PIWI-RNAs Small Noncoding RNAs with Smart Functions: Potential Theranostic Applications in Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
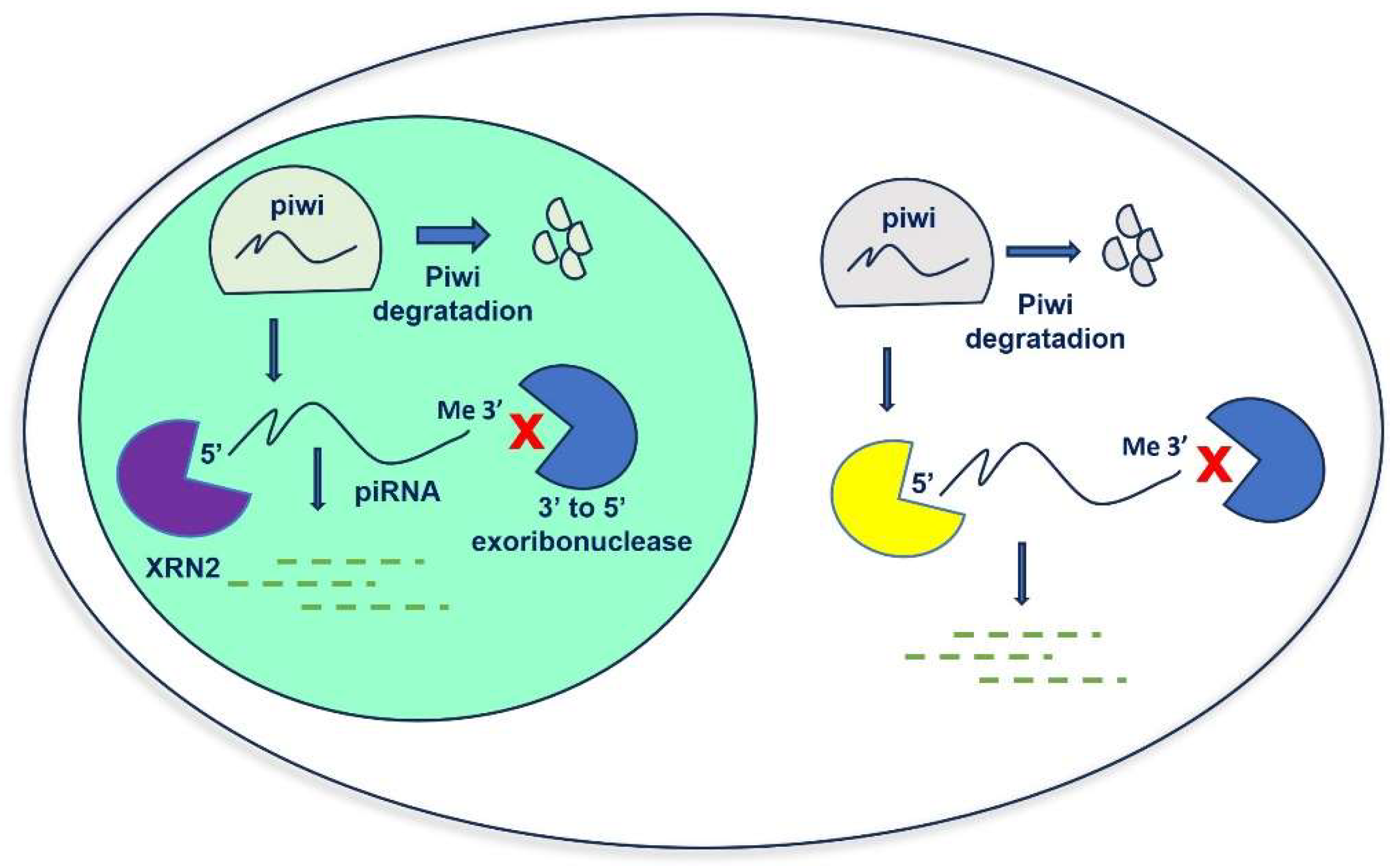
2. Biogenesis of piRNAs
3. Function of piRNAs and PIWI Proteins
4. piRNAs in Cancer
4.1. Role of piRNAs in Cancer Initiation and Progression
4.2. piRNAs as Cancer Biomarkers
5. piRNAs and Liquid Biopsy
5.1. piRNAs in Extracellular Vesicles
5.2. piRNA-Based Therapeutic Approaches in Cancer
6. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dhamija, S.; Menon, M.B. Non-Coding Transcript Variants of Protein-Coding Genes-What Are They Good For? RNA Biol. 2018, 15, 1025–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Gonzalez, E.A.; Rameshwar, P.; Etchegaray, J.-P. Non-Coding RNAs as Mediators of Epigenetic Changes in Malignancies. Cancers 2020, 12, 3657. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Li, J.; Zhao, P. Roles of NcRNAs as CeRNAs in Gastric Cancer. Genes 2021, 12, 1036. [Google Scholar] [CrossRef]
- Cammarata, G.; Barraco, N.; Giusti, I.; Gristina, V.; Dolo, V.; Taverna, S. Extracellular Vesicles-CeRNAs as Ovarian Cancer Biomarkers: Looking into CircRNA-MiRNA-MRNA Code. Cancers 2022, 14, 3404. [Google Scholar] [CrossRef] [PubMed]
- Pucci, M.; Reclusa Asiain, P.; Durendez Saez, E.; Jantus-Lewintre, E.; Malarani, M.; Khan, S.; Fontana, S.; Naing, A.; Passiglia, F.; Raez, L.E.; et al. Extracellular Vesicles As MiRNA Nano-Shuttles: Dual Role in Tumor Progression. Target. Oncol. 2018, 13, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Ameli Mojarad, M.; Ameli Mojarad, M.; Shojaee, B.; Nazemalhosseini-Mojarad, E. PiRNA: A Promising Biomarker in Early Detection of Gastrointestinal Cancer. Pathol. Res. Pract. 2022, 230, 153757. [Google Scholar] [CrossRef] [PubMed]
- Cammarata, G.; de Miguel-Perez, D.; Russo, A.; Peleg, A.; Dolo, V.; Rolfo, C.; Taverna, S. Emerging Noncoding RNAs Contained in Extracellular Vesicles: Rising Stars as Biomarkers in Lung Cancer Liquid Biopsy. Ther. Adv. Med. Oncol. 2022, 14, 17588359221131228. [Google Scholar] [CrossRef]
- Weng, W.; Li, H.; Goel, A. Piwi-Interacting RNAs (PiRNAs) and Cancer: Emerging Biological Concepts and Potential Clinical Implications. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 160–169. [Google Scholar] [CrossRef]
- Koch, B.; Geßner, A.; Farmand, S.; Fuhrmann, D.C.; Chiocchetti, A.G.; Schubert, R.; Baer, P.C. Effects of Hypoxia on RNA Cargo in Extracellular Vesicles from Human Adipose-Derived Stromal/Stem Cells. Int. J. Mol. Sci. 2022, 23, 7384. [Google Scholar] [CrossRef]
- Huang, M.; Peng, X.; Yang, L.; Yang, S.; Li, X.; Tang, S.; Li, B.; Jin, H.; Wu, B.; Liu, J.; et al. Non-Coding RNA Derived from Extracellular Vesicles in Cancer Immune Escape: Biological Functions and Potential Clinical Applications. Cancer Lett. 2021, 501, 234–246. [Google Scholar] [CrossRef]
- Sadoughi, F.; Mirhashemi, S.M.; Asemi, Z. Epigenetic Roles of PIWI Proteins and PiRNAs in Colorectal Cancer. Cancer Cell Int. 2021, 21, 328. [Google Scholar] [CrossRef] [PubMed]
- Houwing, S.; Kamminga, L.M.; Berezikov, E.; Cronembold, D.; Girard, A.; van den Elst, H.; Filippov, D.V.; Blaser, H.; Raz, E.; Moens, C.B.; et al. A Role for Piwi and PiRNAs in Germ Cell Maintenance and Transposon Silencing in Zebrafish. Cell 2007, 129, 69–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Billi, A.C.; Alessi, A.F.; Khivansara, V.; Han, T.; Freeberg, M.; Mitani, S.; Kim, J.K. The Caenorhabditis Elegans HEN1 Ortholog, HENN-1, Methylates and Stabilizes Select Subclasses of Germline Small RNAs. PLoS Genet. 2012, 8, e1002617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Xu, J.; Koppetsch, B.S.; Wang, J.; Tipping, C.; Ma, S.; Weng, Z.; Theurkauf, W.E.; Zamore, P.D. Heterotypic PiRNA Ping-Pong Requires Qin, a Protein with Both E3 Ligase and Tudor Domains. Mol. Cell 2011, 44, 572–584. [Google Scholar] [CrossRef] [Green Version]
- Sato, K.; Iwasaki, Y.W.; Siomi, H.; Siomi, M.C. Tudor-Domain Containing Proteins Act to Make the PiRNA Pathways More Robust in Drosophila. Fly 2015, 9, 86–90. [Google Scholar] [CrossRef] [Green Version]
