Preparing Well for Esophageal Endoscopic Detection Using a Hybrid Model and Transfer Learning
Abstract
:Simple Summary
Abstract
1. Introduction
- Hybrid model results in high accuracy while ensuring compactness when making real-time predictions;
- Transfer learning can handle data shortage, reduce data collection time, and deploy the application for many different custom dataset cases;
- Visualization is provided to a model, especially a model based on the attention mechanism, and support for visual diagnosis using images is provided.
2. Materials and Methods
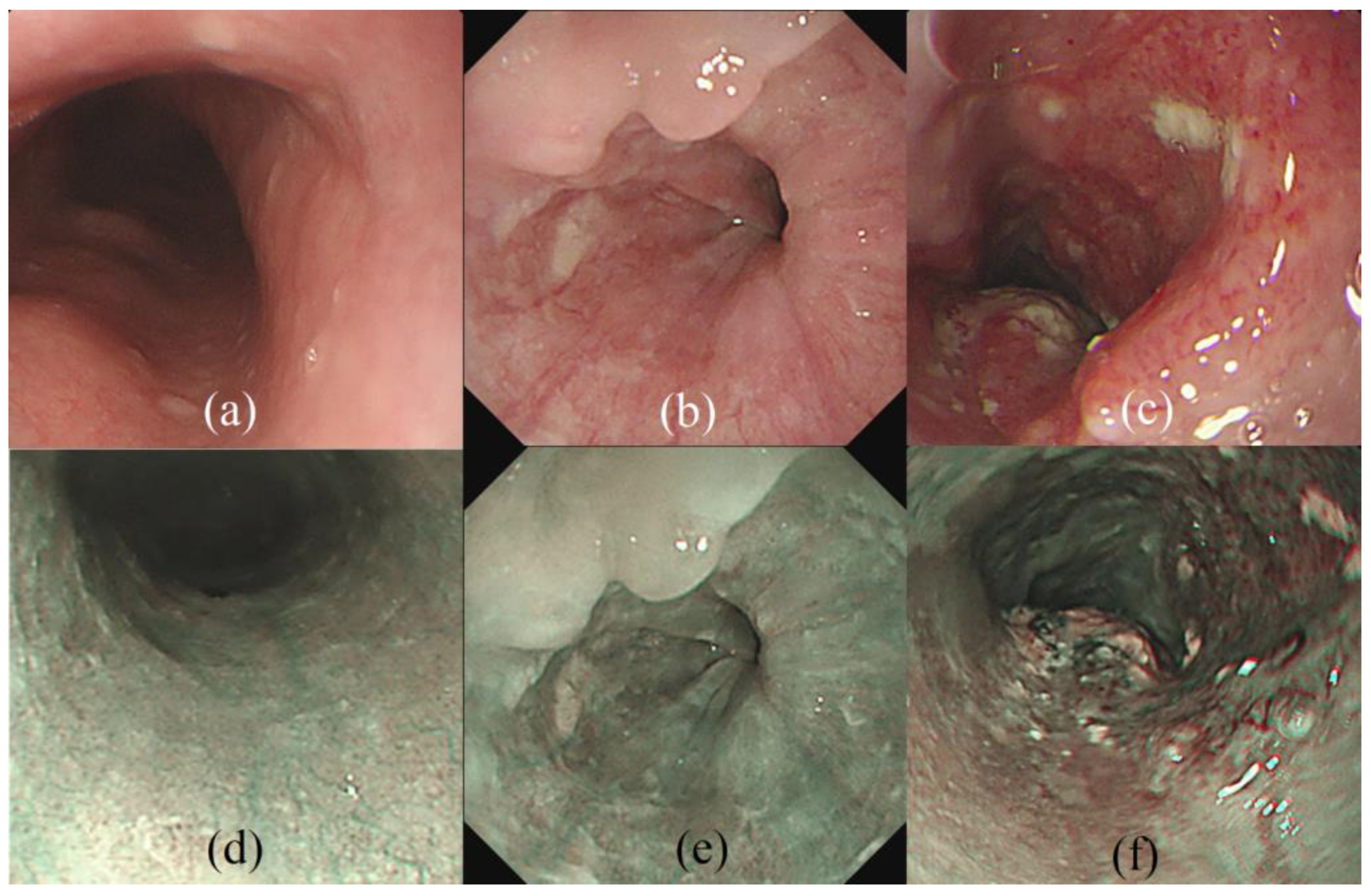
2.1. Data Preparation
2.2. Network Architecture
2.3. Experimental Setups
2.4. Transfer Learning Strategies
3. Results
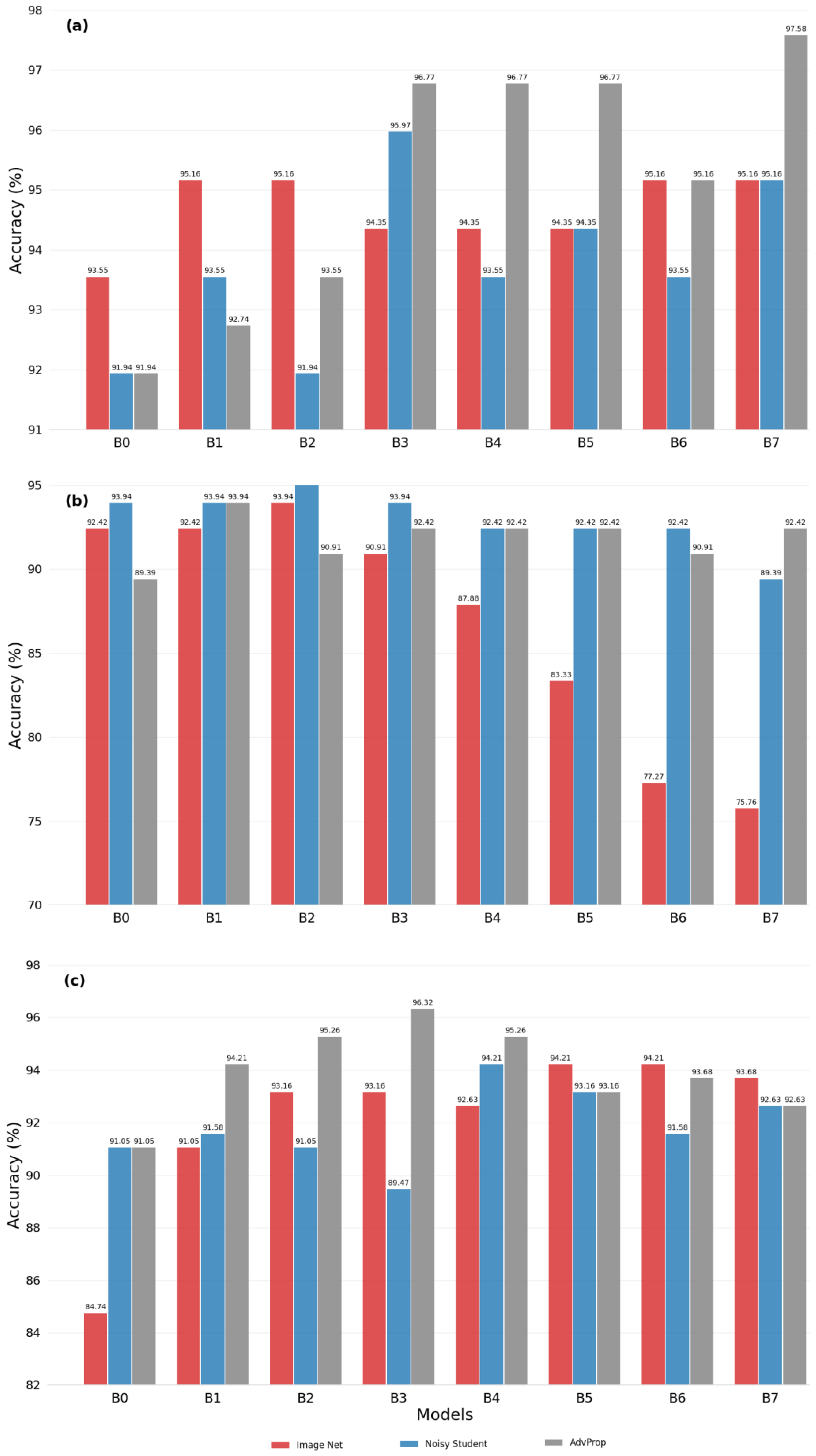
3.1. Evaluating Transfer Learning Strategies
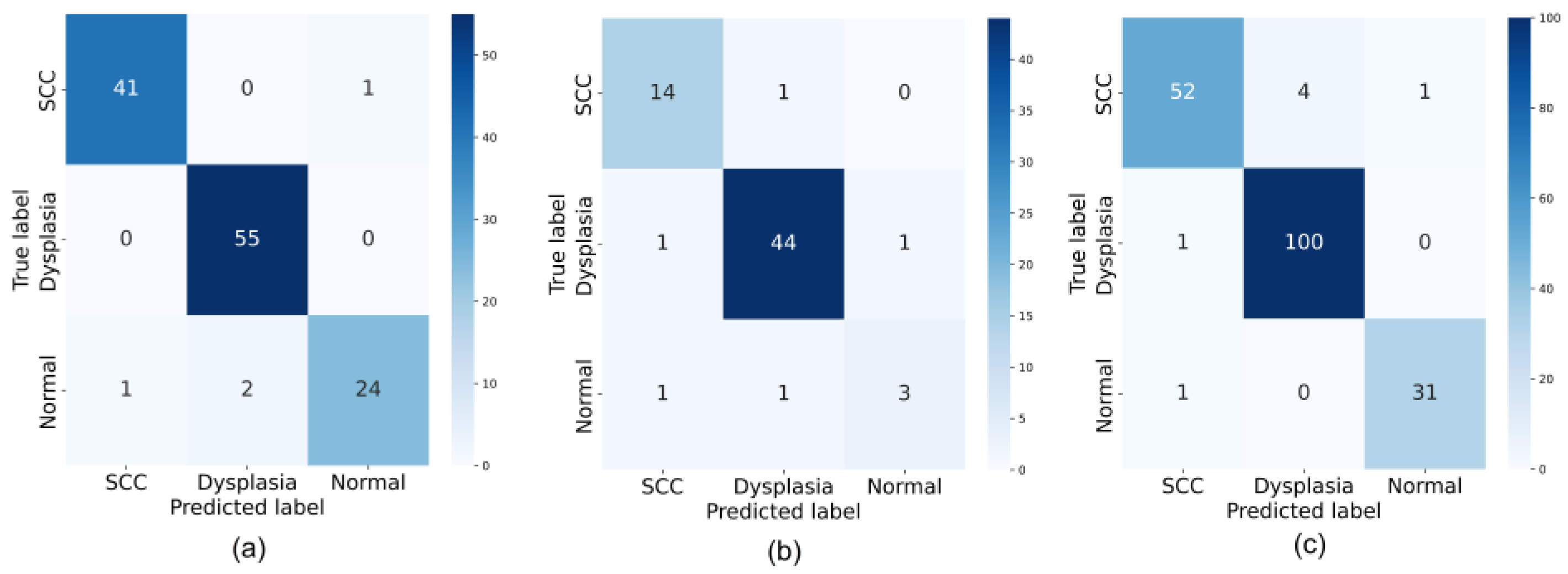
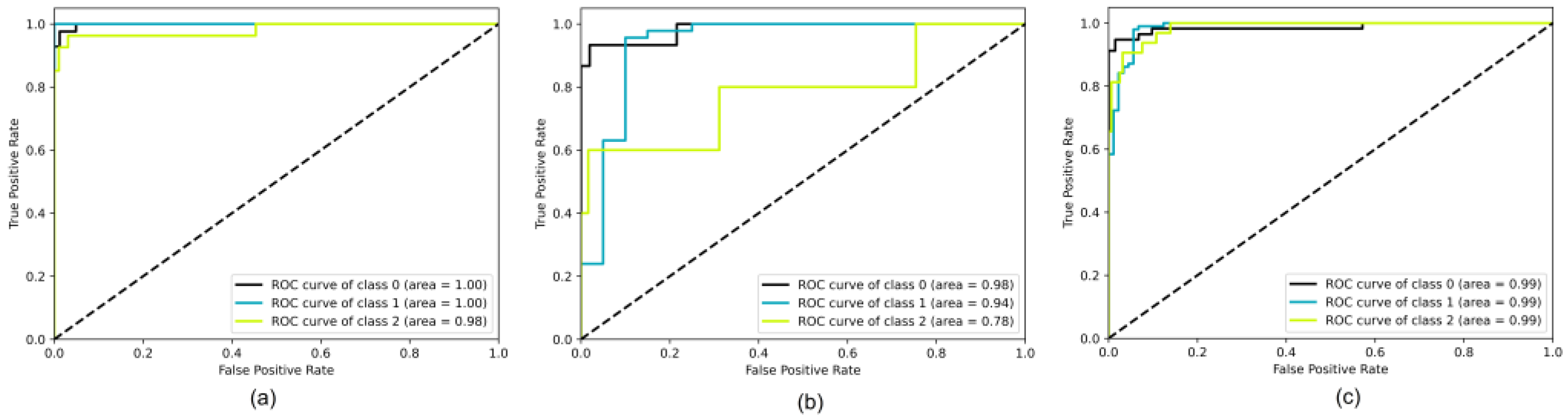
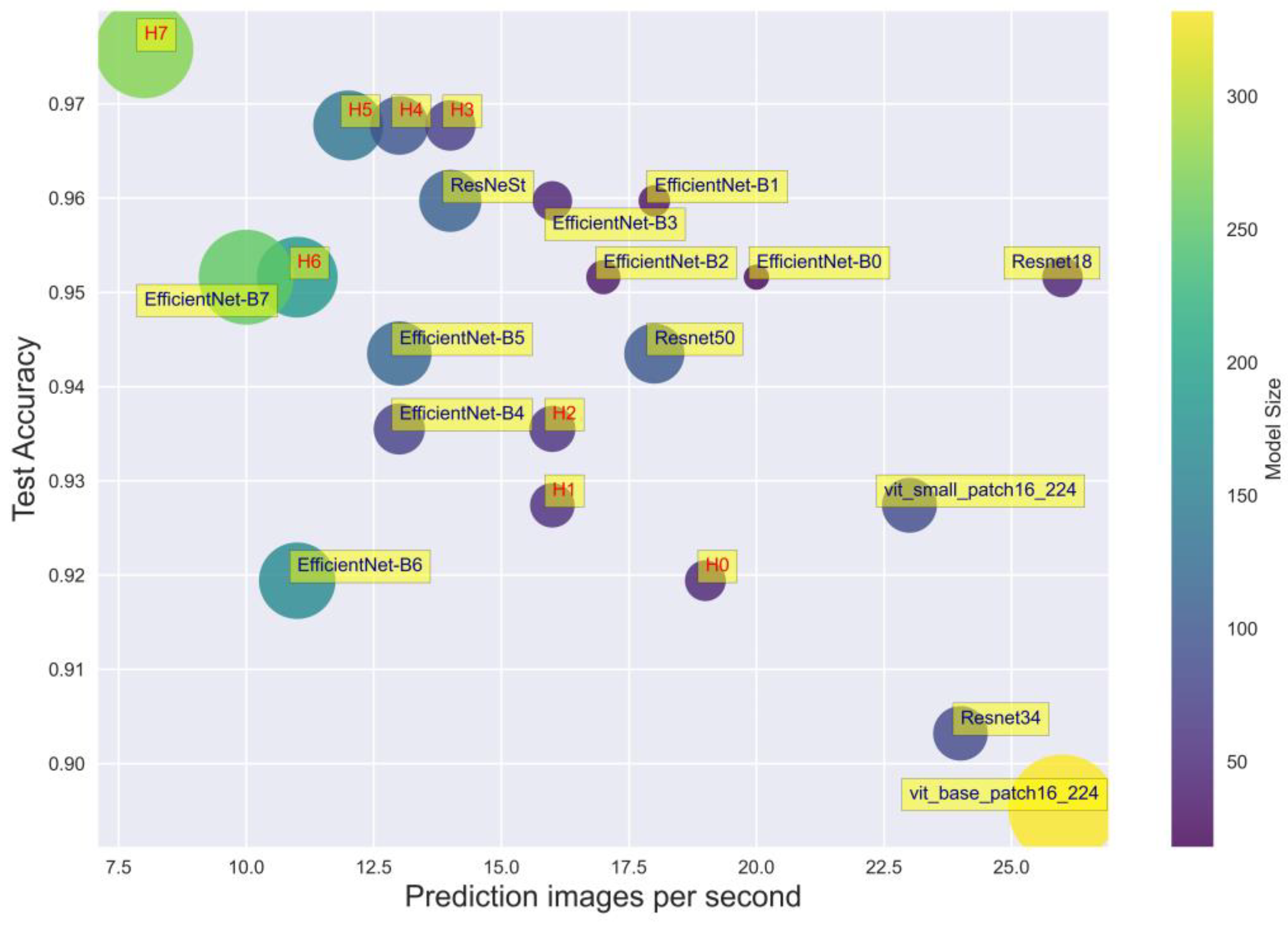
3.2. Model Performance Evaluation
3.3. Visualization of Hybrid Model
3.4. Ablation Study
3.5. Comparison with Other Existing Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shaheen, N.J.; Falk, G.W.; Iyer, P.G.; Souza, R.F.; Yadlapati, R.H.; Sauer, B.G.; Wani, S. Diagnosis and management of Barrett’s esophagus: An updated ACG guideline. Am. J. Gastroenterol. 2022, 117, 559. [Google Scholar] [CrossRef] [PubMed]
- Katzka, D.A.; Pandolfino, J.E.; Kahrilas, P.J. Phenotypes of gastroesophageal reflux disease: Where Rome, Lyon, and Montreal meet. Clin. Gastroenterol. Hepatol. 2020, 18, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, L.A.; Obeid, N.R.; Yang, J.; Zhu, C.; Altieri, M.S.; Spaniolas, K.; Pryor, A.D. Incidence of GERD, esophagitis, Barrett’s esophagus, and esophageal adenocarcinoma after bariatric surgery. Surg. Obes. Relat. Dis. 2020, 16, 1828–1836. [Google Scholar] [CrossRef] [PubMed]
