Histological Subtypes Drive Distinct Prognostic Immune Signatures in Classical Hodgkin Lymphoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. NanoString Protocol: Sample Preparation and Hybridization
2.3. Data Processing and Analysis
3. Results
3.1. Clinical and Histological Features of Patients Included in the Study
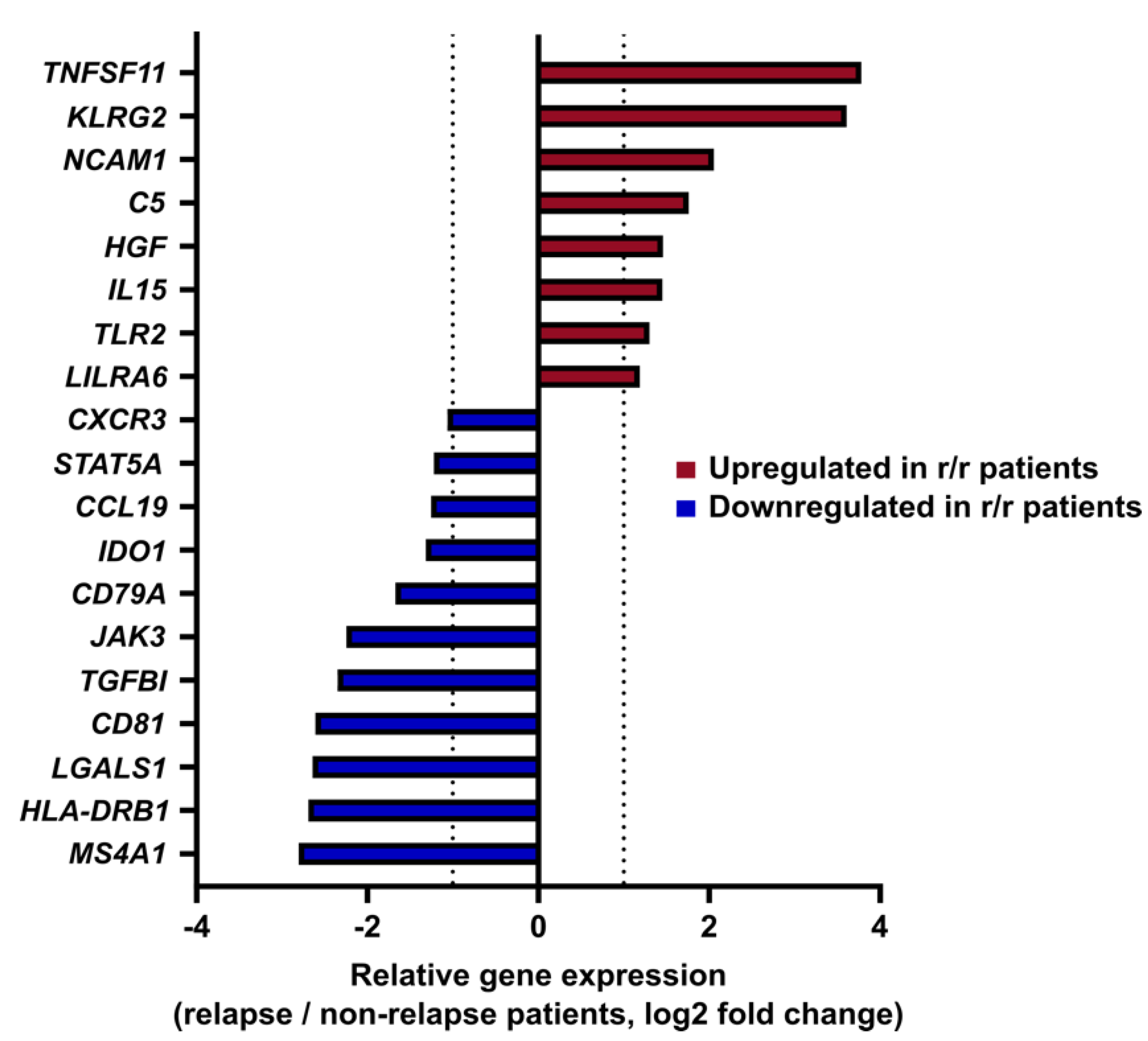
3.2. Gene Expression Profiling Identifies an Immune Signature Predictive of cHL Relapse
