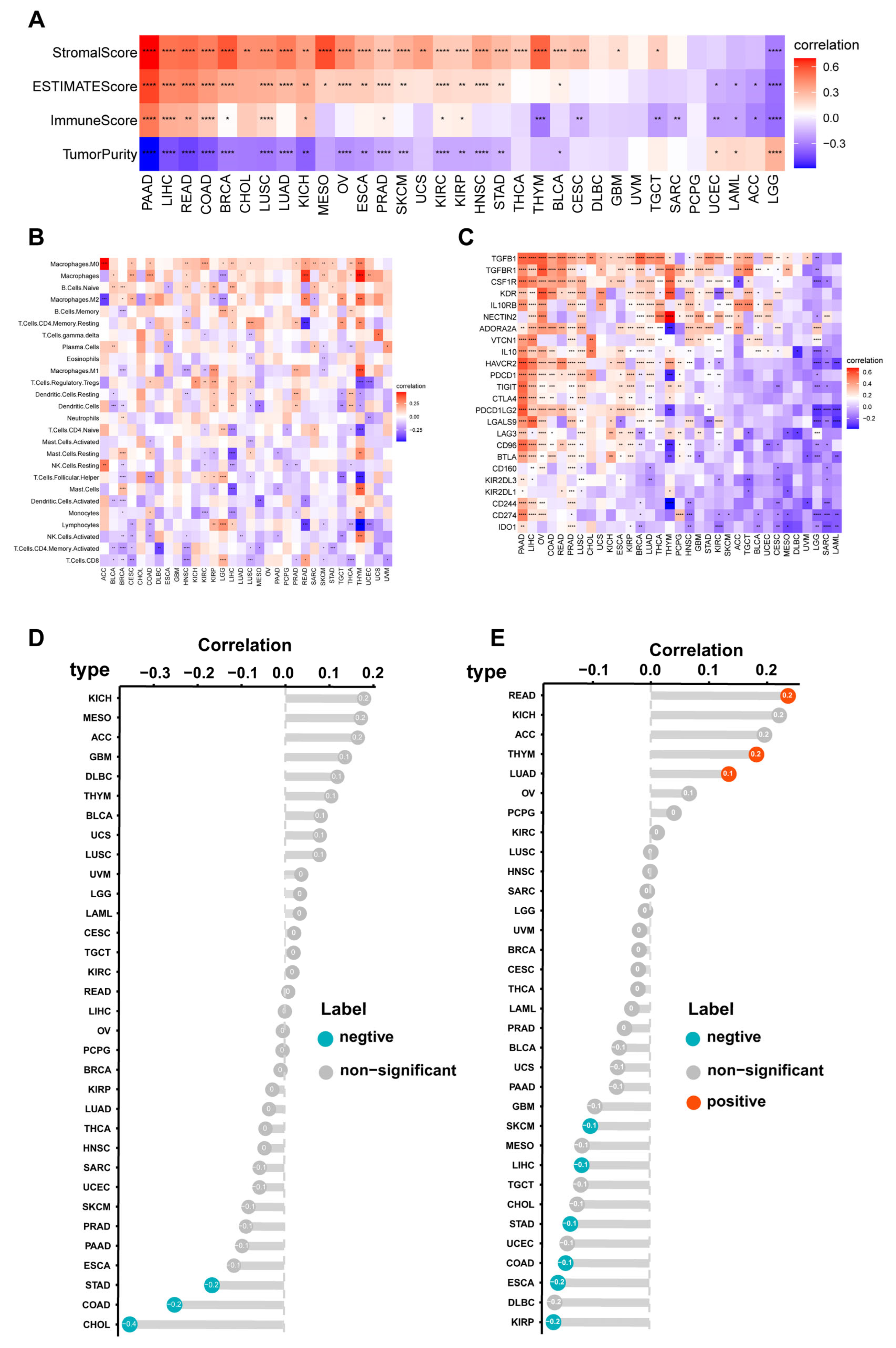
3.4. Immune Cell Infiltration Analyses of CILP2 across the Different Cancers
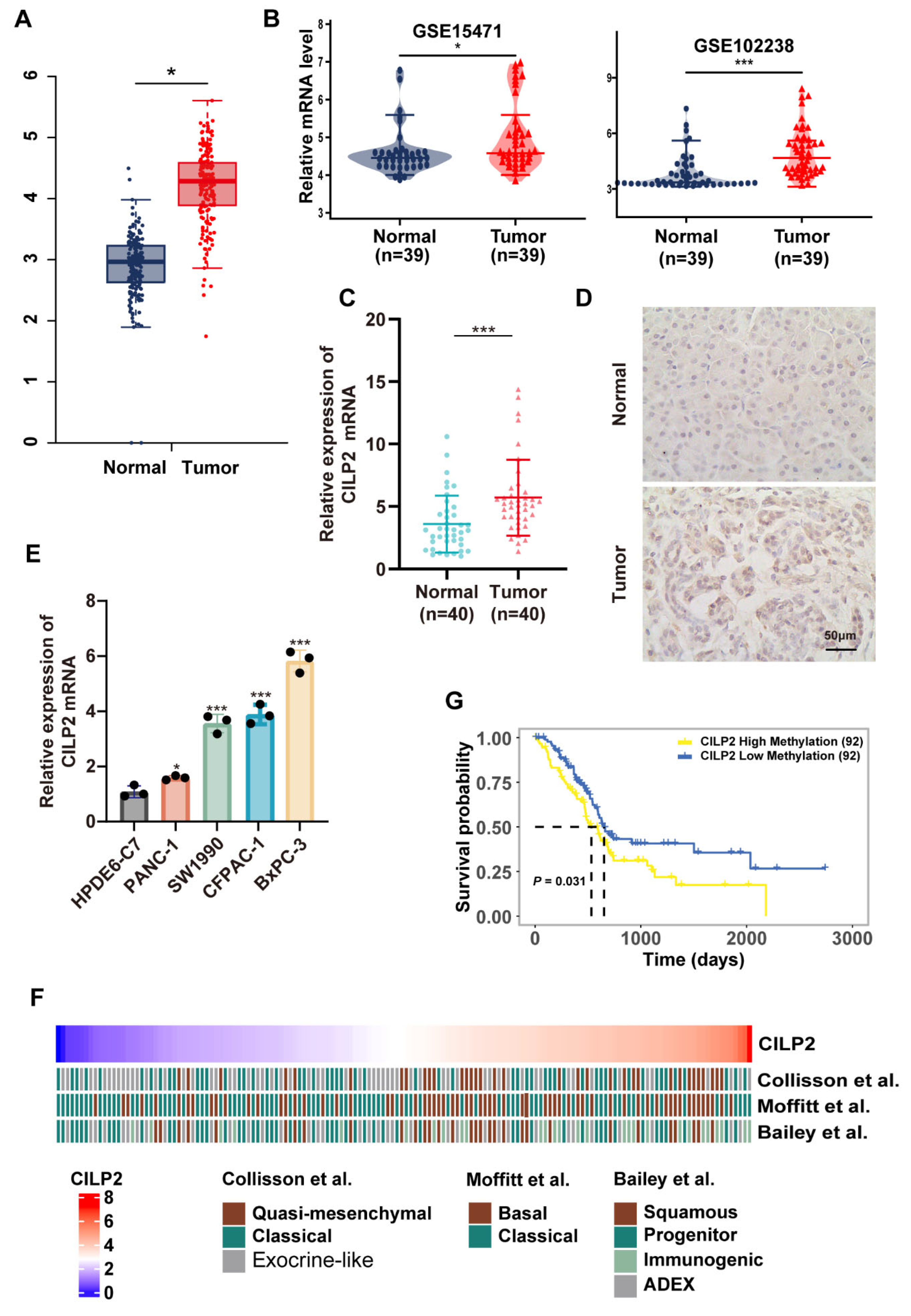
3.5. CILP2 was Highly Expressed in PDAC and was Associated with Subtypes with Poorer Survival
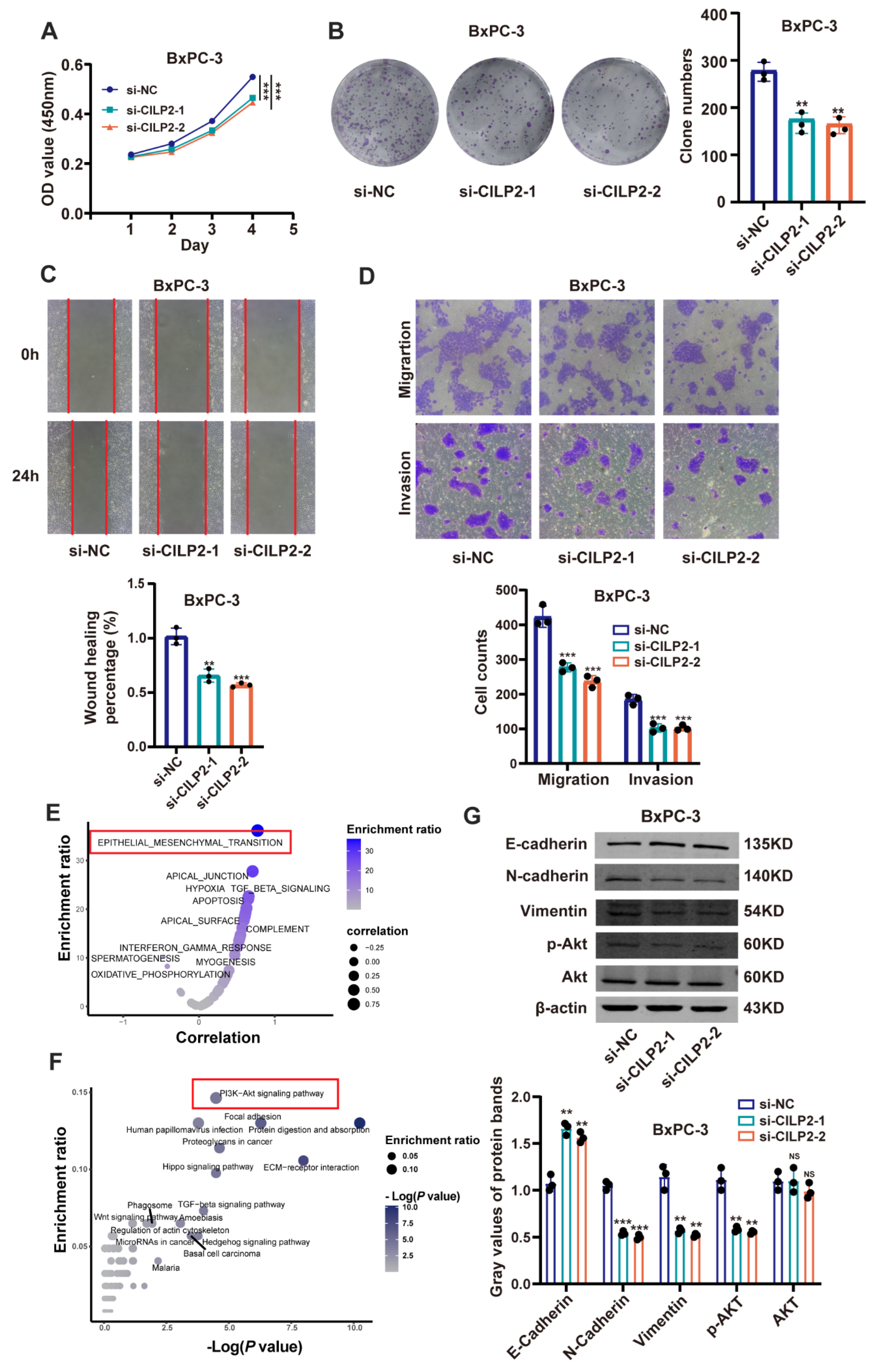
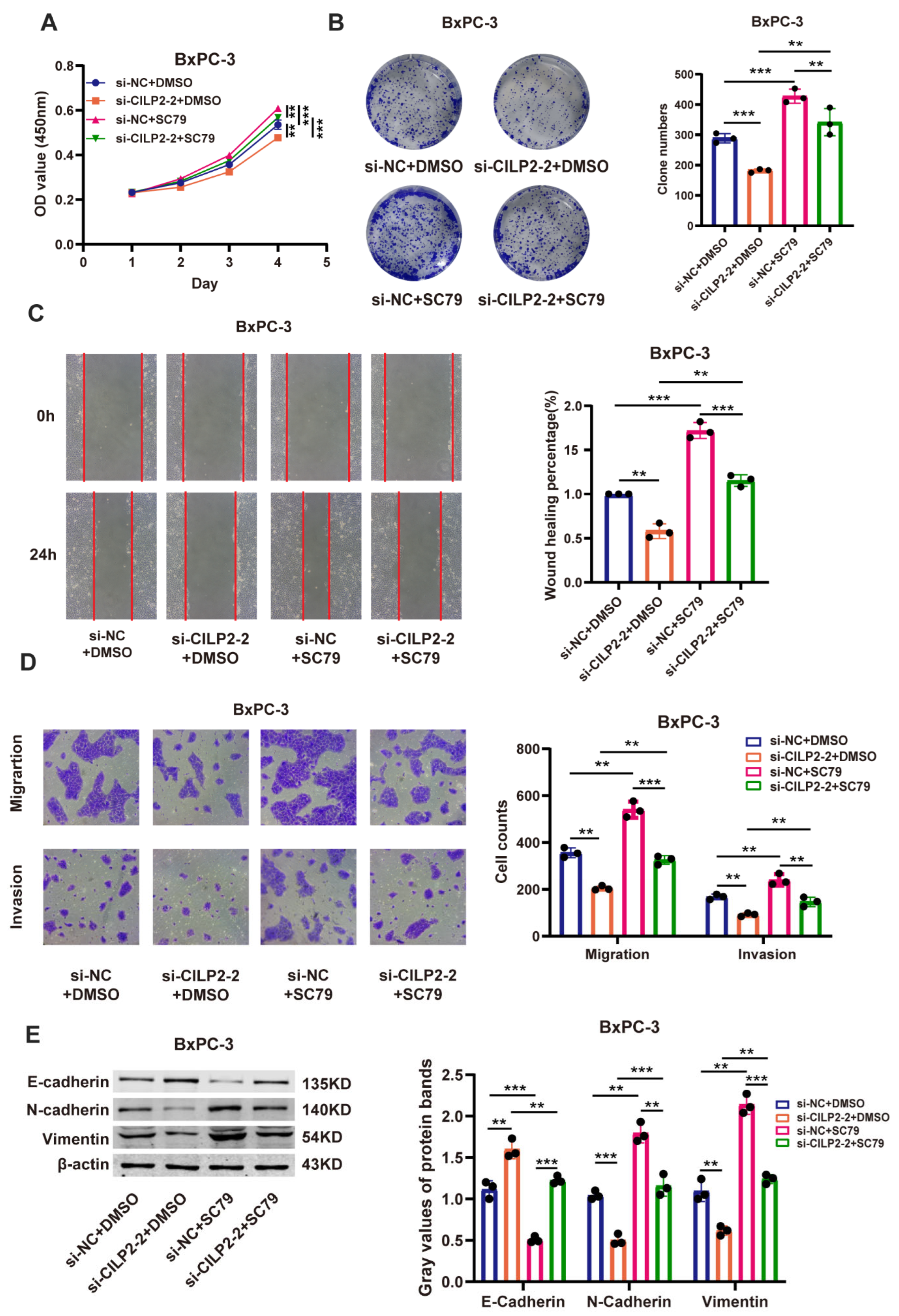
3.6. CILP2 Promoted the Proliferation, Migration, and Invasion of PDAC Cell Lines via the AKT Pathway
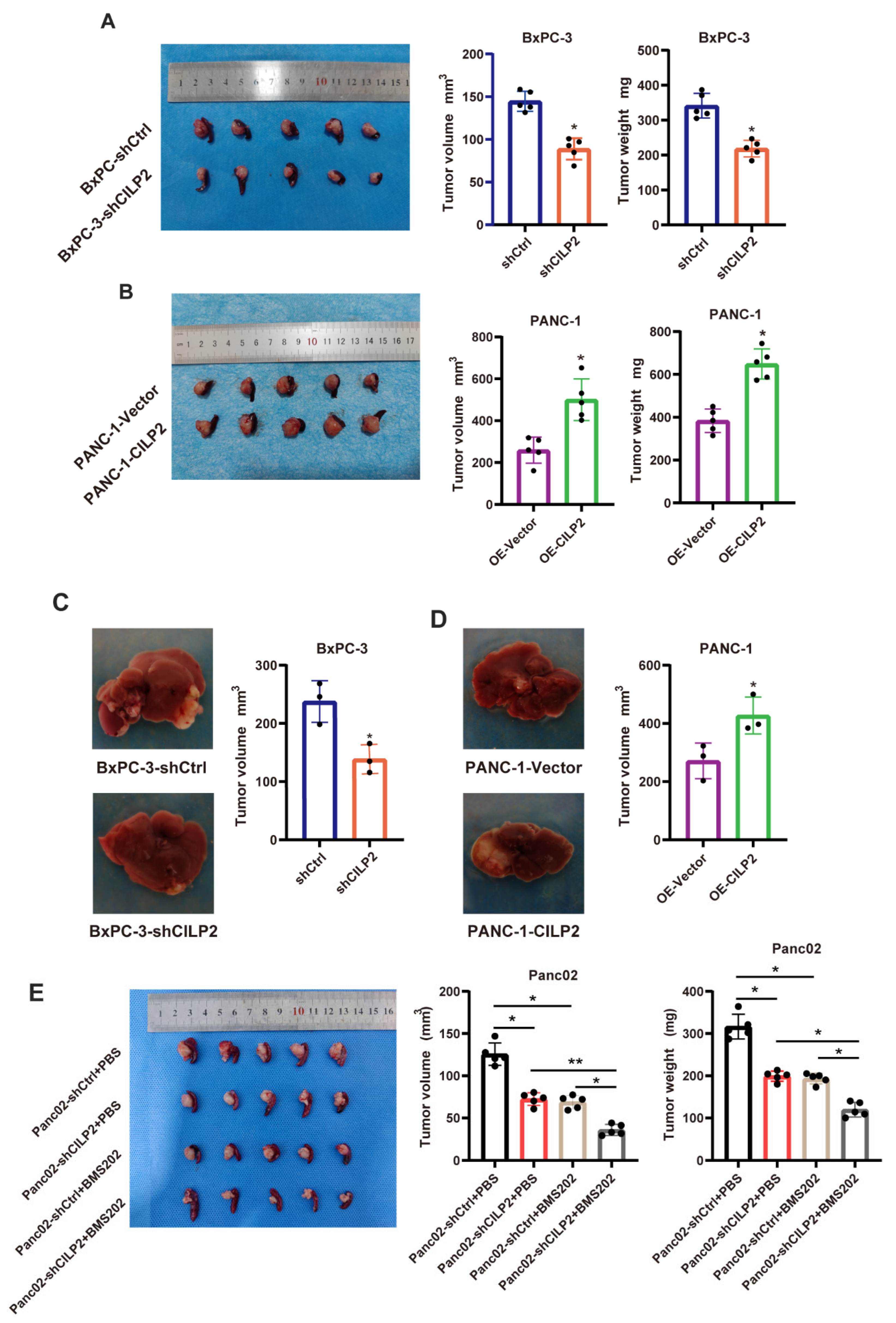
3.7. The Knockdown of CILP2 Inhibited the Progression of Pancreatic Cancer and Synergized with ICIs In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Wahida, A.; Buschhorn, L.; Fröhling, S.; Jost, P.J.; Schneeweiss, A.; Lichter, P.; Kurzrock, R. The coming decade in precision oncology: Six riddles. Nat. Rev. Cancer 2023, 23, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Bedard, P.L.; Hyman, D.M.; Davids, M.S.; Siu, L.L. Small molecules, big impact: 20 years of targeted therapy in oncology. Lancet 2020, 395, 1078–1088. [Google Scholar] [CrossRef]
- Jiang, P.; Sinha, S.; Aldape, K.; Hannenhalli, S.; Sahinalp, C.; Ruppin, E. Big data in basic and translational cancer research. Nat. Rev. Cancer 2022, 22, 625–639. [Google Scholar] [CrossRef]
