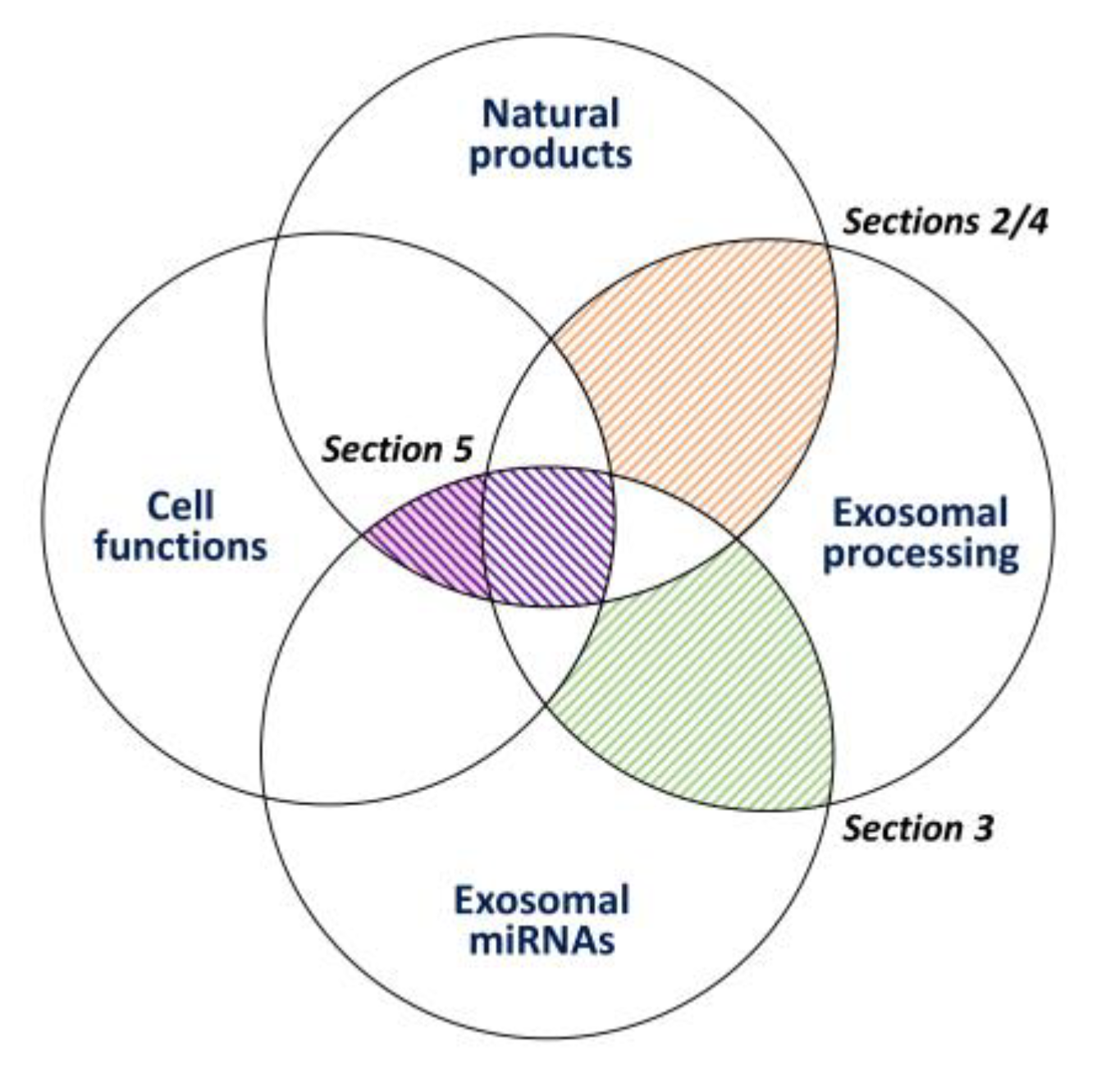
Modulating Effects of Cancer-Derived Exosomal miRNAs and Exosomal Processing by Natural Products
Abstract
Simple Summary
Abstract
1. Introduction
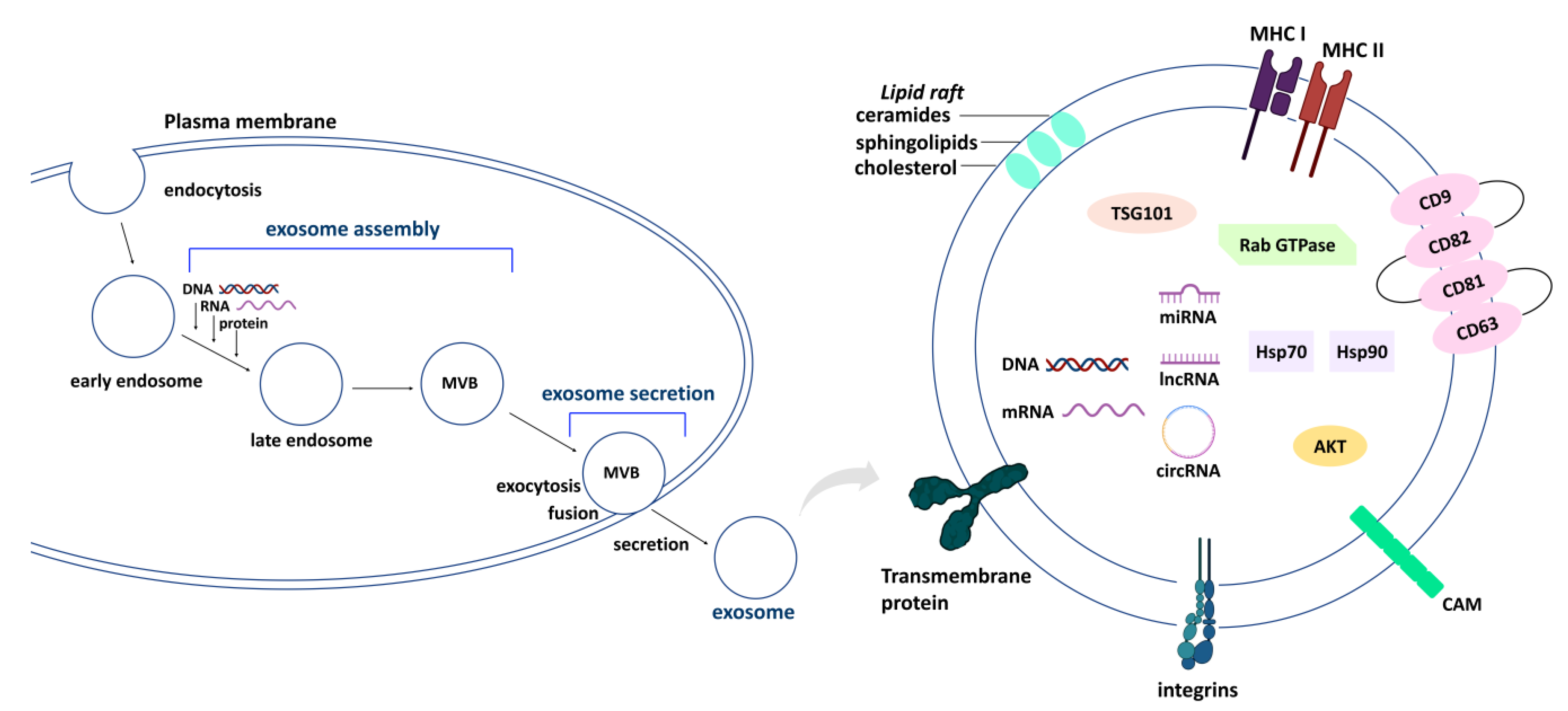
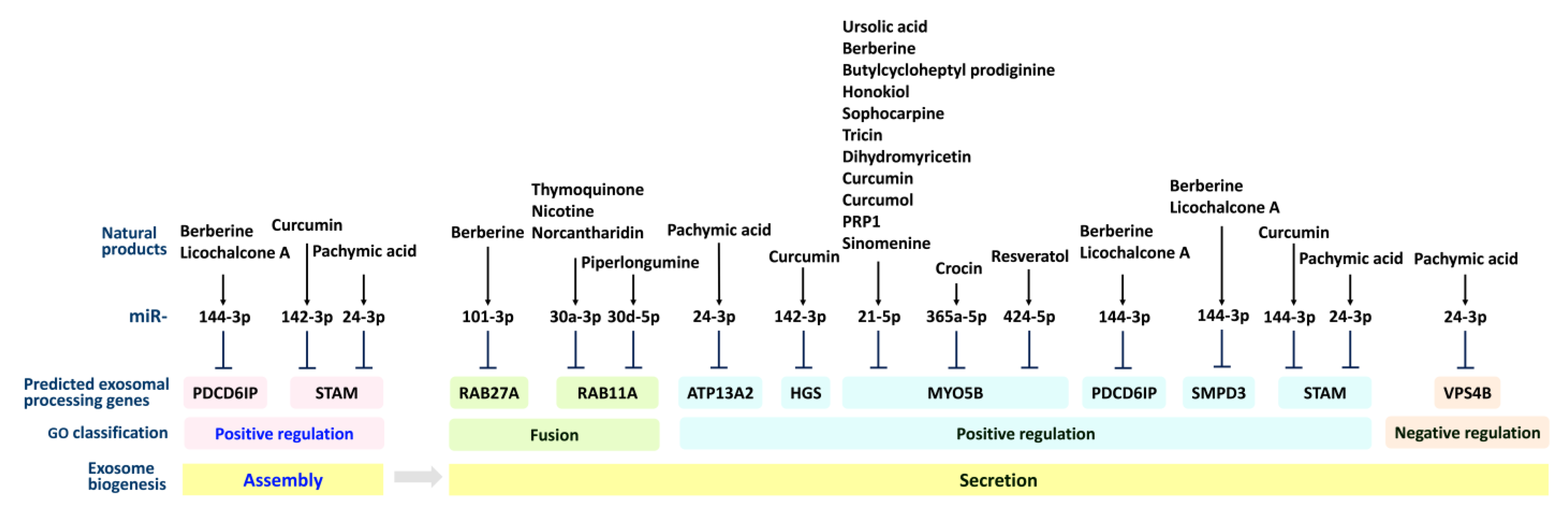
2. Exosome Processing (Secretion and Assembly) and Natural-Product-Modulated Cell Functions
2.1. Exosome Processing (Secretion and Assembly) Genes
2.2. Exosomal Secretion and Assembly Effects of Natural Products in Regulating Cell Functions
2.2.1. Exosomal Secretion Effects of Natural Products in Regulating Cell Functions
| Natural Products | Genes | Cell Functions | Cancer | References |
|---|---|---|---|---|
| Exosomal secretion | ||||
| Methanolic extract of | ↓COPS5 | apoptosis | cervical | [39] |
| Moringa oleifera | ||||
| Rutin | ↓COPS5 | apoptosis | cervical | [40] |
| Hesperidin | ↓RAB11A | hepatoprotective | (rat) | [33] |
| Heteronemin | ↑STEAP3 | ferroptosis | pancreatic | [41] |
| Dihydroartemisinin | ↓STEAP3 | anti-iron uptake | liver | [42] |
| Robustaflavone A | ↓STEAP3 | ferroptosis | breast | [43] |
| Tenuifolin, Schisandrin A, | ↑PRKN | mitophagy | (renal tubular cells) | [35] |
| Celastrol, Salidroside, | ||||
| Carnosic acid | ||||
| Liensinine | ↑RAB7A | anti-autophagy | breast | [44] |
| Sulfisoxazole | ↓VPS4B | antimetastatic | breast | [45] |
| Bavachinin | ↑VPS4B | pro-survival | (hepatocyte) | [38] |
| Squalamine | ↑ATP13A2 | α-synuclein aggregation | neuroblastoma | [46] |
| 7-α-Hydroxyfrullanolide | ↑PDCD6IP | apoptosis | breast | [47] |
| Salvianolic acid B | ↓SDC1 | renal interstitial fibrosis | (proximal tubular cells) | [36] |
| Rutaecarpine | ↓SDC1 | antimigration | glioblastoma | [48] |
| Echinacea angustifolia/ | ↓SDCBP | immunomodulation | (human study) | [37] |
| Zingiber officinale | ||||
| extracts | ||||
| Dioscin | ↑SDCBP | apoptosis, autophagy, | liver ca | [49] |
| DNA damage | ||||
| Sulforaphane | ↑SDCBP | apoptosis | leukemia | [50] |
| Acetyl-11-keto-b-boswellic | ↑SMPD3 | antiproliferation | colon | [51] |
| acid | ||||
| Withanolide D | ↑/↓SMPD3 | apoptosis | leukemia | [52] |
| Resveratrol | ↑TSG101 | antiproliferation | intestinal tumor | [53] |
| Exosomal assembly | ||||
| Astragalus membranaceus | ↑CD34 | angiogenesis | (myocardial infarction) | [54] |
| extract | ||||
| D Rhamnose bhederin | ↓STAM | chemoresistance | breast | [55] |
2.2.2. Exosomal Assembly Effects of Natural Products in Regulating Cell Functions
3. Prediction of the Targeting of Exosome-Processing and AKT-Signaling Genes of Certain Exosome miRNAs
| ATP9A | ATP13A2 | HGS | MYO5B | RAB27A | RAB11A | RAB7A | RAB7B | PDCD6IP | SDC1 | SDCBP | SMPD3 | STAM | TSG101 | VPS4B | AKT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| miR-29a-3p [4,16] | SMPD3 | AKT2/3 | ||||||||||||||
| miR-101-3p [4] | RAB27A | AKT3 | ||||||||||||||
| miR-106a-5p [16,57] | MYO5B | TSG101 | AKT3 | |||||||||||||
| miR-106b-5p [57] | MYO5B | TSG101 | AKT3 | |||||||||||||
| miR-128-3p [57] | STAM | VPS4B | ||||||||||||||
| miR-142-3p [4,16] | HGS | STAM | ||||||||||||||
| miR-200c-3p [4,16] | STAM | |||||||||||||||
| miR-21-5p [4,57] | MYO5B | RAB11A | ||||||||||||||
| miR-223-3p [16] | MYO5B | STAM | ||||||||||||||
| miR-24-3p [4,16] | ATP13A2 | STAM | ||||||||||||||
| miR-32-5p [57] | VPS4B | |||||||||||||||
| miR-365a-3p [57] | MYO5B | AKT3 | ||||||||||||||
| miR-374a-5p [57] | MYO5B | AKT1 | ||||||||||||||
| miR-522-3p [57] | PDCD6IP | |||||||||||||||
| miR-6887-5p [4] | RAB7A | RAB7B | AKT3 | |||||||||||||
| miR-8485 [4] | ATP9A | PDCD6IP | SDC1 | SDCBP | STAM | |||||||||||
| miR-92a-3p [57] | VPS4B | |||||||||||||||
| miR-30a-3p [62] | RAB11A | AKT3 |
4. Natural Products Modulate Exosome Production and Their Exosome Delivery for the Purpose of Cancer Treatment
5. The Role of Natural-Product-Modulating miRNAs and Exosomes in Regulating Cancer Cell Functions
5.1. Antiproliferation by Natural-Product-Modulated Exosomal miRNAs
5.2. Apoptosis by Natural-Product-Modulated Exosomal miRNAs
5.3. Antimigration/Anti-Invasion/Anti-EMT/Anti-Angiogenesis by Natural-Product-Modulated Exosomal miRNAs
5.4. Modulation of Chemo- and Radio-Resistance by Natural-Product-Modulated Exosomal miRNAs
5.5. Potential Modulation Effects of Target Immunotherapy of Cancer by Natural-Product-Modulated Exosomal miRNAs
5.6. Other Cell Functions Influenced by Natural-Product-Modulated Exosomal miRNAs
5.7. Overview of the Natural Products and Their Modulating Exosomal miRNAs That Regulate Exosomal Processing
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhang, C.; Ji, Q.; Yang, Y.; Li, Q.; Wang, Z. Exosome: Function and role in cancer metastasis and drug resistance. Technol. Cancer Res. Treat. 2018, 17, 1533033818763450. [Google Scholar] [CrossRef] [PubMed]
- Dilsiz, N. Role of exosomes and exosomal microRNAs in cancer. Future Sci. OA 2020, 6, FSO465. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, G.; Liu, J.; Zhang, C.; Yao, Y.; Liao, W. Exosomal cargoes in OSCC: Current findings and potential functions. PeerJ 2020, 8, e10062. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Lou, K.; Zou, X.; Zou, J.; Zhang, G. The potential role of exosomal proteins in prostate cancer. Front. Oncol. 2022, 12, 873296. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Duan, L.; Lu, J.; Xia, J. Engineering exosomes for targeted drug delivery. Theranostics 2021, 11, 3183–3195. [Google Scholar] [CrossRef]
- Jan, A.T.; Rahman, S.; Badierah, R.; Lee, E.J.; Mattar, E.H.; Redwan, E.M.; Choi, I. Expedition into exosome biology: A perspective of progress from discovery to therapeutic development. Cancers 2021, 13, 1157. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kang, M.H.; Jeyaraj, M.; Qasim, M.; Kim, J.H. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Farooqi, A.A.; Desai, N.N.; Qureshi, M.Z.; Librelotto, D.R.N.; Gasparri, M.L.; Bishayee, A.; Nabavi, S.M.; Curti, V.; Daglia, M. Exosome biogenesis, bioactivities and functions as new delivery systems of natural compounds. Biotechnol. Adv. 2018, 36, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Teng, F.; Fussenegger, M. Shedding light on extracellular vesicle biogenesis and bioengineering. Adv. Sci. 2020, 8, 2003505. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Liu, Y.; Guan, S.; Qiu, Z.; Liu, D. Natural products exert anti-tumor effects by regulating exosomal ncRNA. Front. Oncol. 2022, 12, 1006114. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Bhome, R.; Del Vecchio, F.; Lee, G.H.; Bullock, M.D.; Primrose, J.N.; Sayan, A.E.; Mirnezami, A.H. Exosomal microRNAs (exomiRs): Small molecules with a big role in cancer. Cancer Lett. 2018, 420, 228–235. [Google Scholar] [CrossRef]
- Lu, Y.; Zheng, Z.; Yuan, Y.; Pathak, J.L.; Yang, X.; Wang, L.; Ye, Z.; Cho, W.C.; Zeng, M.; Wu, L. The emerging role of exosomes in oral squamous cell carcinoma. Front. Cell Dev. Biol. 2021, 9, 628103. [Google Scholar] [CrossRef]
- Dhar, R.; Mallik, S.; Devi, A. Exosomal microRNAs (exoMIRs): Micromolecules with macro impact in oral cancer. 3 Biotech 2022, 12, 155. [Google Scholar] [CrossRef]