- Mastracci, L.; Grillo, F.; Parente, P.; Unti, E.; Battista, S.; Spaggiari, P.; Campora, M.; Scaglione, G.; Fassan, M.; Fiocca, R. Gastro-esophageal reflux disease and Barrett’s esophagus: An overview with an histologic diagnostic approach. Pathologica 2020, 112, 117. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Zhang, C. The Oxidative Damage and Inflammation Mechanisms in GERD-Induced Barrett’s Esophagus. Front. Cell Dev. Biol. 2022, 10, 885537. [Google Scholar] [CrossRef]
- Que, J.; Garman, K.S.; Souza, R.F.; Spechler, S.J. Pathogenesis and cells of origin of Barrett’s esophagus. Gastroenterology 2019, 157, 349–364.e1. [Google Scholar] [CrossRef] [Green Version]
- Krishnamoorthi, R.; Mohan, B.P.; Jayaraj, M.; Wang, K.K.; Katzka, D.A.; Ross, A.; Adler, D.G.; Iyer, P.G. Risk of progression in Barrett’s esophagus indefinite for dysplasia: A systematic review and meta-analysis. Gastrointest. Endosc. 2020, 91, 3–10.e3. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Sun, C.; Wu, Y.; Chen, X.; Kailas, S.; Karadsheh, Z.; Li, G.; Guo, Z.; Yang, H.; Hu, L. Do proton pump inhibitors prevent Barrett’s esophagus progression to high-grade dysplasia and esophageal adenocarcinoma? An updated meta-analysis. J. Cancer Res. Clin. Oncol. 2021, 147, 2681–2691. [Google Scholar] [CrossRef]
- Dam, A.N.; Klapman, J. A narrative review of Barrett’s esophagus in 2020, molecular and clinical update. Ann. Transl. Med. 2020, 8, 1107. [Google Scholar] [CrossRef]
- Uhlenhopp, D.J.; Then, E.O.; Sunkara, T.; Gaduputi, V. Epidemiology of esophageal cancer: Update in global trends, etiology and risk factors. Clin. J. Gastroenterol. 2020, 13, 1010–1021. [Google Scholar] [CrossRef]
