Identification and Characterization of Immunogene-Related Alternative Splicing Patterns and Tumor Microenvironment Infiltration Patterns in Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Data Acquisition and Processing
2.2. Identification of the Differentially Spliced IGAS Events
2.3. Construction and Evaluation of the IGAS Prognostic Model
2.4. Construction of the AS Regulatory Network
2.5. Prediction of Immunotherapeutic and Chemotherapeutic Response
2.6. Identification of the IGAS Clusters
2.7. Correlation of Immune Cell Infiltration between IGAS Prognosis Signatures and Clusters
3. Results
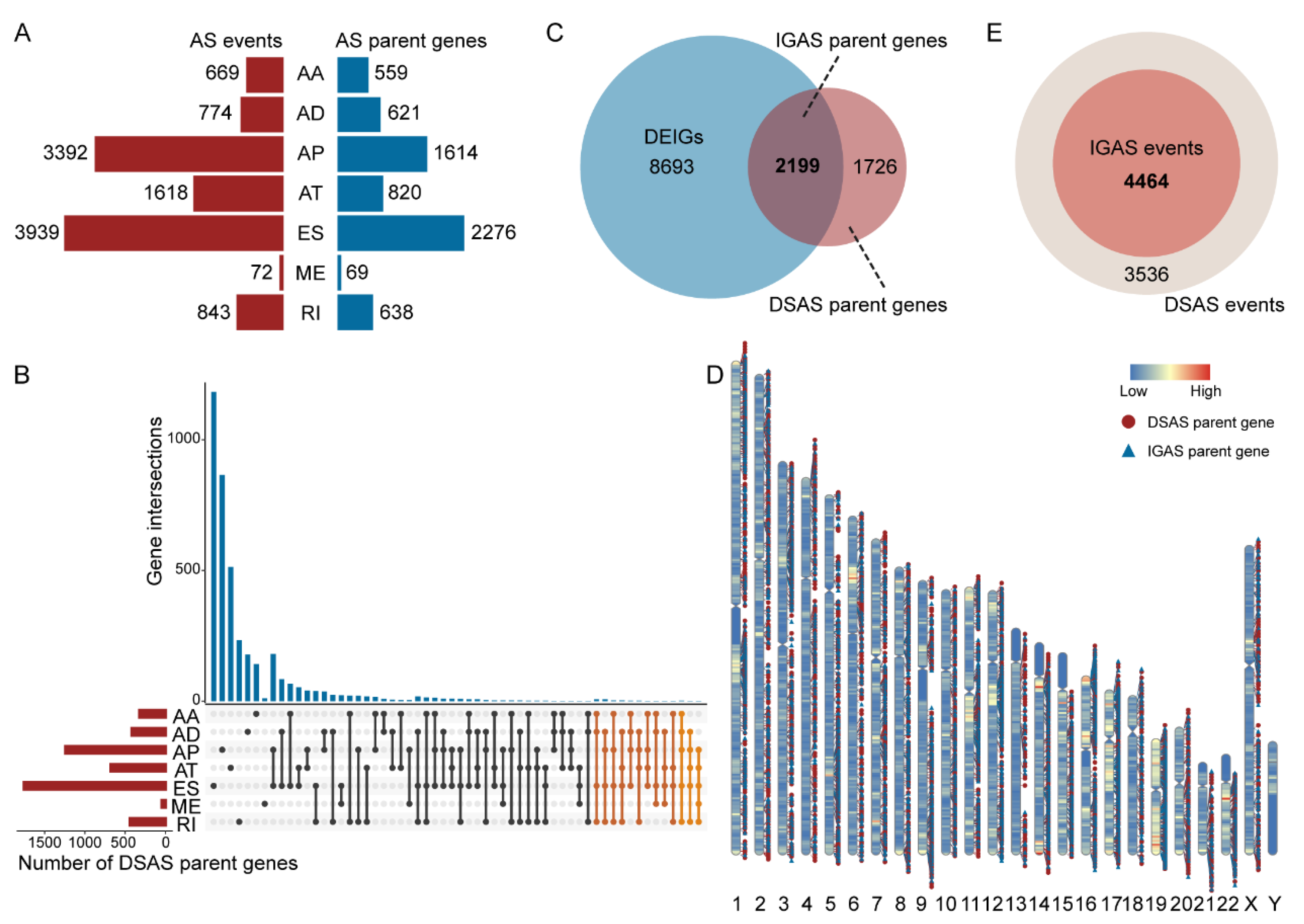
3.1. The Differentially Spliced IGAS Event Is an Important Part of AS
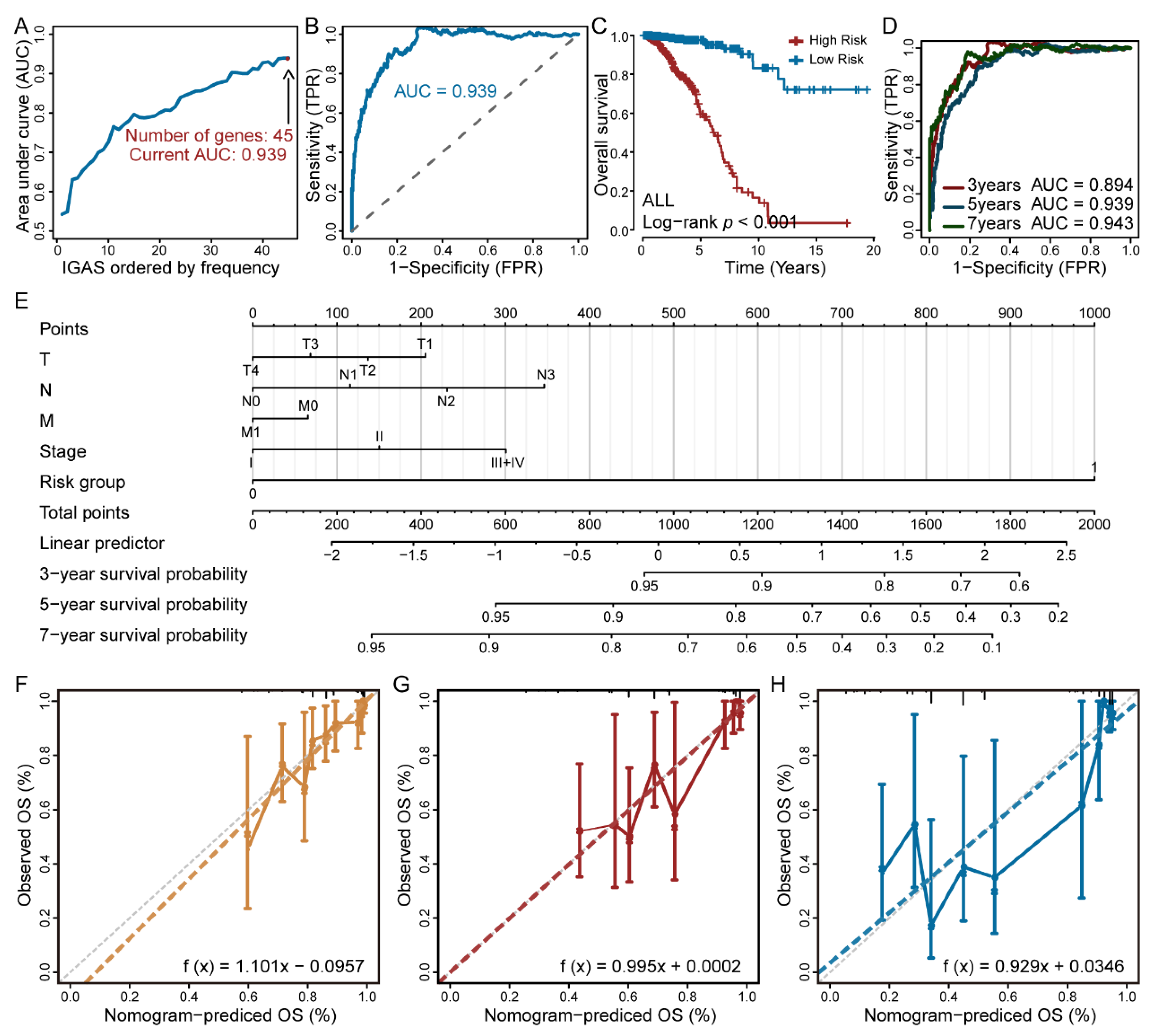
3.2. The IGAS Prognostic Model Is an Independent Prognostic Factor
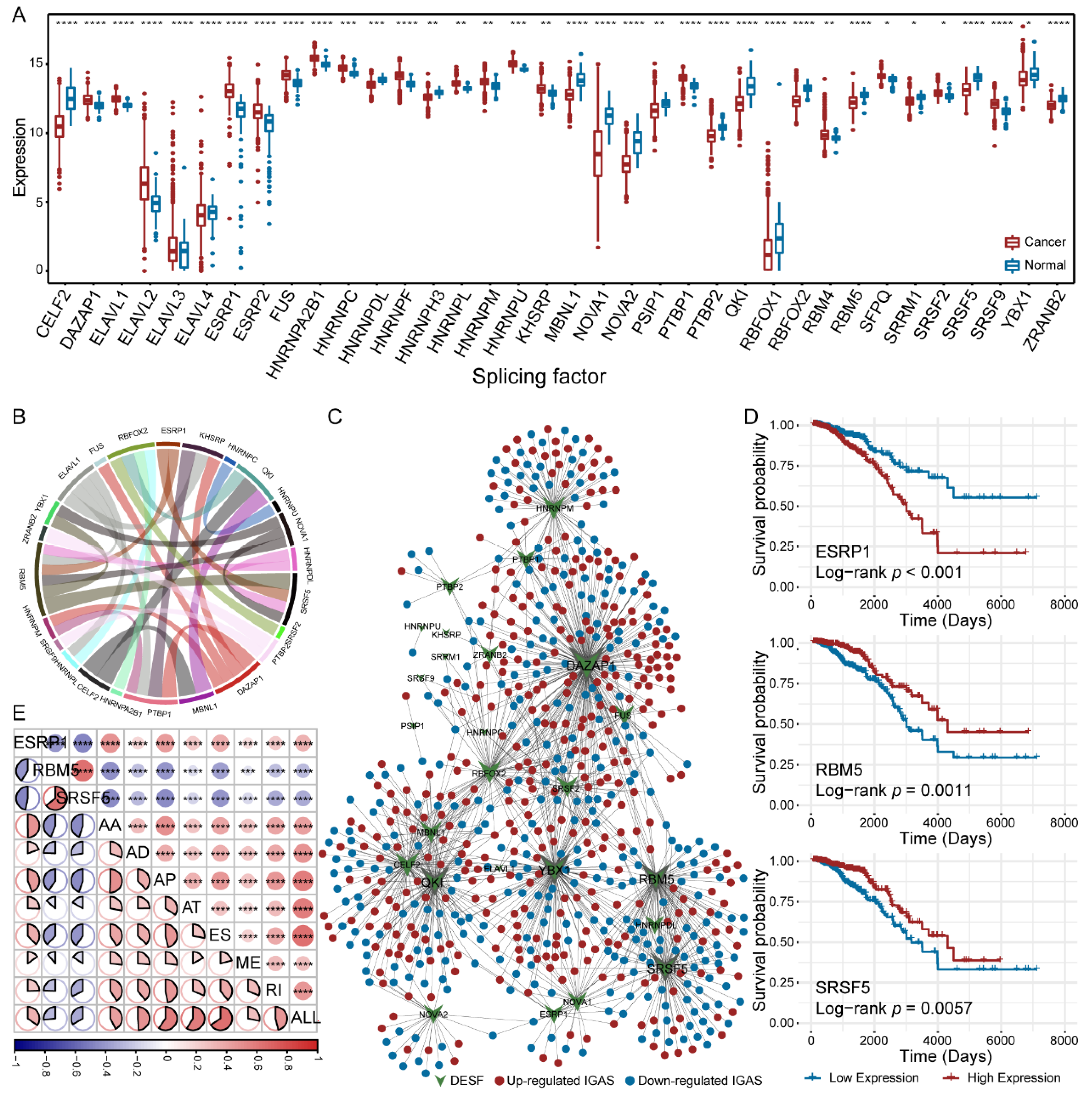
3.3. Diverse Regulatory Patterns of IGAS Events in the AS Regulatory Network
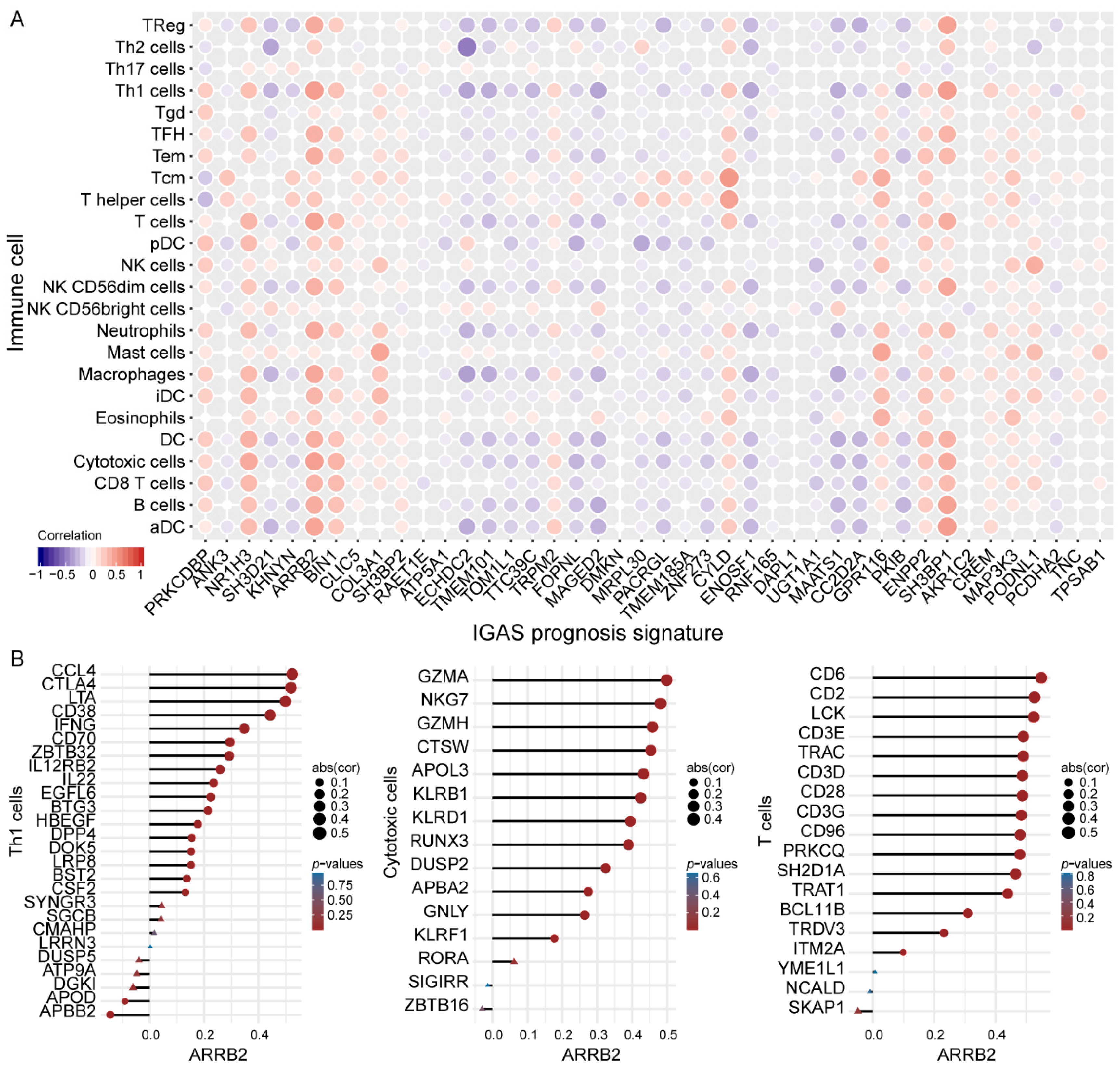
3.4. IGAS Prognosis Signatures and Immune Cells Demonstrated Consistent Trends of Correlation
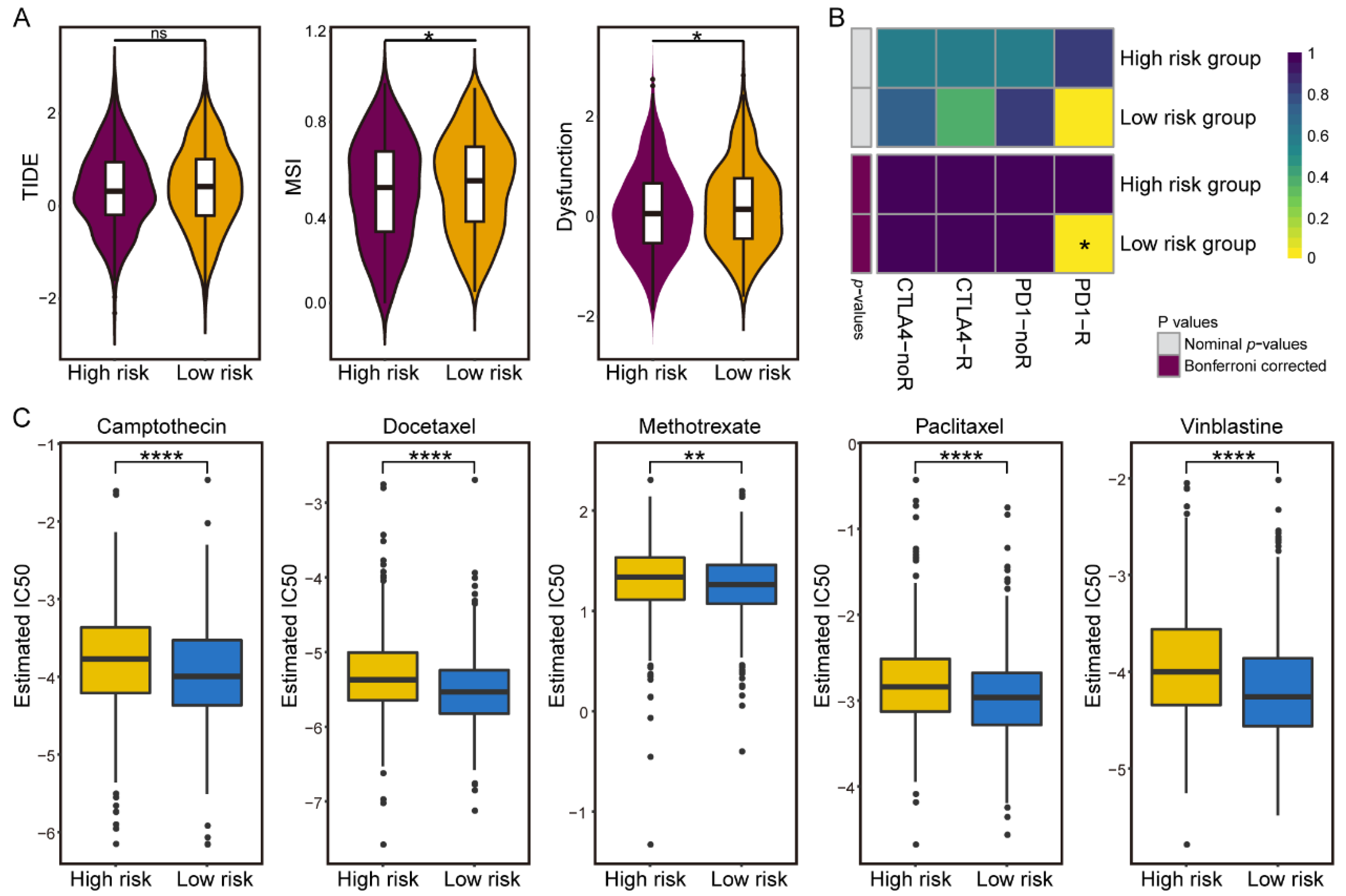
3.5. Differences in Sensitivity of Immunotherapy and Chemotherapy in the IGAS Risk Groups
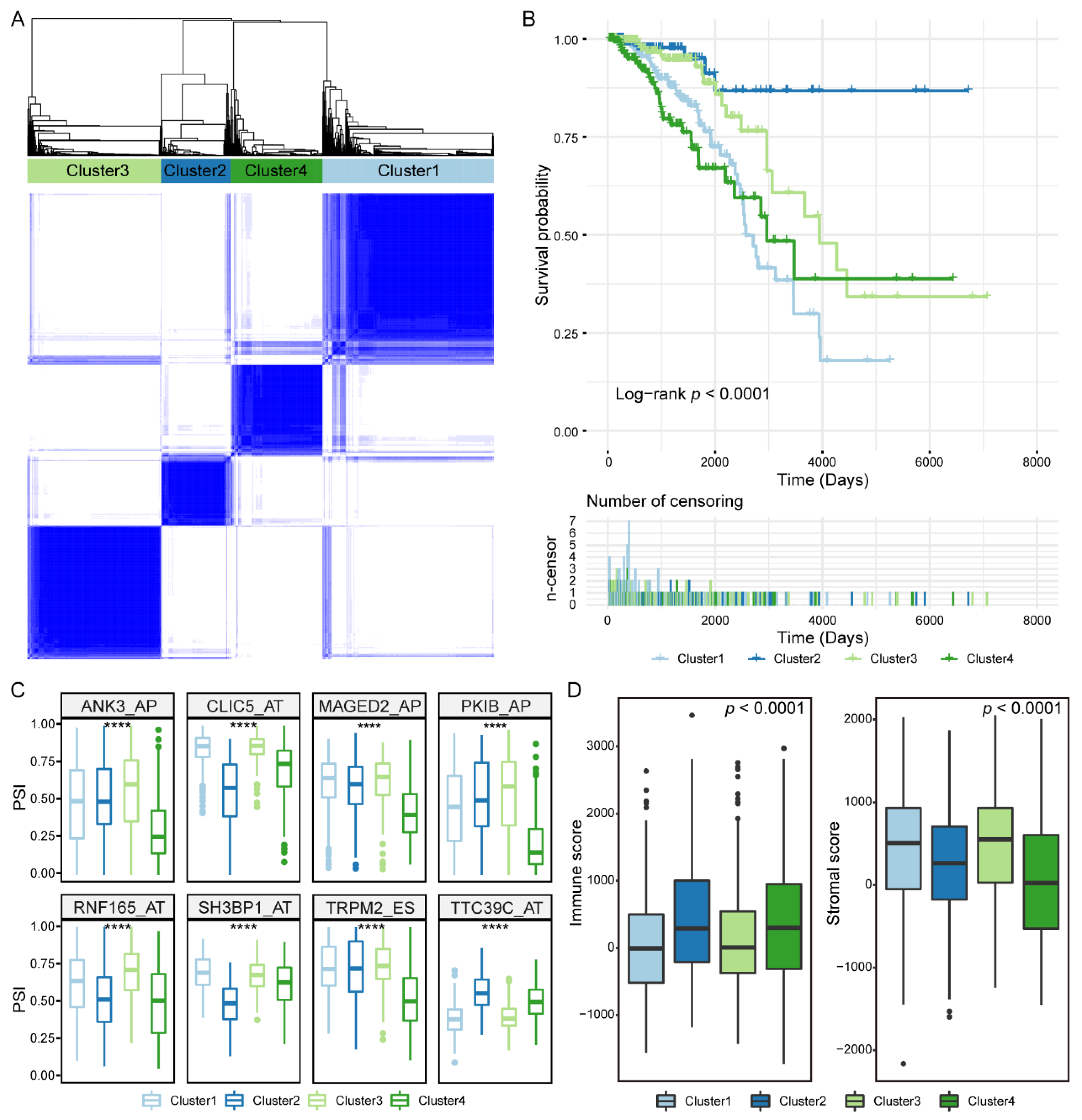
3.6. Prognosis-Related IGAS Clusters Demonstrated the Heterogeneity in Breast Cancer
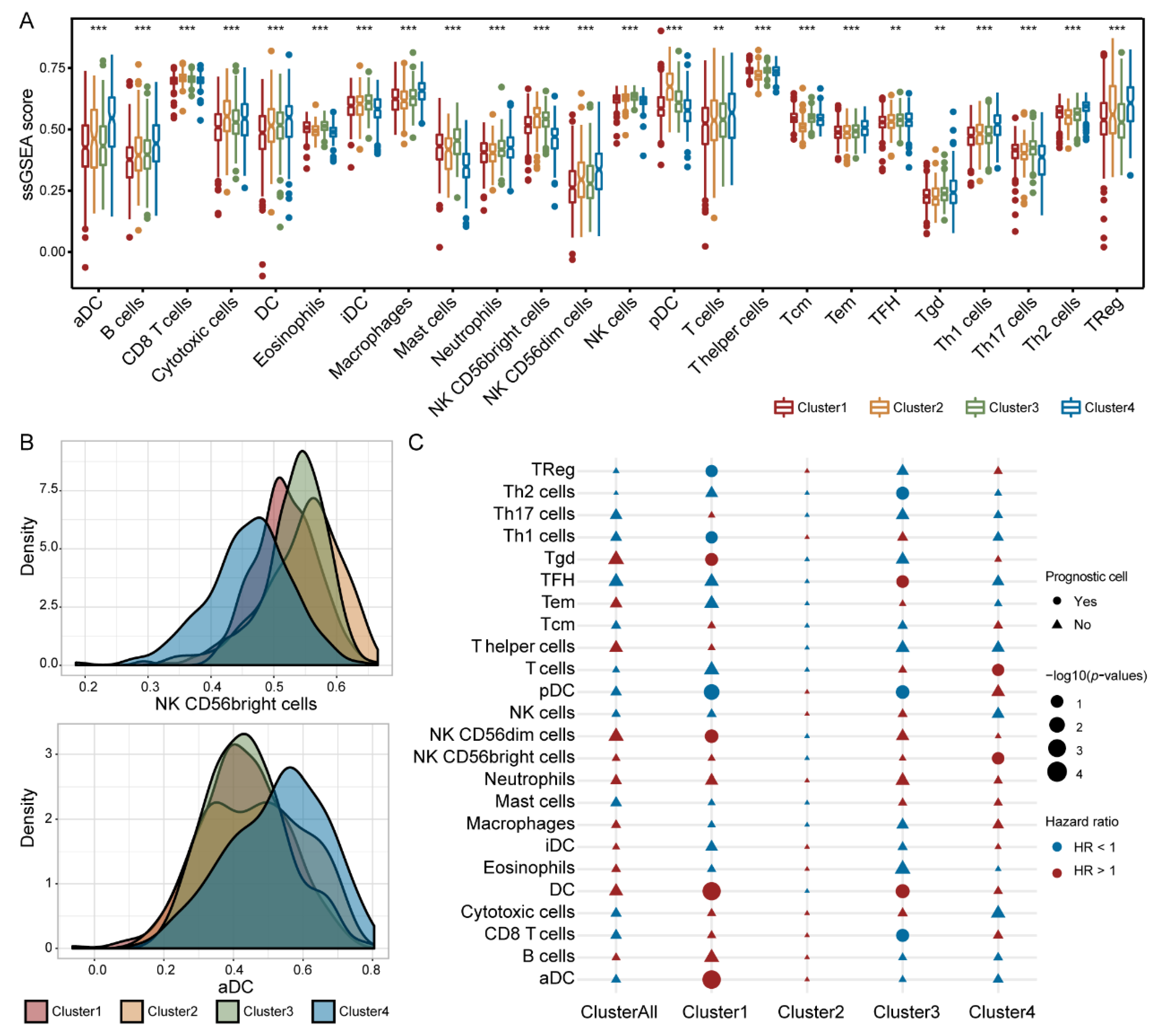