- Li, Y.; Gao, S.; Hu, Q.; Wu, F. Functional properties of cancer epithelium and stroma-derived exosomes in head and neck squamous cell carcinoma. Life 2022, 12, 757. [Google Scholar] [CrossRef]
- St-Denis-Bissonnette, F.; Khoury, R.; Mediratta, K.; El-Sahli, S.; Wang, L.; Lavoie, J.R. Applications of extracellular vesicles in triple-negative breast cancer. Cancers 2022, 14, 451. [Google Scholar] [CrossRef]
- Lorenc, T.; Klimczyk, K.; Michalczewska, I.; Slomka, M.; Kubiak-Tomaszewska, G.; Olejarz, W. Exosomes in prostate cancer diagnosis, prognosis and therapy. Int. J. Mol. Sci. 2020, 21, 2118. [Google Scholar] [CrossRef]
- Li, X.; Jiang, W.; Gan, Y.; Zhou, W. The application of exosomal microRNAs in the treatment of pancreatic cancer and its research progress. Pancreas 2021, 50, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Babaker, M.A.; Aljoud, F.A.; Alkhilaiwi, F.; Algarni, A.; Ahmed, A.; Khan, M.I.; Saadeldin, I.M.; Alzahrani, F.A. The Therapeutic Potential of Milk Extracellular Vesicles on Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 6812. [Google Scholar] [CrossRef] [PubMed]
- Hashemipour, M.; Boroumand, H.; Mollazadeh, S.; Tajiknia, V.; Nourollahzadeh, Z.; Rohani Borj, M.; Pourghadamyari, H.; Rahimian, N.; Hamblin, M.R.; Mirzaei, H. Exosomal microRNAs and exosomal long non-coding RNAs in gynecologic cancers. Gynecol. Oncol. 2021, 161, 314–327. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Fu, S.; Wu, S.; Tu, R. Growing evidence of exosomal microRNA-related metastasis of hepatocellular carcinoma. BioMed Res. Int. 2020, 2020, 4501454. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, R.; Mitra, P.; Gupta, N.; Sharma, S.; Singh, V.K.; Mukerjee, N.; Dhasmana, A.; Gundamaraju, R. Cellular landscaping of exosomal miRNAs in cancer metastasis: From chemoresistance to prognostic markers. Adv. Cancer Biol.-Metastasis 2022, 5, 100050. [Google Scholar] [CrossRef]
- Sorop, A.; Constantinescu, D.; Cojocaru, F.; Dinischiotu, A.; Cucu, D.; Dima, S.O. Exosomal microRNAs as biomarkers and therapeutic targets for hepatocellular carcinoma. Int. J. Mol. Sci. 2021, 22, 4997. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Cao, Z.; Jiang, D. The effect of miRNA-modified exosomes in animal models of spinal cord injury: A meta-analysis. Front. Bioeng Biotechnol 2021, 9, 819651. [Google Scholar] [CrossRef]
- Cetin, Z.; Saygili, E.I.; Gorgisen, G.; Sokullu, E. Preclinical experimental applications of miRNA loaded BMSC extracellular vesicles. Stem Cell Rev. Rep. 2021, 17, 471–501. [Google Scholar] [CrossRef]
- Rezaei, R.; Baghaei, K.; Hashemi, S.M.; Zali, M.R.; Ghanbarian, H.; Amani, D. Tumor-derived exosomes enriched by miRNA-124 promote anti-tumor immune response in CT-26 tumor-bearing mice. Front. Med. 2021, 8, 619939. [Google Scholar] [CrossRef]
- Guglielmi, L.; Nardella, M.; Musa, C.; Cifola, I.; Porru, M.; Cardinali, B.; Iannetti, I.; Di Pietro, C.; Bolasco, G.; Palmieri, V.; et al. Circulating miRNAs in small extracellular vesicles secreted by a human melanoma xenograft in mouse brains. Cancers 2020, 12, 1635. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Bult, C.J.; Blake, J.A.; Smith, C.L.; Kadin, J.A.; Richardson, J.E.; Mouse Genome Database, G. Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 2019, 47, D801–D806. [Google Scholar] [CrossRef] [PubMed]
- Hasanin, A.H.; Matboli, M.; Seleem, H.S. Hesperidin suppressed hepatic precancerous lesions via modulation of exophagy in rats. J. Cell. Biochem. 2020, 121, 1295–1306. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Q.; Zhang, F.; Yu, L.P.; Mu, J.K.; Yang, Y.Q.; Yu, J.; Yang, X.X. Targeting PINK1 using natural products for the treatment of human diseases. BioMed Res. Int. 2021, 2021, 4045819. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Han, H.; Yan, M.; Zhu, S.; Liu, J.; Liu, Z.; He, L.; Tan, J.; Liu, Y.; Liu, H.; et al. PINK1-PRKN/PARK2 pathway of mitophagy is activated to protect against renal ischemia-reperfusion injury. Autophagy 2018, 14, 880–897. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, M.; Pan, Y.; Li, Q.; Xu, L. Salvianolic acid B attenuates renal interstitial fibrosis by regulating the HPSE/SDC1 axis. Mol. Med. Rep. 2020, 22, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Dall’Acqua, S.; Grabnar, I.; Verardo, R.; Klaric, E.; Marchionni, L.; Luidy-Imada, E.; Sut, S.; Agostinis, C.; Bulla, R.; Perissutti, B.; et al. Combined extracts of Echinacea angustifolia DC. and Zingiber officinale Roscoe in softgel capsules: Pharmacokinetics and immunomodulatory effects assessed by gene expression profiling. Phytomedicine Int. J. Phytother. Phytopharm. 2019, 65, 153090. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Zhu, Y.; Wang, S.; Luo, Y.; Lu, S.; Nan, F.; Sun, G.; Sun, X. Bavachinin inhibits cholesterol synthesis enzyme FDFT1 expression via AKT/mTOR/SREBP-2 pathway. Int. Immunopharmacol. 2020, 88, 106865. [Google Scholar] [CrossRef]
- Pandey, P.; Khan, F. Jab1 inhibition by methanolic extract of Moringa oleifera leaves in cervical cancer cells: A potent targeted therapeutic approach. Nutr. Cancer 2021, 73, 2411–2419. [Google Scholar] [CrossRef]
- Pandey, P.; Khan, F.; Alzahrani, F.A.; Qari, H.A.; Oves, M. A novel approach to unraveling the apoptotic potential of rutin (bioflavonoid) via targeting Jab1 in cervical cancer cells. Molecules 2021, 26, 5529. [Google Scholar] [CrossRef]
- Kaftan, G.; Erdoğan, M.; El-Shazly, M.; Lu, M.-C.; Shih, S.-P.; Lin, H.-Y.; Saso, L.; Armagan, G. Heteronemin promotes iron-dependent cell death in pancreatic cancer. Res. Sq. 2022. [Google Scholar] [CrossRef]
- Ba, Q.; Zhou, N.; Duan, J.; Chen, T.; Hao, M.; Yang, X.; Li, J.; Yin, J.; Chu, R.; Wang, H. Dihydroartemisinin exerts its anticancer activity through depleting cellular iron via transferrin receptor-1. PloS ONE 2012, 7, e42703. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhou, X.; Li, J.; Yao, X.C.; Liu, W.L.; Kang, F.H.; Zou, Z.X.; Xu, K.P.; Xu, P.S.; Tan, G.S. Identification of a new natural biflavonoids against breast cancer cells induced ferroptosis via the mitochondrial pathway. Bioorganic Chem. 2021, 109, 104744. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, G.; Zheng, Y.; Shen, H.M.; Hu, X.; Ming, Q.L.; Huang, C.; Li, P.; Gao, N. A novel autophagy/mitophagy inhibitor liensinine sensitizes breast cancer cells to chemotherapy through DNM1L-mediated mitochondrial fission. Autophagy 2015, 11, 1259–1279. [Google Scholar] [CrossRef] [PubMed]
- Im, E.J.; Lee, C.H.; Moon, P.G.; Rangaswamy, G.G.; Lee, B.; Lee, J.M.; Lee, J.C.; Jee, J.G.; Bae, J.S.; Kwon, T.K.; et al. Sulfisoxazole inhibits the secretion of small extracellular vesicles by targeting the endothelin receptor A. Nat. Commun. 2019, 10, 1387. [Google Scholar] [CrossRef] [PubMed]
- Si, J.; Van den Haute, C.; Lobbestael, E.; Martin, S.; van Veen, S.; Vangheluwe, P.; Baekelandt, V. ATP13A2 regulates cellular alpha-synuclein multimerization, membrane association, and externalization. Int. J. Mol. Sci. 2021, 22, 2689. [Google Scholar] [CrossRef]