- Balaratnam, S.; Hoque, M.E.; West, N.; Basu, S. Decay of Piwi-Interacting RNAs in Human Cells Is Primarily Mediated by 5’ to 3’ Exoribonucleases. ACS Chem. Biol. 2022, 17, 1723–1732. [Google Scholar] [CrossRef]
- Fonseca Cabral, G.; Azevedo Dos Santos Pinheiro, J.; Vidal, A.F.; Santos, S.; Ribeiro-Dos-Santos, Â. PiRNAs in Gastric Cancer: A New Approach Towards Translational Research. Int. J. Mol. Sci. 2020, 21, 2126. [Google Scholar] [CrossRef] [Green Version]
- Tóth, K.F.; Pezic, D.; Stuwe, E.; Webster, A. The PiRNA Pathway Guards the Germline Genome Against Transposable Elements. Adv. Exp. Med. Biol. 2016, 886, 51–77. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Tang, X.; Shen, E.-Z. How Mammalian PiRNAs Instruct de Novo DNA Methylation of Transposons. Signal Transduct. Target. Ther. 2020, 5, 190. [Google Scholar] [CrossRef]
- Wilson, A.S.; Power, B.E.; Molloy, P.L. DNA Hypomethylation and Human Diseases. Biochim. Biophys. Acta-Rev. Cancer 2007, 1775, 138–162. [Google Scholar] [CrossRef]
- Baylin, S.B. DNA Methylation and Gene Silencing in Cancer. Nat. Clin. Pract. Oncol. 2005, 2, S4–S11. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Xie, M.; Ma, X.; Song, J.; Wang, Y.; Xue, X. PIWI-Interacting RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Front. Oncol. 2022, 12, 965684. [Google Scholar] [CrossRef] [PubMed]
- Chénais, B. Transposable Elements and Human Cancer: A Causal Relationship? Biochim. Biophys. Acta-Rev. Cancer 2013, 1835, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Fabry, M.H.; Ciabrelli, F.; Munafò, M.; Eastwood, E.L.; Kneuss, E.; Falciatori, I.; Falconio, F.A.; Hannon, G.J.; Czech, B. PiRNA-Guided Co-Transcriptional Silencing Coopts Nuclear Export Factors. Elife 2019, 8, e47999. [Google Scholar] [CrossRef]
- Jia, D.-D.; Jiang, H.; Zhang, Y.-F.; Zhang, Y.; Qian, L.-L.; Zhang, Y.-F. The Regulatory Function of PiRNA/PIWI Complex in Cancer and Other Human Diseases: The Role of DNA Methylation. Int. J. Biol. Sci. 2022, 18, 3358–3373. [Google Scholar] [CrossRef]
- Ali, R.; Laskar, S.A.; Khan, N.J.; Wahab, S.; Khalid, M. Non-Coding RNA’s Prevalence as Biomarkers for Prognostic, Diagnostic, and Clinical Utility in Breast Cancer. Funct. Integr. Genomics 2023, 23, 195. [Google Scholar] [CrossRef]
- Baylin, S.B.; Jones, P.A. A Decade of Exploring the Cancer Epigenome—Biological and Translational Implications. Nat. Rev. Cancer 2011, 11, 726–734. [Google Scholar] [CrossRef]
- Siddiqi, S.; Matushansky, I. Piwis and Piwi-Interacting RNAs in the Epigenetics of Cancer. J. Cell. Biochem. 2012, 113, 373–380. [Google Scholar] [CrossRef]
- Mentis, A.-F.A.; Dardiotis, E.; Romas, N.A.; Papavassiliou, A.G. PIWI Family Proteins as Prognostic Markers in Cancer: A Systematic Review and Meta-Analysis. Cell. Mol. Life Sci. 2020, 77, 2289–2314. [Google Scholar] [CrossRef]
- Zhou, J.; Xie, H.; Liu, J.; Huang, R.; Xiang, Y.; Tian, D.; Bian, E. PIWI-Interacting RNAs: Critical Roles and Therapeutic Targets in Cancer. Cancer Lett. 2023, 562, 216189. [Google Scholar] [CrossRef]
- Li, F.; Yuan, P.; Rao, M.; Jin, C.-H.; Tang, W.; Rong, Y.-F.; Hu, Y.-P.; Zhang, F.; Wei, T.; Yin, Q.; et al. PiRNA-Independent Function of PIWIL1 as a Co-Activator for Anaphase Promoting Complex/Cyclosome to Drive Pancreatic Cancer Metastasis. Nat. Cell Biol. 2020, 22, 425–438. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Huang, L.; Lu, Y.; Jiang, W.; Song, Y.; Qiu, B.; Tao, D.; Liu, Y.; Ma, Y. PIWIL2 Interacting with IKK to Regulate Autophagy and Apoptosis in Esophageal Squamous Cell Carcinoma. Cell Death Differ. 2021, 28, 1941–1954. [Google Scholar] [CrossRef]
- Chen, C.; Liu, J.; Xu, G. Overexpression of PIWI Proteins in Human Stage III Epithelial Ovarian Cancer with Lymph Node Metastasis. Cancer Biomark. 2013, 13, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Gambichler, T.; Kohsik, C.; Höh, A.-K.; Lang, K.; Käfferlein, H.U.; Brüning, T.; Stockfleth, E.; Stücker, M.; Dreißigacker, M.; Sand, M. Expression of PIWIL3 in Primary and Metastatic Melanoma. J. Cancer Res. Clin. Oncol. 2017, 143, 433–437. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, W.-J.; Li, Z.-W.; Wang, X.-Z. Downregulation of Piwil3 Suppresses Cell Proliferation, Migration and Invasion in Gastric Cancer. Cancer Biomark. 2017, 20, 499–509. [Google Scholar] [CrossRef]