- Zhang, Y. Epidemiology of esophageal cancer. World J. Gastroenterol. WJG 2013, 19, 5598. [Google Scholar] [CrossRef]
- Choi, J.; Kim, S.G.; Im, J.P.; Kim, J.S.; Jung, H.C.; Song, I.S. Endoscopic prediction of tumor invasion depth in early gastric cancer. Gastrointest. Endosc. 2011, 73, 917–927. [Google Scholar] [CrossRef]
- Oyama, T.; Inoue, H.; Arima, M.; Momma, K.; Omori, T.; Ishihara, R.; Hirasawa, D.; Takeuchi, M.; Tomori, A.; Goda, K. Prediction of the invasion depth of superficial squamous cell carcinoma based on microvessel morphology: Magnifying endoscopic classification of the Japan Esophageal Society. Esophagus 2017, 14, 105–112. [Google Scholar] [CrossRef] [Green Version]
- Crawford, M.; Liu, X.; Cheng, Y.-S.L.; Tsai, R.Y. Nucleostemin upregulation and STAT3 activation as early events in oral epithelial dysplasia progression to squamous cell carcinoma. Neoplasia 2021, 23, 1289–1299. [Google Scholar] [CrossRef]
- Muto, M.; Minashi, K.; Yano, T.; Saito, Y.; Oda, I.; Nonaka, S.; Omori, T.; Sugiura, H.; Goda, K.; Kaise, M. Early detection of superficial squamous cell carcinoma in the head and neck region and esophagus by narrow band imaging: A multicenter randomized controlled trial. J. Clin. Oncol. 2010, 28, 1566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- di Pietro, M.; Canto, M.I.; Fitzgerald, R.C. Endoscopic management of early adenocarcinoma and squamous cell carcinoma of the esophagus: Screening, diagnosis, and therapy. Gastroenterology 2018, 154, 421–436. [Google Scholar] [CrossRef] [PubMed]
- Barbeiro, S.; Libânio, D.; Castro, R.; Dinis-Ribeiro, M.; Pimentel-Nunes, P. Narrow-band imaging: Clinical application in gastrointestinal endoscopy. GE-Port. J. Gastroenterol. 2019, 26, 40–53. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, M.A.; Dayyeh, B.K.A.; Bhat, Y.M.; Chauhan, S.S.; Gottlieb, K.T.; Hwang, J.H.; Komanduri, S.; Konda, V.; Lo, S.K.; Maple, J.T. Electronic chromoendoscopy. Gastrointest. Endosc. 2015, 81, 249–261. [Google Scholar] [CrossRef]