3.7. Immune Cells Exhibited Different Prognostic Abilities in Different IGAS Clusters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Primers 2019, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Maughan, K.L.; Lutterbie, M.A.; Ham, P.S. Treatment of breast cancer. Am. Fam. Physician 2010, 81, 1339–1346. [Google Scholar] [PubMed]
- Simpson, T.R.; Li, F.; Montalvo-Ortiz, W.; Sepulveda, M.A.; Bergerhoff, K.; Arce, F.; Roddie, C.; Henry, J.Y.; Yagita, H.; Wolchok, J.D.; et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti–CTLA-4 therapy against melanoma. J. Exp. Med. 2013, 210, 1695–1710. [Google Scholar] [CrossRef] [PubMed]
- Garon, E.B.; Rizvi, N.A.; Hui, R.; Leighl, N.; Balmanoukian, A.S.; Eder, J.P.; Patnaik, A.; Aggarwal, C.; Gubens, M.; Horn, L.; et al. Pembrolizumab for the Treatment of Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 372, 2018–2028. [Google Scholar] [CrossRef]
- Laplagne, C.; Domagala, M.; Le Naour, A.; Quemerais, C.; Hamel, D.; Fournié, J.-J.; Couderc, B.; Bousquet, C.; Ferrand, A.; Poupot, M. Latest Advances in Targeting the Tumor Microenvironment for Tumor Suppression. Int. J. Mol. Sci. 2019, 20, 4719. [Google Scholar] [CrossRef] [Green Version]
- Fridman, W.H.; Pagès, F.; Sautes-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef]
- Oltean, S.; Bates, D. Hallmarks of alternative splicing in cancer. Oncogene 2014, 33, 5311–5318. [Google Scholar] [CrossRef] [Green Version]
- Schaub, A.; Glasmacher, E. Splicing in immune cells—mechanistic insights and emerging topics. Int. Immunol. 2017, 29, 173–181. [Google Scholar] [CrossRef]