- Chimplee, S.; Roytrakul, S.; Sukrong, S.; Srisawat, T.; Graidist, P.; Kanokwiroon, K. Anticancer effects and molecular action of 7-alpha-hydroxyfrullanolide in G2/M-phase arrest and apoptosis in triple negative breast cancer cells. Molecules 2022, 27, 407. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, Y.; Zhu, R.; Xu, L.; Xie, H.Q.; Zhao, B. Rutaecarpine inhibits U87 glioblastoma cell migration by activating the aryl hydrocarbon receptor signaling pathway. Front. Mol. Neurosci. 2021, 14, 765712. [Google Scholar] [CrossRef]
- Mao, Z.; Han, X.; Chen, D.; Xu, Y.; Xu, L.; Yin, L.; Sun, H.; Qi, Y.; Fang, L.; Liu, K.; et al. Potent effects of dioscin against hepatocellular carcinoma through regulating TP53-induced glycolysis and apoptosis regulator (TIGAR)-mediated apoptosis, autophagy, and DNA damage. Br. J. Pharmacol. 2019, 176, 919–937. [Google Scholar] [CrossRef]
- Shang, H.S.; Shih, Y.L.; Lee, C.H.; Hsueh, S.C.; Liu, J.Y.; Liao, N.C.; Chen, Y.L.; Huang, Y.P.; Lu, H.F.; Chung, J.G. Sulforaphane-induced apoptosis in human leukemia HL-60 cells through extrinsic and intrinsic signal pathways and altering associated genes expression assayed by cDNA microarray. Environ. Toxicol. 2017, 32, 311–328. [Google Scholar] [CrossRef]
- Shen, Y.; Takahashi, M.; Byun, H.M.; Link, A.; Sharma, N.; Balaguer, F.; Leung, H.C.; Boland, C.R.; Goel, A. Boswellic acid induces epigenetic alterations by modulating DNA methylation in colorectal cancer cells. Cancer Biol. Ther. 2012, 13, 542–552. [Google Scholar] [CrossRef]
- Mondal, S.; Mandal, C.; Sangwan, R.; Chandra, S.; Mandal, C. Withanolide D induces apoptosis in leukemia by targeting the activation of neutral sphingomyelinase-ceramide cascade mediated by synergistic activation of c-Jun N-terminal kinase and p38 mitogen-activated protein kinase. Mol. Cancer 2010, 9, 239. [Google Scholar] [CrossRef] [PubMed]
- Schneider, Y.; Duranton, B.; Gosse, F.; Schleiffer, R.; Seiler, N.; Raul, F. Resveratrol inhibits intestinal tumorigenesis and modulates host-defense-related gene expression in an animal model of human familial adenomatous polyposis. Nutr. Cancer 2001, 39, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Liu, N.; Yang, L.; Mao, Y.; Ye, S. Astragalus membranaceus extract promotes angiogenesis by inducing VEGF, CD34 and eNOS expression in rats subjected to myocardial infarction. Int. J. Clin. Exp. Med. 2016, 9, 5709–5718. [Google Scholar]
- Chen, W.X.; Xu, L.Y.; Qian, Q.; He, X.; Peng, W.T.; Fan, W.Q.; Zhu, Y.L.; Tang, J.H.; Cheng, L. d Rhamnose beta-hederin reverses chemoresistance of breast cancer cells by regulating exosome-mediated resistance transmission. Biosci. Rep. 2018, 38, BSR20180110. [Google Scholar] [CrossRef] [PubMed]
- Perni, M.; Galvagnion, C.; Maltsev, A.; Meisl, G.; Muller, M.B.; Challa, P.K.; Kirkegaard, J.B.; Flagmeier, P.; Cohen, S.I.; Cascella, R.; et al. A natural product inhibits the initiation of alpha-synuclein aggregation and suppresses its toxicity. Proc. Natl. Acad. Sci. USA 2017, 114, E1009–E1017. [Google Scholar] [CrossRef]
- Salehi, M.; Vafadar, A.; Khatami, S.H.; Taheri-Anganeh, M.; Vakili, O.; Savardashtaki, A.; Negahdari, B.; Naeli, P.; Behrouj, H.; Ghasemi, H.; et al. Gastrointestinal cancer drug resistance: The role of exosomal miRNAs. Mol. Biol. Rep. 2022, 49, 2421–2432. [Google Scholar] [CrossRef]
- Shi, L.; Zhu, W.; Huang, Y.; Zhuo, L.; Wang, S.; Chen, S.; Zhang, B.; Ke, B. Cancer-associated fibroblast-derived exosomal microRNA-20a suppresses the PTEN/PI3K-AKT pathway to promote the progression and chemoresistance of non-small cell lung cancer. Clin. Transl. Med. 2022, 12, e989. [Google Scholar] [CrossRef]
- Zhang, L.; Ouyang, P.; He, G.; Wang, X.; Song, D.; Yang, Y.; He, X. Exosomes from microRNA-126 overexpressing mesenchymal stem cells promote angiogenesis by targeting the PIK3R2-mediated PI3K/Akt signalling pathway. J. Cell. Mol. Med. 2021, 25, 2148–2162. [Google Scholar] [CrossRef]
- Sun, J.; Wei, J.; Zhang, Y.; Li, J.; Li, J.; Yan, J.; Guo, M.; Han, J.; Qiao, H. Plasma exosomes transfer miR-885-3p targeting the AKT/NFkappaB signaling pathway to improve the sensitivity of intravenous glucocorticoid therapy against Graves ophthalmopathy. Front. Immunol. 2022, 13, 819680. [Google Scholar] [CrossRef]
- Bae, I.S.; Kim, S.H. Milk exosome-derived microRNA-2478 suppresses melanogenesis through the Akt-GSK3β pathway. Cells 2021, 10, 2848. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Wang, S.; Wei, Y.; Wu, J.; Huang, G.; Chen, J.; Shi, J.; Xia, J. Norcantharidin modulates the miR-30a/Metadherin/AKT signaling axis to suppress proliferation and metastasis of stromal tumor cells in giant cell tumor of bone. Biomed. Pharmacother. 2018, 103, 1092–1100. [Google Scholar] [CrossRef] [PubMed]
- Ali, N.B.; Abdull Razis, A.F.; Ooi, J.; Chan, K.W.; Ismail, N.; Foo, J.B. Theragnostic applications of mammal and plant-derived extracellular vesicles: Latest findings, current technologies, and prospects. Molecules 2022, 27, 3941. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, K.; Yamamoto, Y.; Matsuoka, R.; Ochiya, T. Maintaining good miRNAs in the body keeps the doctor away?: Perspectives on the relationship between food-derived natural products and microRNAs in relation to exosomes/extracellular vesicles. Mol. Nutr. Food Res. 2017. Available online: https://onlinelibrary.wiley.com/doi/10.1002/mnfr.201700080. [CrossRef]
- Song, H.; Liu, B.; Dong, B.; Xu, J.; Zhou, H.; Na, S.; Liu, Y.; Pan, Y.; Chen, F.; Li, L.; et al. Exosome-based delivery of natural products in cancer therapy. Front. Cell Dev. Biol. 2021, 9, 650426. [Google Scholar] [CrossRef]
- Xu, Y.; Feng, K.; Zhao, H.; Di, L.; Wang, L.; Wang, R. Tumor-derived extracellular vesicles as messengers of natural products in cancer treatment. Theranostics 2022, 12, 1683–1714. [Google Scholar] [CrossRef]
- Zheng, K.; Ma, J.; Wang, Y.; He, Z.; Deng, K. Sulforaphane inhibits autophagy and induces exosome-mediated paracrine senescence via regulating mTOR/TFE3. Mol. Nutr. Food Res. 2020, 64, e1901231. [Google Scholar] [CrossRef]
- Komatsu, M.; Waguri, S.; Koike, M.; Sou, Y.S.; Ueno, T.; Hara, T.; Mizushima, N.; Iwata, J.; Ezaki, J.; Murata, S.; et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007, 131, 1149–1163. [Google Scholar] [CrossRef]
- Xu, J.; Camfield, R.; Gorski, S.M. The interplay between exosomes and autophagy-partners in crime. J. Cell Sci. 2018, 131, jcs215210. [Google Scholar] [CrossRef]
- Mitani, F.; Lin, J.; Sakamoto, T.; Uehara, R.; Hikita, T.; Yoshida, T.; Setiawan, A.; Arai, M.; Oneyama, C. Asteltoxin inhibits extracellular vesicle production through AMPK/mTOR-mediated activation of lysosome function. Sci. Rep. 2022, 12, 6674. [Google Scholar] [CrossRef]
- Gu, S.; Song, X.; Xie, R.; Ouyang, C.; Xie, L.; Li, Q.; Su, T.; Xu, M.; Xu, T.; Huang, D.; et al. Berberine inhibits cancer cells growth by suppressing fatty acid synthesis and biogenesis of extracellular vesicles. Life Sci. 2020, 257, 118122. [Google Scholar] [CrossRef]