- Liu, X.; Zheng, J.; Xue, Y.; Yu, H.; Gong, W.; Wang, P.; Li, Z.; Liu, Y. PIWIL3/OIP5-AS1/MiR-367-3p/CEBPA Feedback Loop Regulates the Biological Behavior of Glioma Cells. Theranostics 2018, 8, 1084–1105. [Google Scholar] [CrossRef]
- Li, W.; Martinez-Useros, J.; Garcia-Carbonero, N.; Fernandez-Aceñero, M.J.; Orta, A.; Ortega-Medina, L.; Garcia-Botella, S.; Perez-Aguirre, E.; Diez-Valladares, L.; Celdran, A.; et al. The Clinical Significance of PIWIL3 and PIWIL4 Expression in Pancreatic Cancer. J. Clin. Med. 2020, 9, 1252. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, K.; Kage, H.; Aki, N.; Sano, A.; Kitagawa, H.; Nagase, T.; Yatomi, Y.; Ohishi, N.; Takai, D. The Induction of H3K9 Methylation by PIWIL4 at the P16Ink4a Locus. Biochem. Biophys. Res. Commun. 2007, 359, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Coley, W.; Van Duyne, R.; Carpio, L.; Guendel, I.; Kehn-Hall, K.; Chevalier, S.; Narayanan, A.; Luu, T.; Lee, N.; Klase, Z.; et al. Absence of DICER in Monocytes and Its Regulation by HIV-1. J. Biol. Chem. 2010, 285, 31930–31943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zoch, A.; Auchynnikava, T.; Berrens, R.V.; Kabayama, Y.; Schöpp, T.; Heep, M.; Vasiliauskaitė, L.; Pérez-Rico, Y.A.; Cook, A.G.; Shkumatava, A.; et al. SPOCD1 Is an Essential Executor of PiRNA-Directed de Novo DNA Methylation. Nature 2020, 584, 635–639. [Google Scholar] [CrossRef]
- Sohn, E.J.; Oh, S.-O. P-Element-Induced Wimpy Testis Proteins and P-Element-Induced Wimpy Testis-Interacting RNAs Expression in Ovarian Cancer Stem Cells. Genet. Test. Mol. Biomark. 2023, 27, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, H.J.; Heyn, H.; Garcia del Muro, X.; Vidal, A.; Larriba, S.; Muñoz, C.; Villanueva, A.; Esteller, M. Epigenetic Loss of the PIWI/PiRNA Machinery in Human Testicular Tumorigenesis. Epigenetics 2014, 9, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wang, Q.; Jiang, W.; Bian, Y.; Zhou, Y.; Gou, A.; Zhang, W.; Fu, K.; Shi, W. Emerging Roles of PiRNAs in Cancer: Challenges and Prospects. Aging 2019, 11, 9932–9946. [Google Scholar] [CrossRef]
- Li, D.; Luo, Y.; Gao, Y.; Yang, Y.; Wang, Y.; Xu, Y.; Tan, S.; Zhang, Y.; Duan, J.; Yang, Y. PiR-651 Promotes Tumor Formation in Non-Small Cell Lung Carcinoma through the Upregulation of Cyclin D1 and CDK4. Int. J. Mol. Med. 2016, 38, 927–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Wang, J.; Sun, L.; Li, M.; He, X.; Jiang, J.; Zhou, Q. Piwi-Interacting RNA-651 Promotes Cell Proliferation and Migration and Inhibits Apoptosis in Breast Cancer by Facilitating DNMT1-Mediated PTEN Promoter Methylation. Cell Cycle 2021, 20, 1603–1616. [Google Scholar] [CrossRef]
- Cordeiro, A.; Navarro, A.; Gaya, A.; Díaz-Beyá, M.; Gonzalez-Farré, B.; Castellano, J.J.; Fuster, D.; Martínez, C.; Martínez, A.; Monzó, M. PiwiRNA-651 as Marker of Treatment Response and Survival in Classical Hodgkin Lymphoma. Oncotarget 2016, 7, 46002–46013. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Fan, G.; Song, S.; Jiang, Y.; Qian, C.; Zhang, W.; Su, Q.; Xue, X.; Zhuang, W.; Li, B. PiRNA-30473 Contributes to Tumorigenesis and Poor Prognosis by Regulating M6A RNA Methylation in DLBCL. Blood 2021, 137, 1603–1614. [Google Scholar] [CrossRef]
- Wang, X.; Ramat, A.; Simonelig, M.; Liu, M.-F. Emerging Roles and Functional Mechanisms of PIWI-Interacting RNAs. Nat. Rev. Mol. Cell Biol. 2022, 24, 123–141. [Google Scholar] [CrossRef]
- Peng, L.; Song, L.; Liu, C.; Lv, X.; Li, X.; Jie, J.; Zhao, D.; Li, D. PiR-55490 Inhibits the Growth of Lung Carcinoma by Suppressing MTOR Signaling. Tumour Biol. 2016, 37, 2749–2756. [Google Scholar] [CrossRef]
- Liu, Y.; Dong, Y.; He, X.; Gong, A.; Gao, J.; Hao, X.; Wang, S.; Fan, Y.; Wang, Z.; Li, M.; et al. PiR-Hsa-211106 Inhibits the Progression of Lung Adenocarcinoma Through Pyruvate Carboxylase and Enhances Chemotherapy Sensitivity. Front. Oncol. 2021, 11, 651915. [Google Scholar] [CrossRef]