- Ryan, M.; Wong, W.C.; Brown, R.; Akbani, R.; Su, X.; Broom, B.; Melott, J.; Weinstein, J. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res. 2016, 44, D1018–D1022. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Trevino, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Kao, K.-J.; Chang, K.-M.; Hsu, H.-C.; Huang, A.T. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: Implications for treatment optimization. BMC Cancer 2011, 11, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goeman, J.J. L1Penalized Estimation in the Cox Proportional Hazards Model. Biom. J. 2010, 52, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Rio, D.C. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef] [Green Version]
- Giulietti, M.; Piva, F.; D’Antonio, M.; De Meo, P.D.; Paoletti, D.; Castrignanò, T.; D’Erchia, A.M.; Picardi, E.; Zambelli, F.; Principato, G.; et al. SpliceAid-F: A database of human splicing factors and their RNA-binding sites. Nucleic Acids Res. 2013, 41, D125–D131. [Google Scholar] [CrossRef]
- Piva, F.; Giulietti, M.; Burini, A.B.; Principato, G. SpliceAid 2: A database of human splicing factors expression data and RNA target motifs. Hum. Mutat. 2012, 33, 81–85. [Google Scholar] [CrossRef]
- Spellman, A.; Tang, S.-C. Immunotherapy for breast cancer: Past, present, and future. Cancer Metastasis Rev. 2016, 35, 525–546. [Google Scholar] [CrossRef]
- Topalian, S.L.; Drake, C.G.; Pardoll, D.M. Immune Checkpoint Blockade: A Common Denominator Approach to Cancer Therapy. Cancer Cell 2015, 27, 450–461. [Google Scholar] [CrossRef] [Green Version]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef]