- East, J.E.; Vleugels, J.L.; Roelandt, P.; Bhandari, P.; Bisschops, R.; Dekker, E.; Hassan, C.; Horgan, G.; Kiesslich, R.; Longcroft-Wheaton, G. Advanced endoscopic imaging: European Society of Gastrointestinal Endoscopy (ESGE) technology review. Endoscopy 2016, 48, 1029–1045. [Google Scholar] [CrossRef]
- Yamada, M.; Saito, Y.; Imaoka, H.; Saiko, M.; Yamada, S.; Kondo, H.; Takamaru, H.; Sakamoto, T.; Sese, J.; Kuchiba, A. Development of a real-time endoscopic image diagnosis support system using deep learning technology in colonoscopy. Sci. Rep. 2019, 9, 14465. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.-Y.; Li, P.-C.; Chang, R.-F.; Yao, C.-D.; Chen, Y.-Y.; Chang, W.-Y.; Yen, H.-H. Deep learning-based endoscopic anatomy classification: An accelerated approach for data preparation and model validation. Surg. Endosc. 2021, 36, 3811–3821. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, C.; Zhang, Z.; Zhu, Y.; Lin, H.; Zhang, Z.; Sun, Y.; He, T.; Mueller, J.; Manmatha, R. Resnest: Split-attention networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 2736–2746. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S. An image is worth 16x16 words: Transformers for image recognition at scale. In Proceedings of the 9th International Conference on Learning Representations, Virtual Event, 3–7 May 2021; pp. 1–12. [Google Scholar]
- Wang, C.-C.; Chiu, Y.-C.; Chen, W.-L.; Yang, T.-W.; Tsai, M.-C.; Tseng, M.-H. A deep learning model for classification of endoscopic gastroesophageal reflux disease. Int. J. Environ. Res. Public Health 2021, 18, 2428. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the 3rd International Conference on Learning Representations, DBIP, San Diego, CA, USA, 7–9 May 2015; pp. 1409–1556. [Google Scholar]