- Tan, L.; Mai, D.; Zhang, B.; Jiang, X.; Zhang, J.; Bai, R.; Ye, Y.; Li, M.; Pan, L.; Su, J.; et al. PIWI-Interacting RNA-36712 Restrains Breast Cancer Progression and Chemoresistance by Interaction with SEPW1 Pseudogene SEPW1P RNA. Mol. Cancer 2019, 18, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Chen, Q.; Zhou, Z.; Tian, Z.; Zheng, X.; Wang, K. PiRNA-18 Inhibition Cell Proliferation, Migration and Invasion in Colorectal Cancer. Biochem. Genet. 2023. [Google Scholar] [CrossRef] [PubMed]
- Fu, A.; Jacobs, D.I.; Hoffman, A.E.; Zheng, T.; Zhu, Y. PIWI-Interacting RNA 021285 Is Involved in Breast Tumorigenesis Possibly by Remodeling the Cancer Epigenome. Carcinogenesis 2015, 36, 1094–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busch, J.; Ralla, B.; Jung, M.; Wotschofsky, Z.; Trujillo-Arribas, E.; Schwabe, P.; Kilic, E.; Fendler, A.; Jung, K. Piwi-Interacting RNAs as Novel Prognostic Markers in Clear Cell Renal Cell Carcinomas. J. Exp. Clin. Cancer Res. 2015, 34, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, J.; Deng, H.; Xiao, B.; Zhou, H.; Zhou, F.; Shen, Z.; Guo, J. PiR-823, a Novel Non-Coding Small RNA, Demonstrates in Vitro and in Vivo Tumor Suppressive Activity in Human Gastric Cancer Cells. Cancer Lett. 2012, 315, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Jiang, X.-Y.; Qi, W.; Ji, C.-G.; Xie, X.-L.; Zhang, D.-X.; Cui, Z.-J.; Wang, C.-K.; Bai, Y.; Wang, J.; et al. PiR-823 Contributes to Colorectal Tumorigenesis by Enhancing the Transcriptional Activity of HSF1. Cancer Sci. 2017, 108, 1746–1756. [Google Scholar] [CrossRef] [Green Version]
- Mai, D.; Ding, P.; Tan, L.; Zhang, J.; Pan, Z.; Bai, R.; Li, C.; Li, M.; Zhou, Y.; Tan, W.; et al. PIWI-Interacting RNA-54265 Is Oncogenic and a Potential Therapeutic Target in Colorectal Adenocarcinoma. Theranostics 2018, 8, 5213–5230. [Google Scholar] [CrossRef]
- Law, P.T.-Y.; Qin, H.; Ching, A.K.-K.; Lai, K.P.; Co, N.N.; He, M.; Lung, R.W.-M.; Chan, A.W.-H.; Chan, T.-F.; Wong, N. Deep Sequencing of Small RNA Transcriptome Reveals Novel Non-Coding RNAs in Hepatocellular Carcinoma. J. Hepatol. 2013, 58, 1165–1173. [Google Scholar] [CrossRef]
- Yuan, C.; Qin, H.; Ponnusamy, M.; Chen, Y.; Lin, Z. PIWI-interacting RNA in Cancer: Molecular Mechanisms and Possible Clinical Implications (Review). Oncol. Rep. 2021, 46, 209. [Google Scholar] [CrossRef]
- Zhao, Q.; Qian, L.; Guo, Y.; Lü, J.; Li, D.; Xie, H.; Wang, Q.; Ma, W.; Liu, P.; Liu, Y.; et al. IL11 Signaling Mediates PiR-2158 Suppression of Cell Stemness and Angiogenesis in Breast Cancer. Theranostics 2023, 13, 2337–2349. [Google Scholar] [CrossRef]
- Vychytilova-Faltejskova, P.; Stitkovcova, K.; Radova, L.; Sachlova, M.; Kosarova, Z.; Slaba, K.; Kala, Z.; Svoboda, M.; Kiss, I.; Vyzula, R.; et al. Circulating PIWI-Interacting RNAs PiR-5937 and PiR-28876 Are Promising Diagnostic Biomarkers of Colon Cancer. Cancer Epidemiol. Biomark. Prev. 2018, 27, 1019–1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Miguel-Perez, D.; Russo, A.; Arrieta, O.; Ak, M.; Barron, F.; Gunasekaran, M.; Mamindla, P.; Lara-Mejia, L.; Peterson, C.B.; Er, M.E.; et al. Extracellular Vesicle PD-L1 Dynamics Predict Durable Response to Immune-Checkpoint Inhibitors and Survival in Patients with Non-Small Cell Lung Cancer. J. Exp. Clin. Cancer Res. 2022, 41, 186. [Google Scholar] [CrossRef] [PubMed]
- Califf, R.M. Biomarker Definitions and Their Applications. Exp. Biol. Med. 2018, 243, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Rigogliuso, S.; Donati, C.; Cassarà, D.; Taverna, S.; Salamone, M.; Bruni, P.; Vittorelli, M.L. An Active Form of Sphingosine Kinase-1 Is Released in the Extracellular Medium as Component of Membrane Vesicles Shed by Two Human Tumor Cell Lines. J. Oncol. 2010, 2010, 509329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.-J.; Zhao, R.-M.; Zhao, Q.; Li, B.-Y.; Ma, Q.-Y.; Li, X.; Chen, X. Diagnostic Significance of Elevated Expression of HBME-1 in Papillary Thyroid Carcinoma. Tumour Biol. 2016, 37, 8715–8720. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.J.; Garrido-Navas, M.C.; Diaz Mochon, J.J.; Cristofanilli, M.; Gil-Bazo, I.; Pauwels, P.; Malapelle, U.; Russo, A.; Lorente, J.A.; Ruiz-Rodriguez, A.J.; et al. Precision Prevention and Cancer Interception: The New Challenges of Liquid Biopsy. Cancer Discov. 2020, 10, 1635–1644. [Google Scholar] [CrossRef]