- Hoshida, Y.; Brunet, J.-P.; Tamayo, P.; Golub, T.R.; Mesirov, J.P. Subclass Mapping: Identifying Common Subtypes in Independent Disease Data Sets. PLoS ONE 2007, 2, e1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roh, W.; Chen, P.-L.; Reuben, A.; Spencer, C.N.; Prieto, P.A.; Miller, J.P.; Gopalakrishnan, V.; Wang, F.; Cooper, Z.A.; Reddy, S.M.; et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci. Transl. Med. 2017, 9, 385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013, 41, D955–D961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, W.; Du, Y.; Liang, X.; Yua, C.; Fanga, J.; Lua, W.; Guoa, X.; Tianbcef, J.; Jina, Y.; Zhenga, J. Synergistic triple-combination therapy with hyaluronic acid-shelled PPy/CPT nanoparticles results in tumor regression and prevents tumor recurrence and metastasis in 4T1 breast cancer. Biomaterials 2019, 217, 119264. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.E.; Pazdur, R. Docetaxel. J. Clin. Oncol. 1995, 13, 2643–2655. [Google Scholar] [CrossRef]
- Chen, S.; Cai, J.; Zhang, W.; Zheng, X.; Hu, S.; Lu, J.; Xing, J.; Dong, Y. Proteomic identification of differentially expressed proteins associated with the multiple drug resistance in methotrexate-resistant human breast cancer cells. Int. J. Oncol. 2014, 45, 448–458. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues-Ferreira, S.; Nehlig, A.; Moindjie, H.; Monchecourt, C.; Seiler, C.; Marangoni, E.; Chateau-Joubert, S.; Dujaric, M.-E.; Servant, N.; Asselain, B.; et al. Improving breast cancer sensitivity to paclitaxel by increasing aneuploidy. Proc. Natl. Acad. Sci. USA 2019, 116, 23691–23697. [Google Scholar] [CrossRef]