- Bernardo, B.C.; Belluoccio, D.; Rowley, L.; Little, C.B.; Hansen, U.; Bateman, J.F. Cartilage intermediate layer protein 2 (CILP-2) is expressed in articular and meniscal cartilage and down-regulated in experimental osteoarthritis. J. Biol. Chem. 2011, 286, 37758–37767. [Google Scholar] [CrossRef]
- Zhang, C.L.; Zhao, Q.; Liang, H.; Qiao, X.; Wang, J.Y.; Wu, D.; Wu, L.L.; Li, L. Cartilage intermediate layer protein-1 alleviates pressure overload-induced cardiac fibrosis via interfering TGF-β1 signaling. J. Mol. Cell. Cardiol. 2018, 116, 135–144. [Google Scholar] [CrossRef]
- Huang, F.; Peng, Y.; Ye, Q.; Chen, J.; Li, Y.; Liu, S.; Xu, Y.; Huang, L. CILP2 overexpression correlates with tumor progression and poor prognosis in patients with colorectal cancer in The Cancer Genome Atlas (TCGA) study. World J. Surg. Oncol. 2020, 18, 274. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.; Song, N.; Li, K.; Lei, S.; Wang, J.; Wang, Z.; Zhang, W. CILP2: A prognostic biomarker associated with immune infiltration in colorectal cancer. Heliyon 2023, 9, e15535. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017, 32, 185–203.e13. [Google Scholar] [CrossRef] [PubMed]