- Perera, B.P.U.; Morgan, R.K.; Polemi, K.M.; Sala-Hamrick, K.E.; Svoboda, L.K.; Dolinoy, D.C. PIWI-Interacting RNA (PiRNA) and Epigenetic Editing in Environmental Health Sciences. Curr. Environ. Health Rep. 2022, 9, 650–660. [Google Scholar] [CrossRef]
- Galvano, A.; Taverna, S.; Badalamenti, G.; Incorvaia, L.; Castiglia, M.; Barraco, N.; Passiglia, F.; Fulfaro, F.; Beretta, G.; Duro, G.; et al. Detection of RAS Mutations in Circulating Tumor DNA: A New Weapon in an Old War against Colorectal Cancer. A Systematic Review of Literature and Meta-Analysis. Ther. Adv. Med. Oncol. 2019, 11, 1758835919874653. [Google Scholar] [CrossRef]
- Ray, S.K.; Mukherjee, S. Piwi-Interacting RNAs (PiRNAs) and Colorectal Carcinoma: Emerging Non-Invasive Diagnostic Biomarkers with Potential Therapeutic Target Based Clinical Implications. Curr. Mol. Med. 2023, 23, 300–311. [Google Scholar] [CrossRef]
- Pardini, B.; Sabo, A.A.; Birolo, G.; Calin, G.A. Noncoding RNAs in Extracellular Fluids as Cancer Biomarkers: The New Frontier of Liquid Biopsies. Cancers 2019, 11, 1170. [Google Scholar] [CrossRef] [Green Version]
- He, B.; Cai, Q.; Qiao, L.; Huang, C.-Y.; Wang, S.; Miao, W.; Ha, T.; Wang, Y.; Jin, H. RNA-Binding Proteins Contribute to Small RNA Loading in Plant Extracellular Vesicles. Nat. Plants 2021, 7, 342–352. [Google Scholar] [CrossRef]
- Goh, T.-X.; Tan, S.-L.; Roebuck, M.M.; Teo, S.-H.; Kamarul, T. A Systematic Review of Extracellular Vesicle-Derived Piwi-Interacting RNA in Human Body Fluid and Its Role in Disease Progression. Tissue Eng. Part C. Methods 2022, 28, 511–528. [Google Scholar] [CrossRef]
- Mokarram, P.; Niknam, M.; Sadeghdoust, M.; Aligolighasemabadi, F.; Siri, M.; Dastghaib, S.; Brim, H.; Ashktorab, H. PIWI Interacting RNAs Perspectives: A New Avenues in Future Cancer Investigations. Bioengineered 2021, 12, 10401–10419. [Google Scholar] [CrossRef] [PubMed]
- Zimta, A.-A.; Sigurjonsson, O.E.; Gulei, D.; Tomuleasa, C. The Malignant Role of Exosomes as Nanocarriers of Rare RNA Species. Int. J. Mol. Sci. 2020, 21, 5866. [Google Scholar] [CrossRef] [PubMed]
- Monteleone, F.; Taverna, S.; Alessandro, R.; Fontana, S. SWATH-MS Based Quantitative Proteomics Analysis Reveals That Curcumin Alters the Metabolic Enzyme Profile of CML Cells by Affecting the Activity of MiR-22/IPO7/HIF-1alpha Axis. J. Exp. Clin. Cancer Res. 2018, 37, 170. [Google Scholar] [CrossRef] [Green Version]
- Oliveres, H.; Caglevic, C.; Passiglia, F.; Taverna, S.; Smits, E.; Rolfo, C. Vaccine and Immune Cell Therapy in Non-Small Cell Lung Cancer. J. Thorac. Dis. 2018, 10, S1602–S1614. [Google Scholar] [CrossRef]
- Mai, D.; Zheng, Y.; Guo, H.; Ding, P.; Bai, R.; Li, M.; Ye, Y.; Zhang, J.; Huang, X.; Liu, D.; et al. Serum PiRNA-54265 Is a New Biomarker for Early Detection and Clinical Surveillance of Human Colorectal Cancer. Theranostics 2020, 10, 8468–8478. [Google Scholar] [CrossRef] [PubMed]
- Al-Obeidi, E.; Riess, J.W.; Malapelle, U.; Rolfo, C.; Gandara, D.R. Convergence of Precision Oncology and Liquid Biopsy in Non-Small Cell Lung Cancer. Hematol. Oncol. Clin. N. Am. 2023, 37, 475–487. [Google Scholar] [CrossRef]
- Connors, D.; Allen, J.; Alvarez, J.D.; Boyle, J.; Cristofanilli, M.; Hiller, C.; Keating, S.; Kelloff, G.; Leiman, L.; McCormack, R.; et al. International Liquid Biopsy Standardization Alliance White Paper. Crit. Rev. Oncol. Hematol. 2020, 156, 103112. [Google Scholar] [CrossRef]
- Candela, M.E.; Geraci, F.; Turturici, G.; Taverna, S.; Albanese, I.; Sconzo, G. Membrane Vesicles Containing Matrix Metalloproteinase-9 and Fibroblast Growth Factor-2 Are Released into the Extracellular Space from Mouse Mesoangioblast Stem Cells. J. Cell. Physiol. 2010, 224, 144–151. [Google Scholar] [CrossRef]
- Taverna, S.; Giusti, I.; D’Ascenzo, S.; Pizzorno, L.; Dolo, V. Breast Cancer Derived Extracellular Vesicles in Bone Metastasis Induction and Their Clinical Implications as Biomarkers. Int. J. Mol. Sci. 2020, 21, 3573. [Google Scholar] [CrossRef] [PubMed]