- Sui, M.; Jiang, D.; Hinsch, C.; Fan, W. Fulvestrant (ICI 182,780) sensitizes breast cancer cells expressing estrogen receptor α to vinblastine and vinorelbine. Breast Cancer Res. Treat. 2010, 121, 335–345. [Google Scholar] [CrossRef]
- Geeleher, P.; Cox, N.; Huang, R.S. pRRophetic: An R Package for Prediction of Clinical Chemotherapeutic Response from Tumor Gene Expression Levels. PLoS ONE 2014, 9, e107468. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Waldner, M.; Obenauf, A.C.; Angell, H.; Fredriksen, T.; Lafontaine, L.; Berger, A.; et al. Spatiotemporal Dynamics of Intratumoral Immune Cells Reveal the Immune Landscape in Human Cancer. Immunity 2013, 39, 782–795. [Google Scholar] [CrossRef] [Green Version]
- Lynch, K.W. Consequences of regulated pre-mRNA splicing in the immune system. Nat. Rev. Immunol. 2004, 4, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Loibl, S.; Noske, A.; Roller, M.; Müller, B.M.; Komor, M.; Budczies, J.; Darb-Esfahani, S.; Kronenwett, R.; Hanusch, C.; et al. Tumor-Associated Lymphocytes as an Independent Predictor of Response to Neoadjuvant Chemotherapy in Breast Cancer. J. Clin. Oncol. 2010, 28, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Loi, S.; Michiels, S.; Salgado, R.; Sirtaine, N.; Jose, V.; Fumagalli, D.; Kellokumpu-Lehtinen, P.-L.; Bono, P.; Kataja, V.; Desmedt, C.; et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: Results from the FinHER trial. Ann. Oncol. 