- Collisson, E.A.; Sadanandam, A.; Olson, P.; Gibb, W.J.; Truitt, M.; Gu, S.; Cooc, J.; Weinkle, J.; Kim, G.E.; Jakkula, L.; et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011, 17, 500–503. [Google Scholar] [CrossRef] [PubMed]
- Moffitt, R.A.; Marayati, R.; Flate, E.L.; Volmar, K.E.; Loeza, S.G.H.; Hoadley, K.A.; Rashid, N.U.; Williams, L.A.; Eaton, S.C.; Chung, A.H.; et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015, 47, 1168–1178. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.M.; Gingras, M.C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.I.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innov. Camb. Mass. 2021, 2, 100141. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Cancer Syst. Biol. Methods Protoc. 2018, 1711, 243–259. [Google Scholar] [CrossRef]
- Cheng, C.; Liu, D.; Liu, Z.; Li, M.; Wang, Y.; Sun, B.; Kong, R.; Chen, H.; Wang, G.; Li, L.; et al. Positive feedback regulation of lncRNA TPT1-AS1 and ITGB3 promotes cell growth and metastasis in pancreatic cancer. Cancer Sci. 2022, 113, 2986–3001. [Google Scholar] [CrossRef]
- Geng, X.; Li, L.; Luo, Y.; Yang, W.; Hu, J.; Zhao, Z.; Cheng, C.; Zhang, T.; Zhang, Y.; Liu, L.; et al. Tumor Cell Derived Lnc-FSD2-31:1 Contributes to Cancer-Associated Fibroblasts Activation in Pancreatic Ductal Adenocarcinoma Progression through Extracellular Vesicles Cargo MiR-4736. Adv. Sci. 2023, 10, e2203324. [Google Scholar] [CrossRef]
- Collisson, E.A.; Bailey, P.; Chang, D.K.; Biankin, A.V. Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 207–220. [Google Scholar] [CrossRef]
- Raggatt, L.J.; Partridge, N.C. Cellular and molecular mechanisms of bone remodeling. J. Biol. Chem. 2010, 285, 25103–25108. [Google Scholar] [CrossRef]
- Dehne, T.; Karlsson, C.; Ringe, J.; Sittinger, M.; Lindahl, A. Chondrogenic differentiation potential of osteoarthritic chondrocytes and their possible use in matrix-associated autologous chondrocyte transplantation. Arthritis Res. Ther. 2009, 11, R133. [Google Scholar] [CrossRef] [PubMed]