- Yu, X.; Tang, S.; Cheang, C.F.; Yu, H.H.; Choi, I.C. Multi-task model for esophageal lesion analysis using endoscopic images: Classification with image retrieval and segmentation with attention. Sensors 2022, 22, 283. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Ge, R.; Wen, M.; Liu, G.; Chen, Y.; Zhang, P.; He, X.; Hua, J.; Luo, L.; Li, S. ELNet: Automatic classification and segmentation for esophageal lesions using convolutional neural network. Med. Image Anal. 2021, 67, 101838. [Google Scholar] [CrossRef]
- Pan, W.; Li, X.; Wang, W.; Zhou, L.; Wu, J.; Ren, T.; Liu, C.; Lv, M.; Su, S.; Tang, Y. Identification of Barrett’s esophagus in endoscopic images using deep learning. BMC Gastroenterol. 2021, 21, 479. [Google Scholar] [CrossRef] [PubMed]
- Celik, N.; Ali, S.; Gupta, S.; Braden, B.; Rittscher, J. EndoUDA: A Modality Independent Segmentation Approach for Endoscopy Imaging. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part III 24. Springer: Berlin/Heidelberg, Germany, 2021; pp. 303–312. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the International Conference on Machine Learning, PMLR, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Hussain, Z.; Gimenez, F.; Yi, D.; Rubin, D. Differential Data Augmentation Techniques for Medical Imaging Classification Tasks. AMIA Annu. Symp. Proc. 2017, 2017, 979–984. [Google Scholar] [PubMed]