- Shiau, J.Y.; Chang, Y.Q.; Nakagawa-Goto, K.; Lee, K.H.; Shyur, L.F. Phytoagent deoxyelephantopin and its derivative inhibit triple negative breast cancer cell activity through ROS-mediated exosomal activity and protein functions. Front. Pharmacol. 2017, 8, 398. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Guo, X.J.; Cai, H.; Zhu, Y.H.; Huang, L.Y.; Wang, W.; Luo, L.; Qi, S.H. Momordica charantia-derived extracellular vesicles-like nanovesicles inhibited glioma proliferation, migration, and invasion by regulating the PI3K/AKT signaling pathway. J. Funct. Foods 2022, 90, 104968. [Google Scholar] [CrossRef]
- Lin, L.T.; Shi, Y.C.; Choong, C.Y.; Tai, C.J. The fruits of Paris polyphylla inhibit colorectal cancer cell migration induced by Fusobacterium nucleatum-derived extracellular vesicles. Molecules 2021, 26, 4081. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Rathore, S.; Munshi, A.; Ramesh, R. Organically derived exosomes as carriers of anticancer drugs and imaging agents for cancer treatment. Semin. Cancer Biol. 2022, 86, 80–100. [Google Scholar] [CrossRef]
- Agrawal, A.K.; Aqil, F.; Jeyabalan, J.; Spencer, W.A.; Beck, J.; Gachuki, B.W.; Alhakeem, S.S.; Oben, K.; Munagala, R.; Bondada, S.; et al. Milk-derived exosomes for oral delivery of paclitaxel. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 1627–1636. [Google Scholar] [CrossRef]
- Sun, D.; Zhuang, X.; Xiang, X.; Liu, Y.; Zhang, S.; Liu, C.; Barnes, S.; Grizzle, W.; Miller, D.; Zhang, H.G. A novel nanoparticle drug delivery system: The anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol. Ther. J. Am. Soc. Gene Ther. 2010, 18, 1606–1614. [Google Scholar] [CrossRef]
- Aqil, F.; Kausar, H.; Agrawal, A.K.; Jeyabalan, J.; Kyakulaga, A.H.; Munagala, R.; Gupta, R. Exosomal formulation enhances therapeutic response of celastrol against lung cancer. Exp. Mol. Pathol. 2016, 101, 12–21. [Google Scholar] [CrossRef]
- Zhang, B.; Tian, L.; Xie, J.; Chen, G.; Wang, F. Targeting miRNAs by natural products: A new way for cancer therapy. Biomed. Pharmacother. 2020, 130, 110546. [Google Scholar] [CrossRef]
- Park, Y.; Lee, K.; Kim, S.W.; Lee, M.W.; Kim, B.; Lee, S.G. Effects of induced exosomes from endometrial cancer cells on tumor activity in the presence of Aurea helianthus extract. Molecules 2021, 26, 2207. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y.; Wang, X.; Jiang, C. Ursolic acid inhibits proliferation and induces apoptosis in human glioblastoma cell lines U251 by suppressing TGF-β1/miR-21/PDCD 4 pathway. Basic Clin. Pharmacol. Toxicol. 2012, 111, 106–112. [Google Scholar] [PubMed]
- Zhang, L.; Cai, Q.Y.; Liu, J.; Peng, J.; Chen, Y.Q.; Sferra, T.J.; Lin, J.M. Ursolic acid suppresses the invasive potential of colorectal cancer cells by regulating the TGF-beta1/ZEB1/miR-200c signaling pathway. Oncol. Lett. 2019, 18, 3274–3282. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Ding, J.; Wu, Y. Resveratrol induces apoptosis of bladder cancer cells via miR21 regulation of the Akt/Bcl2 signaling pathway. Mol. Med. Rep. 2014, 9, 1467–1473. [Google Scholar] [CrossRef] [PubMed]
- Alrafas, H.R.; Busbee, P.B.; Nagarkatti, M.; Nagarkatti, P.S. Resveratrol downregulates miR-31 to promote to regulatory cells during prevention of TNBS-induced colitis. Mol. Nutr. Food Res. 2020, 64, e1900633. [Google Scholar] [CrossRef]
- Otsuka, K.; Yamamoto, Y.; Ochiya, T. Regulatory role of resveratrol, a microRNA-controlling compound, in HNRNPA1 expression, which is associated with poor prognosis in breast cancer. Oncotarget 2018, 9, 24718–24730. [Google Scholar] [CrossRef]
- Luo, X.; Gu, J.; Zhu, R.; Feng, M.; Zhu, X.; Li, Y.; Fei, J. Integrative analysis of differential miRNA and functional study of miR-21 by seed-targeting inhibition in multiple myeloma cells in response to berberine. BMC Syst. Biol. 2014, 8, 82. [Google Scholar] [CrossRef]
- Gao, Z.; Tan, C.; Sha, R. Berberine Promotes A549 Cell Apoptosis and Autophagy via miR-144. Nat. Prod. Commun. 2022, 17, 1934578–221124752. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, S. Berberine suppresses growth and metastasis of endometrial cancer cells via miR-101/COX-2. Biomed. Pharmacother. Biomed. Pharmacother. 2018, 103, 1287–1293. [Google Scholar] [CrossRef]
- Matarlo, J.S.; Krumpe, L.R.H.; Heinz, W.F.; Oh, D.; Shenoy, S.R.; Thomas, C.L.; Goncharova, E.I.; Lockett, S.J.; O’Keefe, B.R. The natural product butylcycloheptyl prodiginine binds pre-miR-21, inhibits Dicer-mediated processing of pre-miR-21, and blocks cellular proliferation. Cell Chem. Biol. 2019, 26, 1133–1142.e1134. [Google Scholar] [CrossRef]
- Yang, J.; Zou, Y.; Jiang, D. Honokiol suppresses proliferation and induces apoptosis via regulation of the miR21/PTEN/PI3K/AKT signaling pathway in human osteosarcoma cells. Int. J. Mol. Med. 2018, 41, 1845–1854. [Google Scholar] [CrossRef]
- Avtanski, D.B.; Nagalingam, A.; Bonner, M.Y.; Arbiser, J.L.; Saxena, N.K.; Sharma, D. Honokiol activates LKB1-miR-34a axis and antagonizes the oncogenic actions of leptin in breast cancer. Oncotarget 2015, 6, 29947–29962. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, B.; Chen, G.; Wu, W.; Zhou, L.; Shi, Y.; Zeng, Q.; Li, Y.; Sun, Y.; Deng, X.; et al. Targeting miR-21 with sophocarpine inhibits tumor progression and reverses epithelial-mesenchymal transition in head and neck cancer. Mol. Ther. J. Am. Soc. Gene Ther. 2017, 25, 2129–2139. [Google Scholar] [CrossRef]
- Ghasemi, S.; Lorigooini, Z.; Wibowo, J.; Amini-Khoei, H. Tricin isolated from Allium atroviolaceum potentiated the effect of docetaxel on PC3 cell proliferation: Role of miR-21. Nat. Prod. Res. 2019, 33, 1828–1831. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, Z.S.; Zhou, Y.Z.; Deng, Y.; Jiang, P.; Tan, S.L. Dihydromyricetin inhibits cell proliferation, migration, invasion and promotes apoptosis via regulating miR-21 in human cholangiocarcinoma cells. J. Cancer 2020, 11, 5689–5699. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Hu, C.; Wang, B.; Fan, S.; Jin, W. Curcumin suppresses cell proliferation, migration, and invasion through modulating miR-21-5p/SOX6 axis in hepatocellular carcinoma. Cancer Biother. Radiopharm. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cai, X.; Ma, L. Curcumin modifies epithelial–mesenchymal transition in colorectal cancer through regulation of miR-200c/EPM5. Cancer Manag. Res. 2020, 12, 9405–9415. [Google Scholar] [CrossRef]
- Liu, L.; Fu, Y.; Zheng, Y.; Ma, M.; Wang, C. Curcumin inhibits proteasome activity in triple-negative breast cancer cells through regulating p300/miR-142-3p/PSMB5 axis. Phytomedicine Int. J. Phytother. Phytopharm. 2020, 78, 153312. [Google Scholar] [CrossRef]
- Xu, R.; Li, H.; Wu, S.; Qu, J.; Yuan, H.; Zhou, Y.; Lu, Q. MicroRNA-1246 regulates the radio-sensitizing effect of curcumin in bladder cancer cells via activating P53. Int. Urol. Nephrol. 2019, 51, 1771–1779. [Google Scholar] [CrossRef]