2014, 25, 1544–1550. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.T.; Cristescu, R.; Bass, A.J.; Kim, K.-M.; Odegaard, J.I.; Kim, K.; Liu, X.Q.; Sher, X.; Jung, H.; Lee, M.; et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat. Med. 2018, 24, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Hartkopf, A.D.; Taran, F.-A.; Wallwiener, M.; Walter, C.B.; Krämer, B.; Grischke, E.-M.; Brucker, S.Y. PD-1 and PD-L1 Immune Checkpoint Blockade to Treat Breast Cancer. Breast Care 2016, 11, 385–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shalapour, S.; Font-Burgada, J.; Di Caro, G.; Zhong, Z.; Sanchez-Lopez, E.; Dhar, D.; Willimsky, G.; Ammirante, M.; Strasner, A.; Hansel, D.E.; et al. Immunosuppressive plasma cells impede T-cell-dependent immunogenic chemotherapy. Nature 2015, 521, 94–98. [Google Scholar] [CrossRef]
- Savas, P.; Salgado, R.; Denkert, C.; Sotiriou, C.; Darcy, P.K.; Smyth, M.J.; Loi, S. Clinical relevance of host immunity in breast cancer: From TILs to the clinic. Nat. Rev. Clin. Oncol. 2016, 13, 228–241. [Google Scholar] [CrossRef]
- Dieci, M.V.; Mathieu, M.C.; Guarneri, V.; Conte, P.; Delaloge, S.; Andre, F.; Goubar, A. Prognostic and predictive value of tumor-infiltrating lymphocytes in two phase III randomized adjuvant breast cancer trials. Ann. Oncol. 2015, 26, 1698–1704. [Google Scholar] [CrossRef]
- Joyce, J.A.; Pollard, J.W. Microenvironmental regulation of metastasis. Nat. Rev. Cancer 2009, 9, 239–252. [Google Scholar] [CrossRef]