- Chlap, P.; Min, H.; Vandenberg, N.; Dowling, J.; Holloway, L.; Haworth, A. A review of medical image data augmentation techniques for deep learning applications. J. Med. Imaging Radiat. Oncol. 2021, 65, 545–563. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Yoon, S.; Fuentes, A.; Park, D.S. A comprehensive survey of image augmentation techniques for deep learning. Pattern Recognit. 2023, 137, 109347. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 6000–6010. [Google Scholar]
- Xie, C.; Tan, M.; Gong, B.; Wang, J.; Yuille, A.L.; Le, Q.V. Adversarial examples improve image recognition. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 819–828. [Google Scholar]
- Xie, Q.; Luong, M.-T.; Hovy, E.; Le, Q.V. Self-training with noisy student improves imagenet classification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 10687–10698. [Google Scholar]
- Whiting, P.F.; Rutjes, A.W.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M.; QUADAS-2 Group*. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef]
- Abnar, S.; Zuidema, W. Quantifying attention flow in transformers. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, ACL. Association for Computational Linguistics, Online, 5–10 July 2020; pp. 4190–4197. [Google Scholar]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
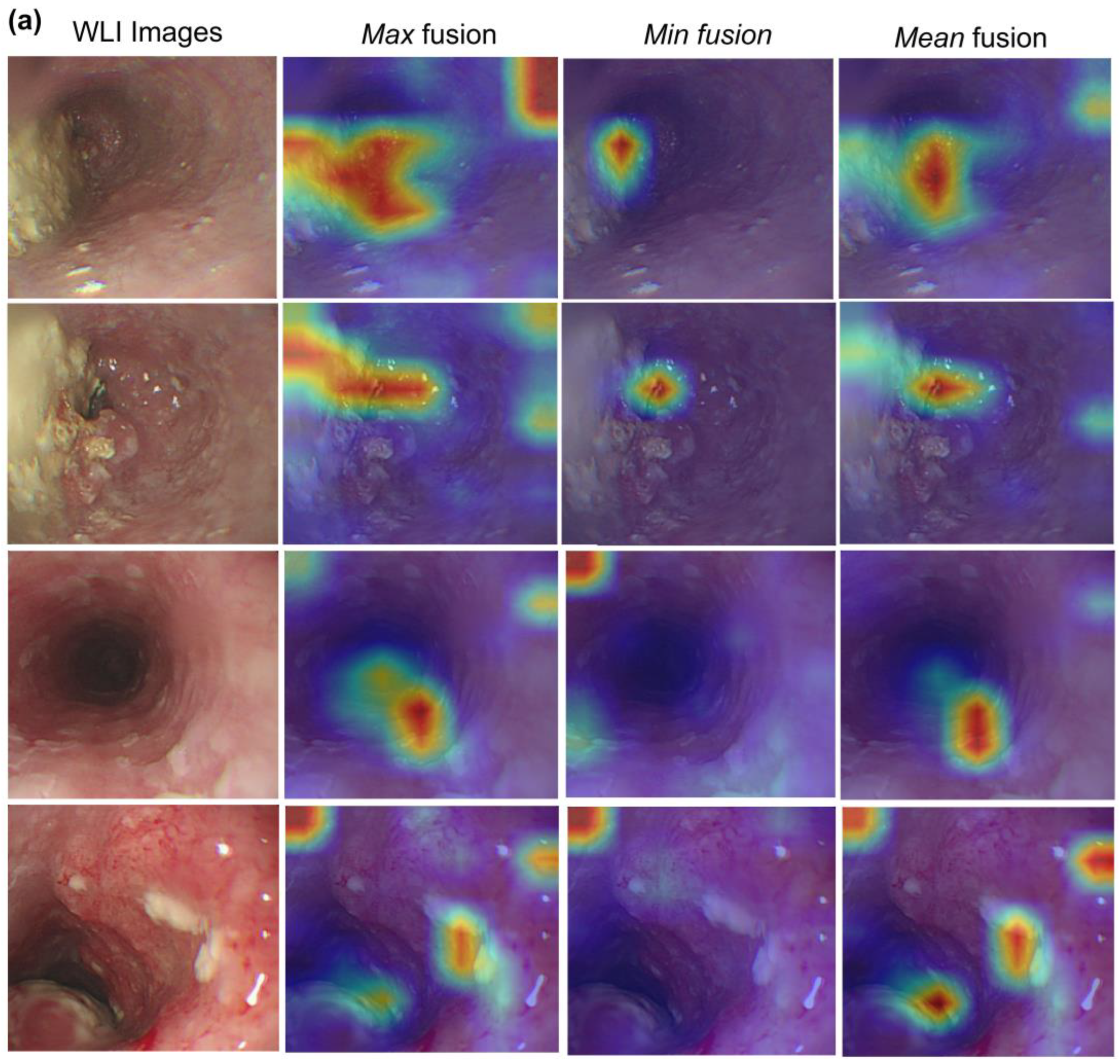
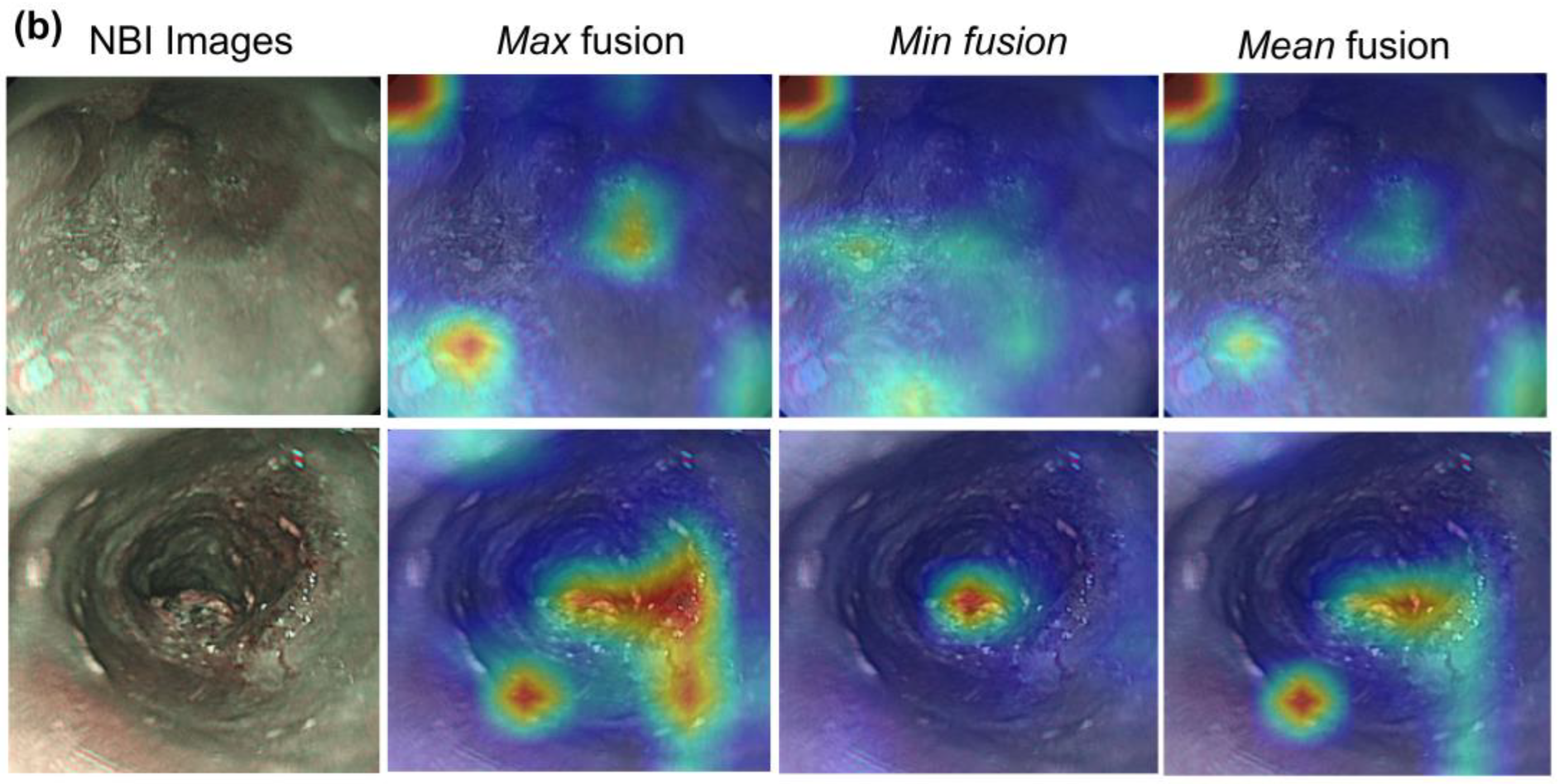
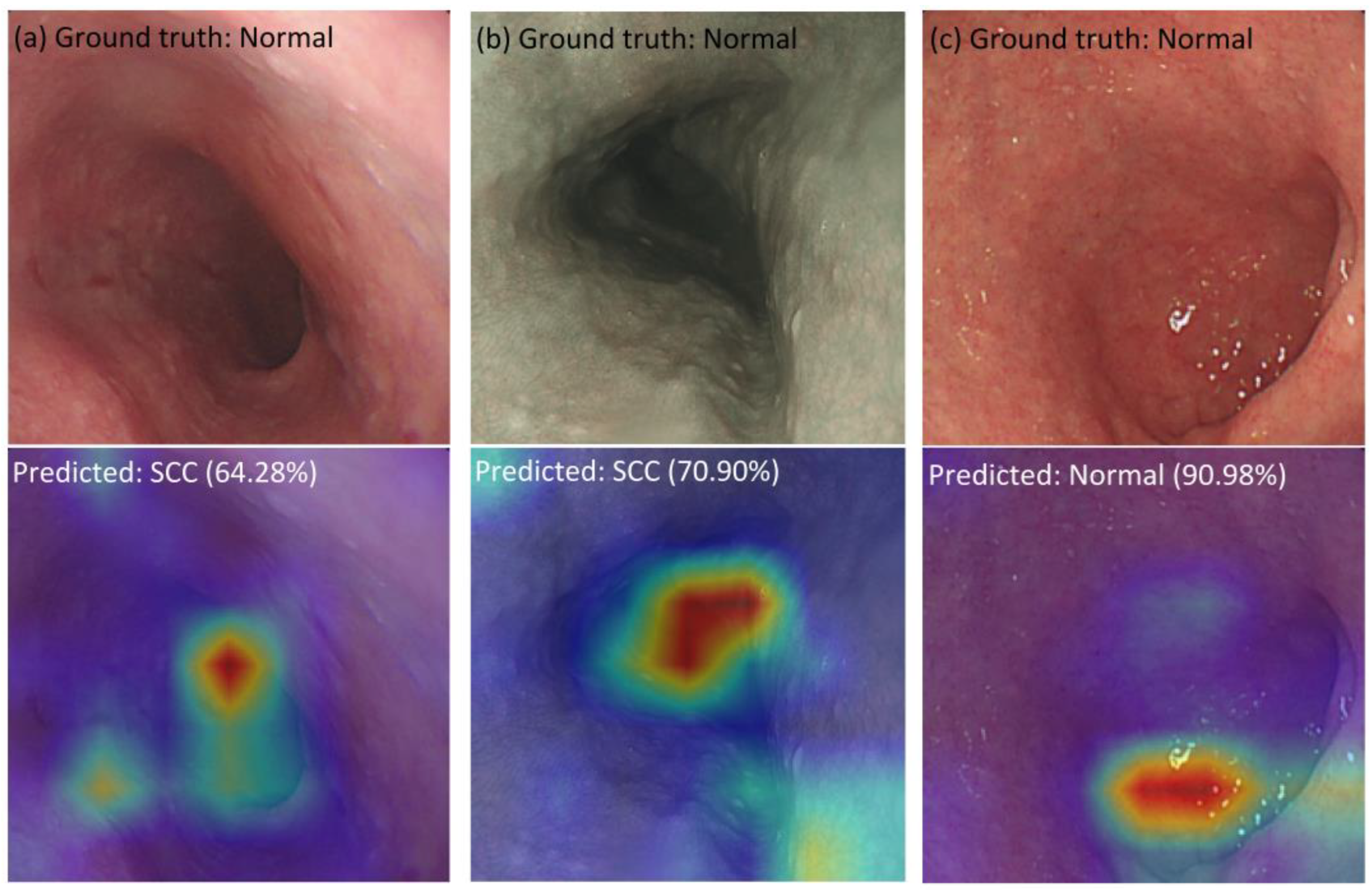
| Metrics | All | Macro-Average | Weighted Average | Class | |||
|---|---|---|---|---|---|---|---|
| Normal | Dysplasia | SCC | |||||
| WLI | Accuracy | 96.77 | |||||
| Precision | 96.70 | 96.77 | 97.62 | 96.49 | 96.00 | ||
| Recall | 95.50 | 96.77 | 97.62 | 100.00 | 88.89 | ||
| f1-score | 96.05 | 96.73 | 97.62 | 98.21 | 92.31 | ||
| NBI | Accuracy | 92.42 | |||||
| Precision | 86.05 | 92.23 | 87.50 | 95.65 | 75.00 | ||
| Recall | 83.00 | 92.42 | 93.33 | 95.65 | 60.00 | ||
| f1-score | 84.21 | 92.25 | 90.32 | 95.65 | 66.67 | ||
| WLI + NBI | Accuracy | 96.32 | |||||