- Liu, H.; Wang, J.; Tao, Y.; Li, X.; Qin, J.; Bai, Z.; Chi, B.; Yan, W.; Chen, X. Curcumol inhibits colorectal cancer proliferation by targeting miR-21 and modulated PTEN/PI3K/Akt pathways. Life Sci. 2019, 221, 354–361. [Google Scholar] [CrossRef]
- He, J.Q.; Zheng, M.X.; Ying, H.Z.; Zhong, Y.S.; Zhang, H.H.; Xu, M.; Yu, C.H. PRP1, a heteropolysaccharide from Platycodonis Radix, induced apoptosis of HepG2 cells via regulating miR-21-mediated PI3K/AKT pathway. Int. J. Biol. Macromol. 2020, 158, 542–551. [Google Scholar] [CrossRef]
- Shen, K.H.; Hung, J.H.; Liao, Y.C.; Tsai, S.T.; Wu, M.J.; Chen, P.S. Sinomenine inhibits migration and invasion of human lung cancer cell through downregulating expression of miR-21 and MMPs. Int. J. Mol. Sci. 2020, 21, 3080. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Ma, X.M.; Wang, T.T.; Yang, Y.; Zhang, N.; Zeng, N.; Bai, Z.G.; Yin, J.; Zhang, J.; Ding, G.Q.; et al. Psoralen suppresses cisplatin-mediated resistance and induces apoptosis of gastric adenocarcinoma by disruption of the miR196a-HOXB7-HER2 axis. Cancer Manag. Res. 2020, 12, 2803–2827. [Google Scholar] [CrossRef] [PubMed]
- Takala, R.; Ramji, D.P.; Andrews, R.; Zhou, Y.; Farhat, M.; Elmajee, M.; Rundle, S.; Choy, E. Pinolenic acid exhibits anti-inflammatory and anti-atherogenic effects in peripheral blood-derived monocytes from patients with rheumatoid arthritis. Sci. Rep. 2022, 12, 8807. [Google Scholar] [CrossRef] [PubMed]
- Younis, N.N.; Salama, A.; Shaheen, M.A.; Eissa, R.G. Pachymic acid attenuated doxorubicin-induced heart failure by suppressing miR-24 and preserving cardiac junctophilin-2 in rats. Int. J. Mol. Sci. 2021, 22, 10710. [Google Scholar] [CrossRef] [PubMed]
- de la Parra, C.; Castillo-Pichardo, L.; Cruz-Collazo, A.; Cubano, L.; Redis, R.; Calin, G.A.; Dharmawardhane, S. Soy isoflavone genistein-mediated downregulation of miR-155 contributes to the anticancer effects of genistein. Nutr. Cancer 2016, 68, 154–164. [Google Scholar] [CrossRef] [PubMed]
- La, X.; Zhang, L.; Li, Z.; Li, H.; Yang, Y. (-)-Epigallocatechin gallate (EGCG) enhances the sensitivity of colorectal cancer cells to 5-FU by inhibiting GRP78/NF-kappaB/miR-155-5p/MDR1 pathway. J. Agric. Food Chem. 2019, 67, 2510–2518. [Google Scholar] [CrossRef] [PubMed]
- Kang, Q.; Zhang, X.; Cao, N.; Chen, C.; Yi, J.; Hao, L.; Ji, Y.; Liu, X.; Lu, J. EGCG enhances cancer cells sensitivity under (60)Cogamma radiation based on miR-34a/Sirt1/p53. Food Chem. Toxicol. 2019, 133, 110807. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, K.; Gailhouste, L.; Yasukawa, K.; Kosaka, N.; Ochiya, T. A robust screening method for dietary agents that activate tumour-suppressor microRNAs. Sci. Rep. 2015, 5, 14697. [Google Scholar] [CrossRef]
- Peng, F.; Xiong, L.; Peng, C. (-)-Sativan inhibits tumor development and regulates miR-200c/PD-L1 in triple negative breast cancer cells. Front. Pharmacol. 2020, 11, 251. [Google Scholar] [CrossRef]
- Peng, F.; Tang, H.; Du, J.; Chen, J.; Peng, C. Isoliquiritigenin suppresses EMT-induced metastasis in triple-negative breast cancer through miR-200c/c-Jun/β-Catenin. Am. J. Chin. Med. 2021, 49, 505–523. [Google Scholar] [CrossRef]
- Geng, W.; Li, C.; Zhan, Y.; Zhang, R.; Zheng, J. Thymoquinone alleviates liver fibrosis via miR-30a-mediated epithelial-mesenchymal transition. J. Cell. Physiol. 2020, 236, 3629–3640. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Yang, K.; Gui, Y.; Wang, X. Nicotine-upregulated miR-30a arrests cell cycle in G1 phase by directly targeting CCNE2 in human periodontal ligament cells. Biochem. Cell Biol. 2020, 98, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Phuah, N.H.; Azmi, M.N.; Awang, K.; Nagoor, N.H. Down-regulation of microRNA-210 confers sensitivity towards 1′s-1′-acetoxychavicol acetate (ACA) in cervical cancer cells by targeting SMAD4. Mol. Cells 2017, 40, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Mollaei, H.; Safaralizadeh, R.; Babaei, E.; Abedini, M.R.; Hoshyar, R. The anti-proliferative and apoptotic effects of crocin on chemosensitive and chemoresistant cervical cancer cells. Biomed. Pharmacother. Biomed. Pharmacother. 2017, 94, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Yang, H.; Yu, J.; Li, Z.; Xu, Q.; Ding, B.; Jia, G. Crocin induces ROS-mediated papillary thyroid cancer cell apoptosis by modulating the miR-34a-5p/PTPN4 axis in vitro. Toxicol. Appl. Pharmacol. 2022, 437, 115892. [Google Scholar] [CrossRef] [PubMed]
- Hsia, S.M.; Yu, C.C.; Shih, Y.H.; Yuanchien Chen, M.; Wang, T.H.; Huang, Y.T.; Shieh, T.M. Isoliquiritigenin as a cause of DNA damage and inhibitor of ataxia-telangiectasia mutated expression leading to G2/M phase arrest and apoptosis in oral squamous cell carcinoma. Head Neck 2016, 38 (Suppl. 1), E360–E371. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Ni, Z.; Yicheng, S.; Pan, H.; Huang, Y.; Xiong, Y.; Liu, T. Anisomycin inhibits angiogenesis in ovarian cancer by attenuating the molecular sponge effect of the lncRNAMeg3/miR421/PDGFRA axis. Int. J. Oncol. 2019, 55, 1296–1312. [Google Scholar] [CrossRef]
- Zhang, F.; Ni, Z.J.; Ye, L.; Zhang, Y.Y.; Thakur, K.; Cespedes-Acuna, C.L.; Han, J.; Zhang, J.G.; Wei, Z.J. Asparanin A inhibits cell migration and invasion in human endometrial cancer via Ras/ERK/MAPK pathway. Food Chem. Toxicol. 2021, 150, 112036. [Google Scholar] [CrossRef]
- Kang, J.; Kim, E.; Kim, W.; Seong, K.M.; Youn, H.; Kim, J.W.; Kim, J.; Youn, B. Rhamnetin and cirsiliol induce radiosensitization and inhibition of epithelial-mesenchymal transition (EMT) by miR-34a-mediated suppression of Notch-1 expression in non-small cell lung cancer cell lines. J. Biol. Chem. 2013, 288, 27343–27357. [Google Scholar] [CrossRef]
- Paccez, J.D.; Duncan, K.; Sekar, D.; Correa, R.G.; Wang, Y.; Gu, X.; Bashin, M.; Chibale, K.; Libermann, T.A.; Zerbini, L.F. Dihydroartemisinin inhibits prostate cancer via JARID2/miR-7/miR-34a-dependent downregulation of Axl. Oncogenesis 2019, 8, 14. [Google Scholar] [CrossRef]
- Liang, X.; Xu, C.; Cao, X.; Wang, W. Isovitexin suppresses cancer stemness property and induces apoptosis of osteosarcoma cells by disruption of the DNMT1/miR-34a/Bcl-2 axis. Cancer Manag. Res. 2019, 11, 8923–8936. [Google Scholar] [CrossRef]
- Bai, J.; Wu, J.; Tang, R.; Sun, C.; Ji, J.; Yin, Z.; Ma, G.; Yang, W. Emodin, a natural anthraquinone, suppresses liver cancer in vitro and in vivo by regulating VEGFR2 and miR-34a. Investig. New Drugs 2020, 38, 229–245. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, L.; Zheng, Y.; Liu, W.; Xu, Y. Kaempferol attenuates the effects of XIST/miR-130a/STAT3 on inflammation and extracellular matrix degradation in osteoarthritis. Future Med. Chem. 2021, 13, 1451–1464. [Google Scholar] [CrossRef]
- Yan, Z.; Huang, Q.-L.; Chen, J.; Liu, F.; Wei, Y.; Chen, S.-L.; Wu, C.-Y.; Li, Z.; Lin, X.-P. Chicoric acid alleviates LPS-induced inflammatory response through miR-130a-3p/IGF-1pathway in human lung A549 epithelial cells. Eur. J. Inflamm. 2021, 19, 20587392211038244. [Google Scholar] [CrossRef]