- Misquitta-Ali, C.M.; Cheng, E.; O’Hanlon, D.; Liu, N.; McGlade, C.J.; Tsao, M.S.; Blencowe, B.J. Global Profiling and Molecular Characterization of Alternative Splicing Events Misregulated in Lung Cancer. Mol. Cell. Biol. 2011, 31, 138–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shakola, F.; Suri, P.; Ruggiu, M. Splicing Regulation of Pro-Inflammatory Cytokines and Chemokines: At the Interface of the Neuroendocrine and Immune Systems. Biomolecules 2015, 5, 2073–2100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Sun, N.; Lu, Z.; Sun, S.; Huang, J.; Chen, Z.; He, J. Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett. 2017, 393, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Guttery, D.S.; Shaw, J.A.; Lloyd, K.; Pringle, J.H.; Walker, R.A. Expression of tenascin-C and its isoforms in the breast. Cancer Metastasis Rev. 2010, 29, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, L.C.; Wang, K.; Robinson, A.G. RBM5 as a putative tumor suppressor gene for lung cancer. J. Thorac. Oncol. 2010, 5, 294–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Coulombe-Huntington, J.; Kang, S.; Sheynkman, G.M.; Hao, T.; Richardson, A.; Sun, S.; Yang, F.; Shen, Y.A.; Murray, R.R.; et al. Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing. Cell 2016, 164, 805–817. [Google Scholar] [CrossRef] [Green Version]
- Soysal, S.; Tzankov, A.; Muenst, S.E. Role of the Tumor Microenvironment in Breast Cancer. Pathobiology 2015, 82, 142–152. [Google Scholar] [CrossRef]