| Precision | 96.44 | 96.32 | 96.30 | 96.15 | 96.88 | ||
| Recall | 95.70 | 96.32 | 91.23 | 99.01 | 96.88 | ||
| f1-score | 96.04 | 96.29 | 93.69 | 97.56 | 96.88 | ||
| Model | Precision | Recall | f1-Score |
|---|---|---|---|
| ViT_small_patch16_224 | 0.9306 | 0.9084 | 0.9177 |
| ViT_base_patch16_224 | 0.9077 | 0.8521 | 0.8652 |
| ViT_large_patch16_224 | 0.8862 | 0.7849 | 0.8028 |
| EfficientNet-B0 | 0.9535 | 0.9492 | 0.9512 |
| Hybrid model (backbone EffiecientNet-B0) | 0.9319 | 0.9352 | 0.9334 |
| EfficientNet-B1 | 0.9511 | 0.9578 | 0.9539 |
| Hybrid model (backbone EffiecientNet-B1) | 0.9473 | 0.9473 | 0.9473 |
| EfficientNet-B2 | 0.9440 | 0.9473 | 0.9455 |
| Hybrid model (backbone EffiecientNet-B2) | 0.9541 | 0.9526 | 0.9520 |
| EfficientNet-B3 | 0.9589 | 0.9559 | 0.9563 |
| Hybrid model (backbone EffiecientNet-B3) | 0.9644 | 0.9570 | 0.9629 |
| Model | Number of Params | Size (MB) | Accuracy | Time Prediction per Image (s) |
|---|---|---|---|---|
| Resnet-18 | 12,084,267 | 46 | 0.7500 | 0.0380 |
| Resnet-34 | 22,192,427 | 84 | 0.8710 | 0.0404 |
| Resnet-50 | 26,738,219 | 102 | 0.9435 | 0.0553 |
| ReStNet | 28,664,427 | 109 | 0.9597 | 0.0697 |
| EfficientNet-B0 | 4,795,519 | 18 | 0.9516 | 0.0481 |
| EfficientNet-B1 | 7,301,155 | 28 | 0.9597 | 0.0538 |
| EfficientNet-B2 | 8,554,501 | 33 | 0.9516 | 0.0576 |
| EfficientNet-B3 | 11,615,275 | 44 | 0.9597 | 0.0617 |
| EfficientNet-B4 | 19,341,616 | 74 | 0.9597 | 0.0745 |
| EfficientNet-B5 | 30,389,784 | 117 | 0.9677 | 0.0741 |
| EfficientNet-B6 | 43,040,704 | 165 | 0.9597 | 0.0836 |
| EfficientNet-B7 | 66,347,960 | 254 | 0.9435 | 0.0925 |
| ViT_small_patch16_224 | 22,379,883 | 85 | 0.9274 | 0.0434 |
| ViT_base_patch16_224 | 87,093,483 | 332 | 0.8952 | 0.0376 |
| ViT_large_patch16_224 | 304,983,531 | 1163 | 0.8468 | 0.0459 |