- Xu, T.; Qin, L.; Zhu, Z.; Wang, X.; Liu, Y.; Fan, Y.; Zhong, S.; Wang, X.; Zhang, X.; Xia, L.; et al. MicroRNA-31 functions as a tumor suppressor and increases sensitivity to mitomycin-C in urothelial bladder cancer by targeting integrin alpha5. Oncotarget 2016, 7, 27445–27457. [Google Scholar] [CrossRef]
- Chen, G.; Ma, Y.; Jiang, Z.; Feng, Y.; Han, Y.; Tang, Y.; Zhang, J.; Ni, H.; Li, X.; Li, N. Lico A causes ER stress and apoptosis via up-regulating miR-144-3p in human lung cancer cell line H292. Front. Pharmacol. 2018, 9, 837. [Google Scholar] [CrossRef]
- Zeng, L.; Sun, Y.; Li, X.; Wang, J.; Yan, L. 10Hydroxycamptothecin induces apoptosis in human fibroblasts by regulating miRNA23b3p expression. Mol. Med. Rep. 2019, 19, 2680–2686. [Google Scholar] [CrossRef]
- Bae, M.; Kim, M.B.; Lee, J.Y. Astaxanthin attenuates the changes in the expression of microRNAs involved in the activation of hepatic stellate cells. Nutrients 2022, 14, 962. [Google Scholar] [CrossRef]
- Jin, Y.; Zhan, X.B.; Zhang, B.; Chen, Y.; Liu, C.F.; Yu, L.L. Polydatin exerts an antitumor effect through regulating the miR-382/PD-L1 axis in colorectal cancer. Cancer Biother. Radiopharm. 2020, 35, 83–91. [Google Scholar] [CrossRef]
- Hu, Y.; Luo, X.; Zhou, J.; Chen, S.; Gong, M.; Deng, Y.; Zhang, H. Piperlongumine inhibits the progression of osteosarcoma by downregulating the SOCS3/JAK2/STAT3 pathway via miR-30d-5p. Life Sci. 2021, 277, 119501. [Google Scholar] [CrossRef]
- Xie, C.; Du, L.Y.; Guo, F.; Li, X.; Cheng, B. Exosomes derived from microRNA-101-3p-overexpressing human bone marrow mesenchymal stem cells suppress oral cancer cell proliferation, invasion, and migration. Mol. Cell. Biochem. 2019, 458, 11–26. [Google Scholar] [CrossRef]
- Xue, P.; Huang, S.; Han, X.; Zhang, C.; Yang, L.; Xiao, W.; Fu, J.; Li, H.; Zhou, Y. Exosomal miR-101-3p and miR-423-5p inhibit medulloblastoma tumorigenesis through targeting FOXP4 and EZH2. Cell Death Differ. 2022, 29, 82–95. [Google Scholar] [CrossRef]
- Tao, L.; Xu, C.; Shen, W.; Tan, J.; Li, L.; Fan, M.; Sun, D.; Lai, Y.; Cheng, H. HIPK3 inhibition by exosomal hsa-miR-101-3p is related to metabolic reprogramming in colorectal cancer. Front. Oncol. 2021, 11, 758336. [Google Scholar] [CrossRef]
- Sakha, S.; Muramatsu, T.; Ueda, K.; Inazawa, J. Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci. Rep. 2016, 6, 38750. [Google Scholar] [CrossRef]
- Zheng, M.; Hou, L.; Ma, Y.; Zhou, L.; Wang, F.; Cheng, B.; Wang, W.; Lu, B.; Liu, P.; Lu, W.; et al. Exosomal let-7d-3p and miR-30d-5p as diagnostic biomarkers for non-invasive screening of cervical cancer and its precursors. Mol Cancer 2019, 18, 76. [Google Scholar] [CrossRef]
- Lin, X.J.; Fang, J.H.; Yang, X.J.; Zhang, C.; Yuan, Y.; Zheng, L.; Zhuang, S.M. Hepatocellular carcinoma cell-secreted exosomal microRNA-210 promotes angiogenesis in vitro and in vivo. Mol. Therapy. Nucleic Acids 2018, 11, 243–252. [Google Scholar] [CrossRef]
- Wang, L.; He, J.; Hu, H.; Tu, L.; Sun, Z.; Liu, Y.; Luo, F. Lung CSC-derived exosomal miR-210-3p contributes to a pro-metastatic phenotype in lung cancer by targeting FGFRL1. J. Cell. Mol. Med. 2020, 24, 6324–6339. [Google Scholar] [CrossRef]
- Ge, L.; Zhou, F.; Nie, J.; Wang, X.; Zhao, Q. Hypoxic colorectal cancer-secreted exosomes deliver miR-210-3p to normoxic tumor cells to elicit a protumoral effect. Exp. Biol. Med. 2021, 246, 1895–1906. [Google Scholar] [CrossRef]
- Min, Q.H.; Wang, X.Z.; Zhang, J.; Chen, Q.G.; Li, S.Q.; Liu, X.Q.; Li, J.; Liu, J.; Yang, W.M.; Jiang, Y.H.; et al. Exosomes derived from imatinib-resistant chronic myeloid leukemia cells mediate a horizontal transfer of drug-resistant trait by delivering miR-365. Exp. Cell Res. 2018, 362, 386–393. [Google Scholar] [CrossRef]
- Mollaei, H.; Hoshyar, R.; Abedini, M.R.; Safaralizadeh, R. Crocin enhances cisplatin-induced chemosensitivity in human cervical cancer cell line. Int. J. Cancer Manag. 2019, 12, e94909. [Google Scholar]
- Wang, X.; Hao, R.; Wang, F.; Wang, F. ZFAS1 promotes cisplatin resistance via suppressing miR-421 expression in oral squamous cell carcinoma. Cancer Manag. Res. 2020, 12, 7251–7262. [Google Scholar] [CrossRef] [PubMed]
- Jingyue, S.; Xiao, W.; Juanmin, Z.; Wei, L.; Daoming, L.; Hong, X. TFAP2E methylation promotes 5fluorouracil resistance via exosomal miR106a5p and miR421 in gastric cancer MGC803 cells. Mol. Med. Rep. 2019, 20, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Tao, Y.W.; Gao, S.; Li, P.; Zheng, J.M.; Zhang, S.E.; Liang, J.; Zhang, Y. Cancer-associated fibroblasts contribute to oral cancer cells proliferation and metastasis via exosome-mediated paracrine miR-34a-5p. EBioMedicine 2018, 36, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Zuo, L.; Tao, H.; Xu, H.; Li, C.; Qiao, G.; Guo, M.; Cao, S.; Liu, M.; Lin, X. Exosomes-coated miR-34a displays potent antitumor activity in pancreatic cancer both in vitro and in vivo. Drug Des. Dev. Ther. 2020, 14, 3495–3507. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, D.; Li, L.; Xin, T.; Zhao, Y.; Ma, R.; Du, J. Exosomal microRNA-144 from bone marrow-derived mesenchymal stem cells inhibits the progression of non-small cell lung cancer by targeting CCNE1 and CCNE2. Stem Cell Res. Ther. 2020, 11, 87. [Google Scholar] [CrossRef]
- Tian, X.; Liu, Y.; Wang, Z.; Wu, S. miR-144 delivered by nasopharyngeal carcinoma-derived EVs stimulates angiogenesis through the FBXW7/HIF-1alpha/VEGF-A axis. Mol. Ther. Nucleic Acids 2021, 24, 1000–1011. [Google Scholar] [CrossRef]
- Zhou, C.; Chen, Y.; He, X.; Zheng, Z.; Xue, D. Functional implication of exosomal miR-217 and miR-23b-3p in the progression of prostate cancer. OncoTargets Ther. 2020, 13, 11595–11606. [Google Scholar] [CrossRef]
- Chen, D.; Wu, X.; Xia, M.; Wu, F.; Ding, J.; Jiao, Y.; Zhan, Q.; An, F. Upregulated exosomic miR23b3p plays regulatory roles in the progression of pancreatic cancer. Oncol. Rep. 2017, 38, 2182–2188. [Google Scholar] [CrossRef]
- Hou, C.X.; Sun, N.N.; Han, W.; Meng, Y.; Wang, C.X.; Zhu, Q.H.; Tang, Y.T.; Ye, J.H. Exosomal microRNA-23b-3p promotes tumor angiogenesis and metastasis by targeting PTEN in salivary adenoid cystic carcinoma. Carcinogenesis 2022, 43, 682–692. [Google Scholar] [CrossRef]
- Sun, L.P.; Xu, K.; Cui, J.; Yuan, D.Y.; Zou, B.; Li, J.; Liu, J.L.; Li, K.Y.; Meng, Z.; Zhang, B. Cancer associated fibroblast derived exosomal miR3825p promotes the migration and invasion of oral squamous cell carcinoma. Oncol. Rep. 2019, 42, 1319–1328. [Google Scholar] [CrossRef]