- Stanton, S.E.; Disis, M.L. Clinical significance of tumor-infiltrating lymphocytes in breast cancer. J. Immunother. Cancer 2016, 4, 59. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, S.; Wang, X.; Zhou, H.; Gao, Y.; Wang, P.; Zhi, H.; Sun, Y.; Hao, Y.; Gan, J.; Zhang, Y.; et al. Identification and Characterization of Immunogene-Related Alternative Splicing Patterns and Tumor Microenvironment Infiltration Patterns in Breast Cancer. Cancers 2022, 14, 595. https://doi.org/10.3390/cancers14030595
Guo S, Wang X, Zhou H, Gao Y, Wang P, Zhi H, Sun Y, Hao Y, Gan J, Zhang Y, et al. Identification and Characterization of Immunogene-Related Alternative Splicing Patterns and Tumor Microenvironment Infiltration Patterns in Breast Cancer. Cancers. 2022; 14(3):595. https://doi.org/10.3390/cancers14030595
Chicago/Turabian StyleGuo, Shuang, Xinyue Wang, Hanxiao Zhou, Yue Gao, Peng Wang, Hui Zhi, Yue Sun, Yangyang Hao, Jing Gan, Yakun Zhang, and et al. 2022. "Identification and Characterization of Immunogene-Related Alternative Splicing Patterns and Tumor Microenvironment Infiltration Patterns in Breast Cancer" Cancers 14, no. 3: 595. https://doi.org/10.3390/cancers14030595
APA StyleGuo, S., Wang, X., Zhou, H., Gao, Y., Wang, P., Zhi, H., Sun, Y., Hao, Y., Gan, J., Zhang, Y., Sun, J., Zheng, W., Zhao, X., Xiao, Y., & Ning, S. (2022). Identification and Characterization of Immunogene-Related Alternative Splicing Patterns and Tumor Microenvironment Infiltration Patterns in Breast Cancer. Cancers, 14(3), 595. https://doi.org/10.3390/cancers14030595