| Hybrid model (backbone EffiecientNet-B0) | 12,234,239 | 47 | 0.9194 | 0.0514 |
| Hybrid model (backbone EffiecientNet-B1) | 14,739,875 | 56 | 0.9274 | 0.0623 |
| Hybrid model (backbone EffiecientNet-B2) | 15,851,653 | 60 | 0.9355 | 0.0624 |
| Hybrid model (backbone EffiecientNet-B3) | 18,762,667 | 72 | 0.9677 | 0.0670 |
| Hybrid model (backbone EffiecientNet-B4) | 25,422,027 | 97 | 0.9677 | 0.0767 |
| Hybrid model (backbone EffiecientNet-B5) | 35,988,403 | 138 | 0.9677 | 0.0810 |
| Hybrid model (backbone EffiecientNet-B6) | 48,124,763 | 184 | 0.9516 | 0.0847 |
| Hybrid model (backbone EffiecientNet-B7) | 70,884,691 | 272 | 0.9758 | 0.1182 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chou, C.-K.; Nguyen, H.-T.; Wang, Y.-K.; Chen, T.-H.; Wu, I.-C.; Huang, C.-W.; Wang, H.-C. Preparing Well for Esophageal Endoscopic Detection Using a Hybrid Model and Transfer Learning. Cancers 2023, 15, 3783. https://doi.org/10.3390/cancers15153783
Chou C-K, Nguyen H-T, Wang Y-K, Chen T-H, Wu I-C, Huang C-W, Wang H-C. Preparing Well for Esophageal Endoscopic Detection Using a Hybrid Model and Transfer Learning. Cancers. 2023; 15(15):3783. https://doi.org/10.3390/cancers15153783
Chicago/Turabian StyleChou, Chu-Kuang, Hong-Thai Nguyen, Yao-Kuang Wang, Tsung-Hsien Chen, I-Chen Wu, Chien-Wei Huang, and Hsiang-Chen Wang. 2023. "Preparing Well for Esophageal Endoscopic Detection Using a Hybrid Model and Transfer Learning" Cancers 15, no. 15: 3783. https://doi.org/10.3390/cancers15153783
APA StyleChou, C.-K., Nguyen, H.-T., Wang, Y.-K., Chen, T.-H., Wu, I.-C., Huang, C.-W., & Wang, H.-C. (2023). Preparing Well for Esophageal Endoscopic Detection Using a Hybrid Model and Transfer Learning. Cancers, 15(15), 3783. https://doi.org/10.3390/cancers15153783