- Johnell, O.; Kanis, J.A. An estimate of the worldwide prevalence and disability associated with osteoporotic fractures. Osteoporos. Int. 2006, 17, 1726–1733. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; McDonald, J.M. Disorders of bone remodeling. Annu. Rev. Pathol. 2011, 6, 121–145. [Google Scholar] [CrossRef] [PubMed]
- Pazianas, M.; Miller, P.D. Osteoporosis and Chronic Kidney Disease-Mineral and Bone Disorder (CKD-MBD): Back to Basics. Am. J. Kidney Dis. 2021, 78, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Marahleh, A.; Kitaura, H.; Ohori, F.; Noguchi, T.; Mizoguchi, I. The osteocyte and its osteoclastogenic potential. Front. Endocrinol. 2023, 14, 1121727. [Google Scholar] [CrossRef] [PubMed]
- Deng, G.X.; Yin, R.X.; Guan, Y.Z.; Liu, C.X.; Zheng, P.F.; Wei, B.L.; Wu, J.Z.; Miao, L. Association of the NCAN-TM6SF2-CILP2-PBX4-SUGP1-MAU2 SNPs and gene-gene and gene-environment interactions with serum lipid levels. Aging 2020, 12, 11893–11913. [Google Scholar] [CrossRef] [PubMed]
- Carlberg, K.; Korotkova, M.; Larsson, L.; Catrina, A.I.; Ståhl, P.L.; Malmström, V. Exploring inflammatory signatures in arthritic joint biopsies with Spatial Transcriptomics. Sci. Rep. 2019, 9, 18975. [Google Scholar] [CrossRef]