- Jiang, Y.; Ji, X.; Liu, K.; Shi, Y.; Wang, C.; Li, Y.; Zhang, T.; He, Y.; Xiang, M.; Zhao, R. Exosomal miR-200c-3p negatively regulates the migraion and invasion of lipopolysaccharide (LPS)-stimulated colorectal cancer (CRC). BMC Mol. Cell. Biol. 2020, 21, 48. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, C.; Wang, S.; Wang, Z.; Jiang, J.; Wang, W.; Li, X.; Chen, J.; Liu, K.; Li, C.; et al. Exosomes derived from hypoxic oral squamous cell carcinoma cells deliver miR-21 to normoxic cells to elicit a prometastatic phenotype. Cancer Res. 2016, 76, 1770–1780. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Chen, G.; Sun, D.; Lei, M.; Li, Y.; Zhou, C.; Li, X.; Xue, W.; Wang, H.; Liu, C.; et al. Exosomes containing miR-21 transfer the characteristic of cisplatin resistance by targeting PTEN and PDCD4 in oral squamous cell carcinoma. Acta Biochim. Biophys. Sin. 2017, 49, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.S.; Zhang, H.P.; Yang, C.Q.; Li, L.N.; Shen, Y.; Zhang, Y.Q. Exosomal miR-155-5p promotes proliferation and migration of gastric cancer cells by inhibiting TP53INP1 expression. Pathol. Res. Pract. 2020, 216, 152986. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, Z.; Ni, Y.; Bian, C.; Huang, J.; Chen, L.; Xie, X.; Wang, J. Tumor-associated macrophages secret exosomal miR-155 and miR-196a-5p to promote metastasis of non-small-cell lung cancer. Transl. Lung Cancer Res. 2021, 10, 1338–1354. [Google Scholar] [CrossRef]
- Gu, W.; Gong, L.; Wu, X.; Yao, X. Hypoxic TAM-derived exosomal miR-155-5p promotes RCC progression through HuR-dependent IGF1R/AKT/PI3K pathway. Cell Death Discov. 2021, 7, 147. [Google Scholar] [CrossRef]
- Wang, D.; Wang, X.; Song, Y.; Si, M.; Sun, Y.; Liu, X.; Cui, S.; Qu, X.; Yu, X. Exosomal miR-146a-5p and miR-155-5p promote CXCL12/CXCR7-induced metastasis of colorectal cancer by crosstalk with cancer-associated fibroblasts. Cell Death Dis. 2022, 13, 380. [Google Scholar] [CrossRef]
- Kirave, P.; Gondaliya, P.; Kulkarni, B.; Rawal, R.; Garg, R.; Jain, A.; Kalia, K. Exosome mediated miR-155 delivery confers cisplatin chemoresistance in oral cancer cells via epithelial-mesenchymal transition. Oncotarget 2020, 11, 1157–1171. [Google Scholar] [CrossRef]
- Xing, Y.; Ruan, G.; Ni, H.; Qin, H.; Chen, S.; Gu, X.; Shang, J.; Zhou, Y.; Tao, X.; Zheng, L. Tumor immune microenvironment and its related miRNAs in tumor progression. Front. Immunol. 2021, 12, 624725. [Google Scholar] [CrossRef]
- Yuan, Y.; Wang, Z.; Chen, M.; Jing, Y.; Shu, W.; Xie, Z.; Li, Z.; Xu, J.; He, F.; Jiao, P.; et al. Macrophage-derived exosomal miR-31-5p promotes oral squamous cell carcinoma tumourigenesis through the large tumor suppressor 2-mediated Hippo signalling pathway. J. Biomed. Nanotechnol. 2021, 17, 822–837. [Google Scholar] [CrossRef]
- Yu, F.; Liang, M.; Huang, Y.; Wu, W.; Zheng, B.; Chen, C. Hypoxic tumor-derived exosomal miR-31-5p promotes lung adenocarcinoma metastasis by negatively regulating SATB2-reversed EMT and activating MEK/ERK signaling. J. Exp. Clin. Cancer Res. CR 2021, 40, 179. [Google Scholar] [CrossRef] [PubMed]
- He, J.; He, J.; Min, L.; He, Y.; Guan, H.; Wang, J.; Peng, X. Extracellular vesicles transmitted miR-31-5p promotes sorafenib resistance by targeting MLH1 in renal cell carcinoma. Int. J. Cancer. J. Int. Cancer 2020, 146, 1052–1063. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.H.; Kuo, W.W.; Shih, H.N.; Cheng, S.F.; Yang, C.K.; Chen, M.C.; Tu, C.C.; Viswanadha, V.P.; Liao, P.H.; Huang, C.Y. FOXC1 regulation of miR-31-5p confers oxaliplatin resistance by targeting LATS2 in colorectal cancer. Cancers 2019, 11, 1576. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Qin, X.; Yan, M.; Shi, J.; Xu, Q.; Li, Z.; Yang, W.; Zhang, J.; Chen, W. Loss of exosomal miR-3188 in cancer-associated fibroblasts contributes to HNC progression. J. Exp. Clin. Cancer Res. CR 2019, 38, 151. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Luo, R.; Liu, Y.; Gao, L.; Fu, Z.; Fu, Q.; Luo, X.; Chen, Y.; Deng, X.; Liang, Z.; et al. miR-3188 regulates nasopharyngeal carcinoma proliferation and chemosensitivity through a FOXO1-modulated positive feedback loop with mTOR-p-PI3K/AKT-c-JUN. Nat. Commun. 2016, 7, 11309. [Google Scholar] [CrossRef]
- Wang, C.; Liu, E.; Li, W.; Cui, J.; Li, T. MiR-3188 inhibits non-small cell lung cancer cell proliferation through FOXO1-mediated mTOR-p-PI3K/AKT-c-JUN signaling pathway. Front. Pharmacol. 2018, 9, 1362. [Google Scholar] [CrossRef]
- He, L.; Ping, F.; Fan, Z.; Zhang, C.; Deng, M.; Cheng, B.; Xia, J. Salivary exosomal miR-24-3p serves as a potential detective biomarker for oral squamous cell carcinoma screening. Biomed. Pharmacother. 2020, 121, 109553. [Google Scholar] [CrossRef]
- Zhang, H.W.; Shi, Y.; Liu, J.B.; Wang, H.M.; Wang, P.Y.; Wu, Z.J.; Li, L.; Gu, L.P.; Cao, P.S.; Wang, G.R.; et al. Cancer-associated fibroblast-derived exosomal microRNA-24-3p enhances colon cancer cell resistance to MTX by down-regulating CDX2/HEPH axis. J. Cell. Mol. Med. 2021, 25, 3699–3713. [Google Scholar] [CrossRef]
- Kulkarni, B.; Gondaliya, P.; Kirave, P.; Rawal, R.; Jain, A.; Garg, R.; Kalia, K. Exosome-mediated delivery of miR-30a sensitize cisplatin-resistant variant of oral squamous carcinoma cells via modulating Beclin1 and Bcl2. Oncotarget 2020, 11, 1832–1845. [Google Scholar] [CrossRef]
- Tao, K.; Liu, J.; Liang, J.; Xu, X.; Xu, L.; Mao, W. Vascular endothelial cell-derived exosomal miR-30a-5p inhibits lung adenocarcinoma malignant progression by targeting CCNE2. Carcinogenesis 2021, 42, 1056–1067. [Google Scholar] [CrossRef]
- Du, Q.; Ye, X.; Lu, S.R.; Li, H.; Liu, H.Y.; Zhai, Q.; Yu, B. Exosomal miR-30a and miR-222 derived from colon cancer mesenchymal stem cells promote the tumorigenicity of colon cancer through targeting MIA3. J. Gastrointest. Oncol. 2021, 12, 52–68. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Zhang, J.; Li, J.; Shao, J.; Fang, L. MiR-130a-3p inhibits migration and invasion by regulating RAB5B in human breast cancer stem cell-like cells. Biochem. Biophys. Res. Commun. 2018, 501, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Yin, G.; Kong, W.; Zheng, S.; Shan, Y.; Zhang, J.; Ying, R.; Wu, H. Exosomal miR-130a-3p promotes the progression of differentiated thyroid cancer by targeting insulin-like growth factor 1. Oncol. Lett. 2021, 21, 283. [Google Scholar] [CrossRef]
- Dickman, C.T.; Lawson, J.; Jabalee, J.; MacLellan, S.A.; LePard, N.E.; Bennewith, K.L.; Garnis, C. Selective extracellular vesicle exclusion of miR-142-3p by oral cancer cells promotes both internal and extracellular malignant phenotypes. Oncotarget 2017, 8, 15252–15266. [Google Scholar] [CrossRef] [PubMed]
- Plousiou, M.; De Vita, A.; Miserocchi, G.; Bandini, E.; Vannini, I.; Melloni, M.; Masalu, N.; Fabbri, F.; Serra, P. Growth inhibition of retinoblastoma cell line by exosome-mediated transfer of miR-142-3p. Cancer Manag. Res. 2022, 14, 2119–2131. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhang, H.; Liu, W.; Yin, Y.; Jiang, J.; Yan, C.; Wang, Y.; Li, L. Mechanism of HBV-positive liver cancer cell exosomal miR-142-3p by inducing ferroptosis of M1 macrophages to promote liver cancer progression. Transl. Cancer Res. 2022, 11, 1173–1187. [Google Scholar] [CrossRef]
- Lou, C.; Shi, J.; Xu, Q. Exosomal miR-626 promotes the malignant behavior of oral cancer cells by targeting NFIB. Mol. Biol. Rep. 2022, 49, 4829–4840. [Google Scholar] [CrossRef]