- Reclusa, P.; Verstraelen, P.; Taverna, S.; Gunasekaran, M.; Pucci, M.; Pintelon, I.; Claes, N.; de Miguel-Pérez, D.; Alessandro, R.; Bals, S.; et al. Improving Extracellular Vesicles Visualization: From Static to Motion. Sci. Rep. 2020, 10, 6494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalluri, R.; LeBleu, V.S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Li, J.; Wang, N.; Zhang, F.; Jin, S.; Dong, Y.; Dong, X.; Chen, Y.; Kong, X.; Tong, Y.; Mi, Q.; et al. PIWI-Interacting RNAs Are Aberrantly Expressed and May Serve as Novel Biomarkers for Diagnosis of Lung Adenocarcinoma. Thorac. Cancer 2021, 12, 2468–2477. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dong, Y.; Zhao, S.; Gao, J.; Hao, X.; Wang, Z.; Li, M.; Wang, M.; Liu, Y.; Yu, X.; et al. Serum-Derived PiR-Hsa-164586 of Extracellular Vesicles as a Novel Biomarker for Early Diagnosis of Non-Small Cell Lung Cancer. Front. Oncol. 2022, 12, 850363. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shi, B.; Zhang, X.; Shen, P.; He, Q.; Yin, M.; Pan, Y.; Ma, J. Exosomal Hsa-PiR1089 Promotes Proliferation and Migration in Neuroblastoma via Targeting KEAP1. Pathol. Res. Pract. 2023, 241, 154240. [Google Scholar] [CrossRef]
- Yan, H.; Wu, Q.-L.; Sun, C.-Y.; Ai, L.-S.; Deng, J.; Zhang, L.; Chen, L.; Chu, Z.-B.; Tang, B.; Wang, K.; et al. PiRNA-823 Contributes to Tumorigenesis by Regulating de Novo DNA Methylation and Angiogenesis in Multiple Myeloma. Leukemia 2015, 29, 196–206. [Google Scholar] [CrossRef]
- Li, B.; Hong, J.; Hong, M.; Wang, Y.; Yu, T.; Zang, S.; Wu, Q. PiRNA-823 Delivered by Multiple Myeloma-Derived Extracellular Vesicles Promoted Tumorigenesis through Re-Educating Endothelial Cells in the Tumor Environment. Oncogene 2019, 38, 5227–5238. [Google Scholar] [CrossRef]
- Ai, L.; Mu, S.; Sun, C.; Fan, F.; Yan, H.; Qin, Y.; Cui, G.; Wang, Y.; Guo, T.; Mei, H.; et al. Myeloid-Derived Suppressor Cells Endow Stem-like Qualities to Multiple Myeloma Cells by Inducing PiRNA-823 Expression and DNMT3B Activation. Mol. Cancer 2019, 18, 88. [Google Scholar] [CrossRef]
- Feng, J.; Yang, M.; Wei, Q.; Song, F.; Zhang, Y.; Wang, X.; Liu, B.; Li, J. Novel Evidence for Oncogenic PiRNA-823 as a Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. J. Cell. Mol. Med. 2020, 24, 9028–9040. [Google Scholar] [CrossRef]
- Gu, X.; Wang, C.; Deng, H.; Qing, C.; Liu, R.; Liu, S.; Xue, X. Exosomal PiRNA Profiling Revealed Unique Circulating PiRNA Signatures of Cholangiocarcinoma and Gallbladder Carcinoma. Acta Biochim. Biophys. Sin. 2020, 52, 475–484. [Google Scholar] [CrossRef]
- Li, G.; Yi, X.; Du, S.; Gong, L.; Wu, Q.; Cai, J.; Sun, S.; Cao, Y.; Chen, L.; Xu, L.; et al. Tumour-Derived Exosomal PiR-25783 Promotes Omental Metastasis of Ovarian Carcinoma by Inducing the Fibroblast to Myofibroblast Transition. Oncogene 2023, 42, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Zhang, N.; Li, D.; Wu, Y.; Wang, H.; Wang, J. Circulating Exosomal Small RNAs Are Promising Non-Invasive Diagnostic Biomarkers for Gastric Cancer. J. Cell. Mol. Med. 2020, 24, 14502–14513. [Google Scholar] [CrossRef] [PubMed]
- Bajo-Santos, C.; Brokāne, A.; Zayakin, P.; Endzeliņš, E.; Soboļevska, K.; Belovs, A.; Jansons, J.; Sperga, M.; Llorente, A.; Radoviča-Spalviņa, I.; et al. Plasma and Urinary Extracellular Vesicles as a Source of RNA Biomarkers for Prostate Cancer in Liquid Biopsies. Front. Mol. Biosci. 2023, 10, 980433. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Chiu, P.K.-F.; Wong, C.Y.-P.; Cheng, C.K.-L.; Teoh, J.Y.-C.; Ng, C.-F. Identification of PiRNA Targets in Urinary Extracellular Vesicles for the Diagnosis of Prostate Cancer. Diagnostics 2021, 11, 1828. [Google Scholar] [CrossRef] [PubMed]
- Sabo, A.A.; Birolo, G.; Naccarati, A.; Dragomir, M.P.; Aneli, S.; Allione, A.; Oderda, M.; Allasia, M.; Gontero, P.; Sacerdote, C.; et al. Small Non-Coding RNA Profiling in Plasma Extracellular Vesicles of Bladder Cancer Patients by Next-Generation Sequencing: Expression Levels of MiR-126-3p and PiR-5936 Increase with Higher Histologic Grades. Cancers 2020, 12, 1507. [Google Scholar] [CrossRef]
- Rayford, K.J.; Cooley, A.; Rumph, J.T.; Arun, A.; Rachakonda, G.; Villalta, F.; Lima, M.F.; Pratap, S.; Misra, S.; Nde, P.N. PiRNAs as Modulators of Disease Pathogenesis. Int. J. Mol. Sci. 2021, 22, 2373. [Google Scholar] [CrossRef]
- Bobis-Wozowicz, S.; Marbán, E. Editorial: Extracellular Vesicles as Next Generation Therapeutics. Front. Cell Dev. Biol. 2022, 10, 919426. [Google Scholar] [CrossRef]