- Hu, W.; Li, K.; Han, H.; Geng, S.; Zhou, B.; Fan, X.; Xu, S.; Yang, M.; Liu, H.; Yang, G.; et al. Circulating Levels of CILP2 Are Elevated in Coronary Heart Disease and Associated with Atherosclerosis. Oxid. Med. Cell. Longev. 2020, 2020, 1871984. [Google Scholar] [CrossRef]
- Wu, T.; Zhang, Q.; Wu, S.; Hu, W.; Zhou, T.; Li, K.; Liu, D.; Gu, H.F.; Zheng, H.; Zhu, Z.; et al. CILP-2 is a novel secreted protein and associated with insulin resistance. J. Mol. Cell Biol. 2019, 11, 1083–1094. [Google Scholar] [CrossRef]

| Low (N = 75) | High (N = 75) | p | |
|---|---|---|---|
| Subtype (Moffitt et al.): | 0.003 | ||
| Basal | 23 (30.7%) | 42 (56.0%) | |
| Classical | 52 (69.3%) | 33 (44.0%) | |
| Subtype (Collison et al.): | <0.001 | ||
| QM(Quasi-mesenchymal) | 7 (9.33%) | 27 (36.0%) | |
| Classical | 28 (37.3%) | 26 (34.7%) | |
| Exocrine | 40 (53.3%) | 22 (29.3%) | |
| Subtype (Bailey et al.): | <0.001 | ||
| Squamous | 10 (13.3%) | 21 (28.0%) | |
| Progenitor | 36 (48.0%) | 17 (22.7%) | |
| Immunogenic | 5 (6.67%) | 23 (30.7%) | |