- Singh, R.; Pochampally, R.; Watabe, K.; Lu, Z.; Mo, Y.Y. Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol. Cancer 2014, 13, 256. [Google Scholar] [CrossRef]
- Yan, T.; Wang, X.; Wei, G.; Li, H.; Hao, L.; Liu, Y.; Yu, X.; Zhu, W.; Liu, P.; Zhu, Y.; et al. Exosomal miR-10b-5p mediates cell communication of gastric cancer cells and fibroblasts and facilitates cell proliferation. J. Cancer 2021, 12, 2140–2150. [Google Scholar] [CrossRef]
- Higaki, M.; Shintani, T.; Hamada, A.; Rosli, S.N.Z.; Okamoto, T. Eldecalcitol (ED-71)-induced exosomal miR-6887-5p suppresses squamous cell carcinoma cell growth by targeting heparin-binding protein 17/fibroblast growth factor-binding protein-1 (HBp17/FGFBP-1). Vitr. Cell. Dev. Biol. Anim. 2020, 56, 222–233. [Google Scholar] [CrossRef]
- Chen, T.; Liu, Y.; Chen, J.; Zheng, H.; Chen, Q.; Zhao, J. Exosomal miR-3180-3p inhibits proliferation and metastasis of non-small cell lung cancer by downregulating FOXP4. Thorac. Cancer 2021, 12, 372–381. [Google Scholar] [CrossRef] [PubMed]
| Natural Products | miRNAs | Cell Functions | Cancer | References |
|---|---|---|---|---|
| Ursolic acid | ↓miR-21-5p | apoptosis | glioblastoma | [81] |
| ↑miR-200c-3p | apoptosis, anti-invasion | colon | [82] | |
| Resveratrol | ↓miR-21-5p | apoptosis | bladder | [83] |
| ↓miR-31-5p | anticolitis | (T cells) | [84] | |
| ↑miR-424-5p | antiproliferation | breast | [85] | |
| Berberine | ↓miR-21-5p | antiproliferation | myeloma | [86] |
| ↑miR-144-3p | apoptosis, autophagy | lung | [87] | |
| ↑miR-101-3p | antiproliferation, antimigration | endometrial | [88] | |
| Butylcycloheptyl prodiginine | ↓miR-21-5p | antiproliferation | colon | [89] |
| Honokiol | ↓miR-21-5p | apoptosis | osteosarcoma | [90] |
| ↑miR-34a-5p | anti-EMT | breast | [91] | |
| Sophocarpine | ↓miR-21-5p | antiproliferation, anti-EMT | head/neck | [92] |
| Tricin | ↓miR-21-5p | chemosensitization | prostate | [93] |
| Dihydromyricetin | ↓miR-21-5p | antiproliferation, antimigration | cholangiocarcinoma | [94] |
| Curcumin | ↓miR-21-5p | antiproliferation, antimigration | liver | [95] |
| ↑miR-200c-3p | anti-EMT | colon | [96] | |
| ↑miR-142-3p | 20S proteasome suppression | breast | [97] | |
| ↓miR-1246 | antiproliferation | bladder | [98] | |
| Curcumol | ↓miR-21-5p | antiproliferation | colon | [99] |
| PRP1 | ↓miR-21-5p | apoptosis | liver | [100] |
| Sinomenine | ↓miR-21-5p | antimigration | lung | [101] |
| Psoralen | ↑miR-196a-5p | apoptosis | gastric | [102] |
| Pinolenic acid | ↑miR-3188 | anti-inflammation | (rheumatoid arthritis) | [103] |
| Pachymic acid | ↓miR-24-3p | anti-heart failure | (left ventricle) | [104] |
| Genistein | ↓miR-155-5p | antiproliferation | (cardiac) | [105] |
| (−)-Epigallocatechin gallate | ↓miR-155-5p | chemosensitization | colon | [106] |
| ↑miR-34a-5p | radiosensitization | liver | [107] | |
| Enoxolone, Magnolol, | ↑miR-200c-3p | anti-invasion | breast | [108] |
| Palmatine chloride | ||||
| (−)-Sativan | ↑miR-200c-3p | apoptosis, antimigration | breast | [109] |
| Isoliquiritigenin | ↑miR-200c-3p | antimigration | breast | [110] |
| Thymoquinone | ↑miR-30a-5p | anti-liver fibrosis | (liver) | [111] |
| Nicotine | ↑miR-30a-5p | G1 arrest | (periodontal ligament) | [112] |
| Norcantharidin | ↑miR-30a-5p | antiproliferation, antimigration | giant cell tumor of bone | [62] |
| 1΄S-1΄-acetoxychavicol acetate | ↓miR-210-3p | apoptosis | cervical | [113] |
| Crocin | ↓miR-365a-3p | apoptosis | cervical | [114] |
| ↓miR-34a-5p | apoptosis | papillary thyroid | [115] | |
| Isoliquiritigenin | ↓miR-421 | apoptosis, DNA damage | oral | [116] |
| Anisomycin | ↑miR-421 | anti-angiogenesis | ovarian | [117] |
| Asparanin A | ↓miR-421 | antimigration | endometrial | [118] |
| Rhamnetin, Cirsiliol | ↑miR-34a-5p | radiosensitization, anti-EMT | lung | [119] |
| Dihydroartemisinin | ↑miR-34a-5p | apoptosis, antimigration | prostate | [120] |
| Isovitexin | ↑miR-34a-5p | apoptosis | osteosarcoma | [121] |
| Emodin | ↑miR-34a-5p | antiproliferation | liver | [122] |
| Kaempferol | ↑miR-130a-3p | cytokine reduction | (chondrocyte) | [123] |
| Chicoric acid | ↓miR-130a-3p | anti-inflammation | lung | [124] |
| Mitomycin C | ↑miR-31-5p | chemosensitization | bladder | [125] |
| Licochalcone A | ↑miR-144-3p | ER stress, apoptosis | lung | [126] |
| 10-Hydroxycamptothecin | ↑miR-23b-3p | apoptosis | (fibroblast) | [127] |
| Astaxanthin | ↓miR-382-5p | anti-liver fibrosis | (liver) | [128] |
| Polydatin | ↑miR-382-5p | apoptosis | colon | [129] |
| Piperlongumine | ↓miR-30d-5p | antiproliferation | osteosarcoma | [130] |
| miRNAs | miRNAs Status in TIME | miRNA Effects of Natural Products * |
|---|---|---|
| miR-21-5p | ↑TAM [159] ↑MDSC [159] | miR-21-5p (Ursolic acid [81], Resveratrol [83], Berberine [86], Butylcycloheptyl prodiginine [89], Honokiol [90], Sophocarpine [92], Tricin [93], Dihydromyricetin [94], Curcumin [95], Curcumol [99], PRP1 [100], Sinomenine [101]) |
| miR-200c-3p | ↑MDSC [159] | miR-200c-3p (Urolic acid [82], Curcumin [96], Enoxolone, Magnolol, Palmatine chloride [108], (−)-Sativan [109], Isoliquiritigenin [110]) |
| miR-155-5p | ↑TAM,↑MDSC,↑NK [159] | miR-155-5p (Genistein [105], (−)-Epigallocatechin gallate [106]) |
| miR-30a-5p | ↑TAM,↑MDSC [159] | miR-30a-5p (Thymoquinone [111], Nicotine [112], Norcantharidin [62]) |
| miR-34a-5p | ↓TAM [159] | miR-34a-5p (Honokiol [91], (−)-Epigallocatechin gallate [107], Rhamnetin, Cirsiliol [119], Dihydroartemisinin [120], Isovitexin [121], Emodin [122]) |
| miR-34a-5p (Crocin [115]) | ||
| miR-130a-3p | ↑NK [159] | miR-130a-3p (Kaempferol [123]) |
| miR-130a-3p (Chicoric acid [124]) | ||
| miR-101-3p | ↑TAM [159] | miR-101-3p (Berberine [88]) |
| miR-142-3p | ↑TAM [159] | miR-142-3p (Curcumin [97]) |
| miR-24-3p | ↑NK [159] | miR-24-3p (Pachymic acid [104]) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chuang, Y.-T.; Tang, J.-Y.; Shiau, J.-P.; Yen, C.-Y.; Chang, F.-R.; Yang, K.-H.; Hou, M.-F.; Farooqi, A.A.; Chang, H.-W. Modulating Effects of Cancer-Derived Exosomal miRNAs and Exosomal Processing by Natural Products. Cancers 2023, 15, 318. https://doi.org/10.3390/cancers15010318
Chuang Y-T, Tang J-Y, Shiau J-P, Yen C-Y, Chang F-R, Yang K-H, Hou M-F, Farooqi AA, Chang H-W. Modulating Effects of Cancer-Derived Exosomal miRNAs and Exosomal Processing by Natural Products. Cancers. 2023; 15(1):318. https://doi.org/10.3390/cancers15010318
Chicago/Turabian StyleChuang, Ya-Ting, Jen-Yang Tang, Jun-Ping Shiau, Ching-Yu Yen, Fang-Rong Chang, Kun-Han Yang, Ming-Feng Hou, Ammad Ahmad Farooqi, and Hsueh-Wei Chang. 2023. "Modulating Effects of Cancer-Derived Exosomal miRNAs and Exosomal Processing by Natural Products" Cancers 15, no. 1: 318. https://doi.org/10.3390/cancers15010318
APA StyleChuang, Y.-T., Tang, J.-Y., Shiau, J.-P., Yen, C.-Y., Chang, F.-R., Yang, K.-H., Hou, M.-F., Farooqi, A. A., & Chang, H.-W. (2023). Modulating Effects of Cancer-Derived Exosomal miRNAs and Exosomal Processing by Natural Products. Cancers, 15(1), 318. https://doi.org/10.3390/cancers15010318