- Armstrong, J.P.K.; Stevens, M.M. Strategic Design of Extracellular Vesicle Drug Delivery Systems. Adv. Drug Deliv. Rev. 2018, 130, 12–16. [Google Scholar] [CrossRef]
- Huang, J.; Xiao, K. Nanoparticles-Based Strategies to Improve the Delivery of Therapeutic Small Interfering RNA in Precision Oncology. Pharmaceutics 2022, 14, 1586. [Google Scholar] [CrossRef]
- Liu, J.; Peng, X.; Yang, S.; Li, X.; Huang, M.; Wei, S.; Zhang, S.; He, G.; Zheng, H.; Fan, Q.; et al. Extracellular Vesicle PD-L1 in Reshaping Tumor Immune Microenvironment: Biological Function and Potential Therapy Strategies. Cell Commun. Signal. 2022, 20, 14. [Google Scholar] [CrossRef] [PubMed]
- Taverna, S.; Pucci, M.; Alessandro, R. Extracellular Vesicles: Small Bricks for Tissue Repair/Regeneration. Ann. Transl. Med. 2017, 5, 83. [Google Scholar] [CrossRef] [Green Version]
- Kadota, T.; Yoshioka, Y.; Fujita, Y.; Kuwano, K.; Ochiya, T. Extracellular Vesicles in Lung Cancer-From Bench to Bedside. Semin. Cell Dev. Biol. 2017, 67, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Giancaterino, S.; Boi, C. Alternative Biological Sources for Extracellular Vesicles Production and Purification Strategies for Process Scale-Up. Biotechnol. Adv. 2023, 63, 108092. [Google Scholar] [CrossRef]
- Rome, S. Biological Properties of Plant-Derived Extracellular Vesicles. Food Funct. 2019, 10, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Liu, K. Plant-Derived Extracellular Vesicles as Oral Drug Delivery Carriers. J. Control. Release 2022, 350, 389–400. [Google Scholar] [CrossRef]
- Chronopoulos, A.; Kalluri, R. Emerging Role of Bacterial Extracellular Vesicles in Cancer. Oncogene 2020, 39, 6951–6960. [Google Scholar] [CrossRef] [PubMed]
- Carobolante, G.; Mantaj, J.; Ferrari, E.; Vllasaliu, D. Cow Milk and Intestinal Epithelial Cell-Derived Extracellular Vesicles as Systems for Enhancing Oral Drug Delivery. Pharmaceutics 2020, 12, 226. [Google Scholar] [CrossRef] [Green Version]
- Munagala, R.; Aqil, F.; Jeyabalan, J.; Gupta, R.C. Bovine Milk-Derived Exosomes for Drug Delivery. Cancer Lett. 2016, 371, 48–61. [Google Scholar] [CrossRef] [Green Version]
- Maghraby, M.K.; Li, B.; Chi, L.; Ling, C.; Benmoussa, A.; Provost, P.; Postmus, A.C.; Abdi, A.; Pierro, A.; Bourdon, C.; et al. Extracellular Vesicles Isolated from Milk Can Improve Gut Barrier Dysfunction Induced by Malnutrition. Sci. Rep. 2021, 11, 7635. [Google Scholar] [CrossRef]
- Tosar, J.P.; García-Silva, M.R.; Cayota, A. Circulating SNORD57 Rather than PiR-54265 Is a Promising Biomarker for Colorectal Cancer: Common Pitfalls in the Study of Somatic PiRNAs in Cancer. RNA 2021, 27, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Tosar, J.P.; Rovira, C.; Cayota, A. Non-Coding RNA Fragments Account for the Majority of Annotated PiRNAs Expressed in Somatic Non-Gonadal Tissues. Commun. Biol. 2018, 1, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Piwi Protein | Cancer type | Expression | Function | References |
|---|---|---|---|---|
| PIWIL1 | Gasric | Upregulated | Migration proliferation invasion | [32] |
| Mieloma Multiple | Upregulated | Chemoresistance | [30] | |
| Lung | Upregulated | proliferation migration | [31] | |
| Breast | Upregulated | DNA methylation | [33] | |
| PIWIL2 | Colon Rectal | Uperegulation | Chemoresistance Proliferation | [19,22] |
| Gastric | Downregulated | Self renewal properties | [19] | |
| Ovarian | Upregulated | Apoptosis | [20] | |
| Breast | Upregulated | Chemoresistance | [19] | |
| PIWIL3 | Breast | Upregulated | Proliferation Cell survival | [24] |
| Ovarian | Metastasis | Metastasis | [20] | |
| Mieloma Multiple | Upregulated | Apoptosis | [21] | |
| Gastric | Upregulated | proliferation migration invasion | [22] | |
| Glioma | Downregulated | Tumor immune escape | [23] | |
| PIWIL4 | Lung | Upregulated | Proliferation | [39] |
| Liver | Upregulated | Metastasis | [27] | |
| Gastric | Upregulated | Apoptosis | [28] | |
| Testicular | Downpregulated | Proliferation | [29] | |
| Ovarian | Upregulated | Metastasis | [20] |
| Cancer Type | piRNA | Expression | Sample Type | Target | Potential Clinical Application | References |