| ADEX | 24 (32.0%) | 14 (18.7%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, D.; He, C.; Liu, Z.; Xu, L.; Li, J.; Zhao, Z.; Hu, X.; Chen, H.; Sun, B.; Wang, Y. The Prognostic and Immune Significance of CILP2 in Pan-Cancer and Its Relationship with the Progression of Pancreatic Cancer. Cancers 2023, 15, 5842. https://doi.org/10.3390/cancers15245842
Liu D, He C, Liu Z, Xu L, Li J, Zhao Z, Hu X, Chen H, Sun B, Wang Y. The Prognostic and Immune Significance of CILP2 in Pan-Cancer and Its Relationship with the Progression of Pancreatic Cancer. Cancers. 2023; 15(24):5842. https://doi.org/10.3390/cancers15245842
Chicago/Turabian StyleLiu, Danxi, Cong He, Zonglin Liu, Licheng Xu, Jiacheng Li, Zhongjie Zhao, Xuewei Hu, Hua Chen, Bei Sun, and Yongwei Wang. 2023. "The Prognostic and Immune Significance of CILP2 in Pan-Cancer and Its Relationship with the Progression of Pancreatic Cancer" Cancers 15, no. 24: 5842. https://doi.org/10.3390/cancers15245842
APA StyleLiu, D., He, C., Liu, Z., Xu, L., Li, J., Zhao, Z., Hu, X., Chen, H., Sun, B., & Wang, Y. (2023). The Prognostic and Immune Significance of CILP2 in Pan-Cancer and Its Relationship with the Progression of Pancreatic Cancer. Cancers, 15(24), 5842. https://doi.org/10.3390/cancers15245842