|---|---|---|---|---|---|---|
| Lung | piR-651 | Upregulation | Cell | CDK4 Cyclin D1 MDM2 PTEN DNMT1 | Diagnosis | [34,51] |
| piR-211106 | Upregulation | Cell | Pyruvate carboxylase | Therapy | [56] | |
| piR-55490 | Downregulation | Cell | AKT mTOR | Therapy | [55] | |
| piR-5444 | Upregulation | EV | Diagnosis Prognosis Therapy | [61] | ||
| piR-26925 | Upregulation | EV | Diagnosis Prognosis Therapy | [61] | ||
| piR-164586 | Upregulation | EV | Diagnosis | [62] | ||
| Gastric | piR-823 | Downregulation | Cell Plasma | Diagnosis | [63] | |
| piR-019308 | Upregulation | EV | Diagnosis | [64] | ||
| piR-004918 | Upregulation | EV | Diagnosis | [64] | ||
| piR-018569 | Upregulation | EV | Diagnosis | [64] | ||
| Breast | piR-2158 | Downregulation | Stem cell | IL11 | Therapy | [65] |
| piR-823 | Upregulation | Stem cell | DNMT | Therapy | [66] | |
| piR-36712 | Upregulation | Cell | SEPW1 p53 p21 | Prognosis | [57,58] | |
| piR-021285 | Upregulation | Cell | ARHGAP11A | Therapy | [53,61] | |
| Colon Rectal | piR-823 | Downregulation | Cell | HSF1 | [67] | |
| piR-54265 | Upregulation | Tissue Serum | STAT3 | Prognosis | [35] | |
| piR-5937 | Upregulation | Serum | Diagnosis | [68] | ||
| piR-28876 | Upregulation | Serum | Diagnosis | [68] | ||
| piR-18 | Upregulation | Tissue Cell | Diagnosis Therapy | [58] | ||
| Liver | piR-Hep1 | Upregulation | Tissue Cell | AKT | Diagnosis | [69,70] |
| Neuroblastoma | piR-1089 | Upregulation | EV | KEAP1 | Prognosis | [71] |
| Lymphoma | piR-651 | Downregulation | Serum | Prognosis | [52] | |
| Renal | piR-38756 | Upregulation | Tissue | Prognosis | [60] | |
| piR-57125 | Upregulation | Tissue | Prognosis | [60] | ||
| piR-30924 | Upregulation | Tissue | Prognosis | [60] | ||
| B-cell lymphoma | piR-30473 | Upregulation | Serum | WTAP HK2 | Prognosis | [53] |
| Bladder | piR-5936 | Upregulation | Plasma EV | Diagnosis | [72,73] | |
| Ovarian | piR-25783 | Upregulation | Plasma EV | Diagnosis Prognosis | [74] | |
| Thyroid | piR-13643 | Upregulation | Tissue | Diagnosis | [75] | |
| piR-21238 | Upregulation | Tissue | Diagnosis | [75] | ||
| Cholangio Gallbladder | piR-2660989 | Upregulation | EV | Diagnosis Prognosis | [76] | |
| piR-10506469 | Upregulation | EV | Diagnosis Prognosis | [76] | ||
| piR-20548188 | Upregulation | EV | Diagnosis Prognosis | [76] | ||
| piR-10822895 | Upregulation | EV | Diagnosis Prognosis | [76] | ||
| piR-23209 | Upregulation | EV | Diagnosis Prognosis | [76] | ||
| piR-18044111 | Upregulation | EV | Diagnosis Prognosis | [76] |
| Cancer Type | piRNA | EV Origin | Target/Role | References |
|---|---|---|---|---|
| Lung | piR-26925 piR-5444 piR-164586 | Serum | Biomarker | [61,62] |
| Ovarian | piR-25783 | CM | TGF-β/SMAD2 /SMAD3 pathway | [74] |
| Gastric | piR-019308 piR-004918 piR-018569 | Serum | Biomarker | [64] |
| Prostate | piR-349843 piR-382289 piR-158533 piR-002468 | Urine | Biomarker | [72] |
| Neuroblastoma | piR-1089 | Plasma | KEAP1 | [71] |
| Multiple Mieloma | piR-823 | CM, Plasma | IL-6, VEGF ICAM-1 | [105] |
| Cholangio Gallbladder carcinoma | piR-2660989 piR-10506469 piR-20548188 piR-10822895 piR23209 piR-18044111 | Plasma | Biomarker | [76] |
| Bladder | piR-5936 | Plasma | Biomarker | [73] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taverna, S.; Masucci, A.; Cammarata, G. PIWI-RNAs Small Noncoding RNAs with Smart Functions: Potential Theranostic Applications in Cancer. Cancers 2023, 15, 3912. https://doi.org/10.3390/cancers15153912
Taverna S, Masucci A, Cammarata G. PIWI-RNAs Small Noncoding RNAs with Smart Functions: Potential Theranostic Applications in Cancer. Cancers. 2023; 15(15):3912. https://doi.org/10.3390/cancers15153912
Chicago/Turabian StyleTaverna, Simona, Anna Masucci, and Giuseppe Cammarata. 2023. "PIWI-RNAs Small Noncoding RNAs with Smart Functions: Potential Theranostic Applications in Cancer" Cancers 15, no. 15: 3912. https://doi.org/10.3390/cancers15153912
APA StyleTaverna, S., Masucci, A., & Cammarata, G. (2023). PIWI-RNAs Small Noncoding RNAs with Smart Functions: Potential Theranostic Applications in Cancer. Cancers, 15(15), 3912. https://doi.org/10.3390/cancers15153912