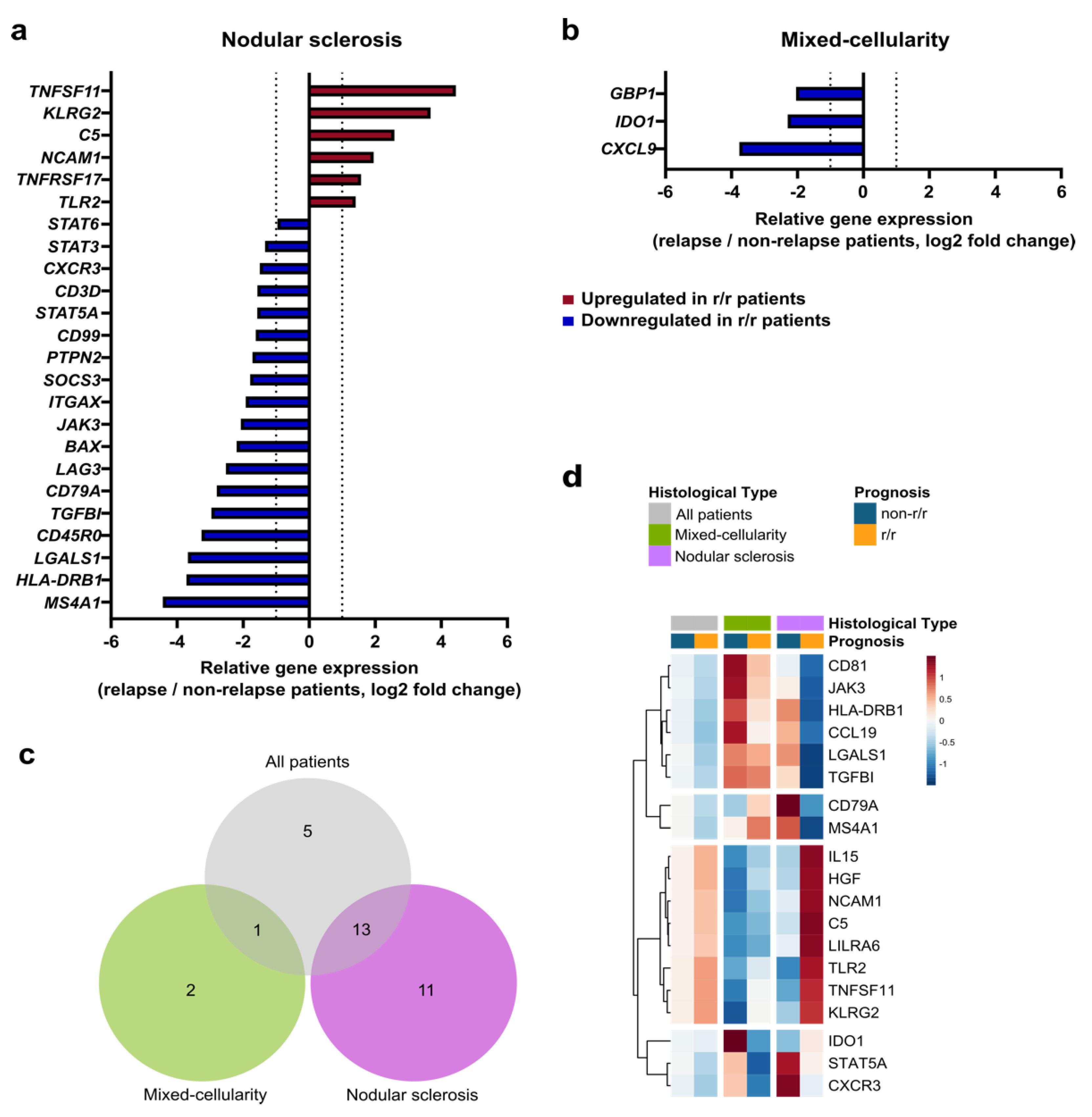
3.3. The Immune Prognostic Signature Depends on the cHL Histological Subtype
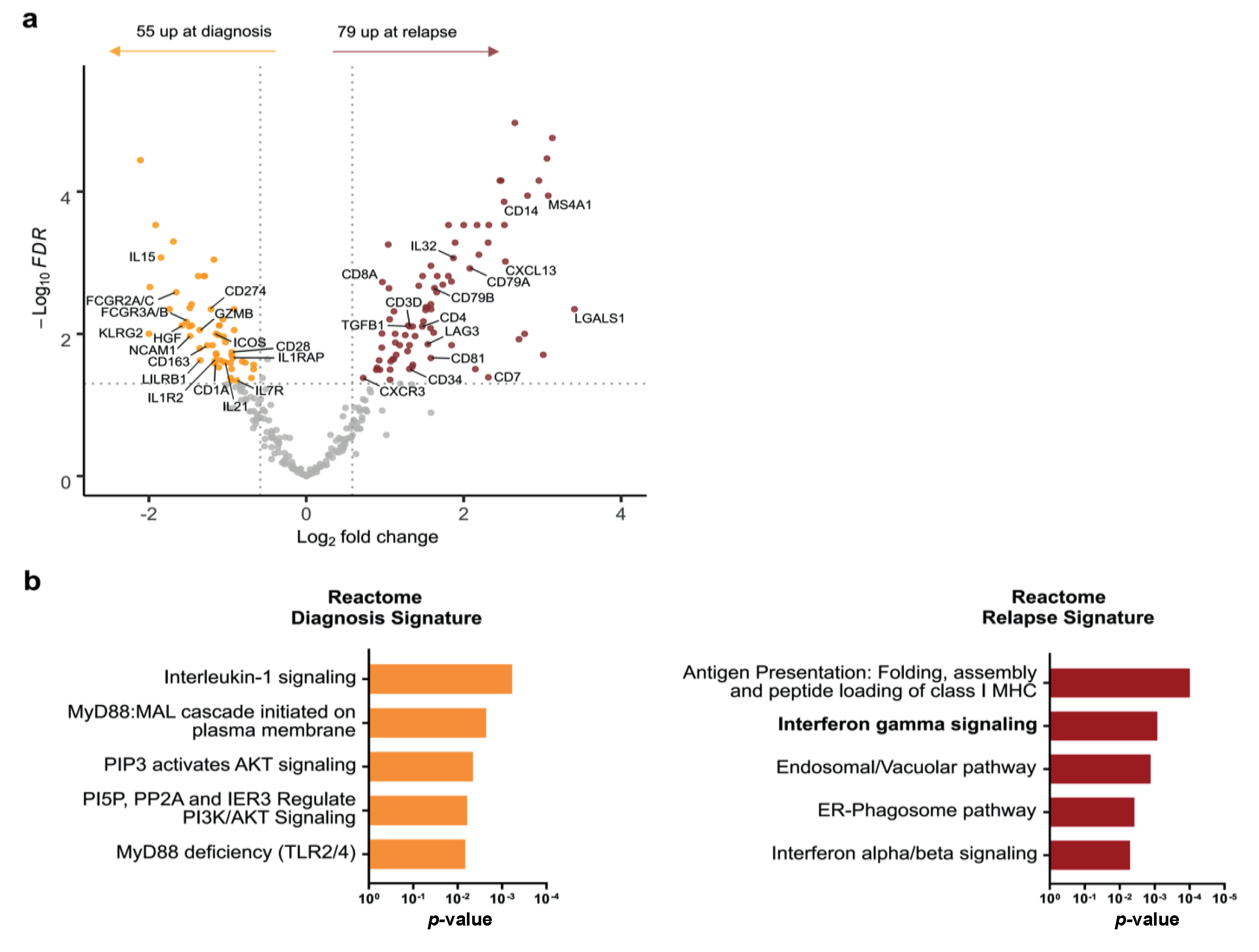
3.4. Comparative Analysis of Paired Diagnosis and Relapse Samples Reveals Distinct Immune Profiles
3.5. Changes in cHL Immune Environment at Relapse Depend on Histological Subtype
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stein, H.; Marafioti, T.; Foss, H.-D.; Laumen, H.; Hummel, M.; Anagnostopoulos, I.; Wirth, T.; Demel, G.; Falini, B. Down-Regulation of BOB.1/OBF.1 and Oct2 in Classical Hodgkin Disease but Not in Lymphocyte Predominant Hodgkin Disease Correlates with Immunoglobulin Transcription. Blood 2001, 97, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 Revision of the World Health Organization Classification of Lymphoid Neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [PubMed]
- Mani, H.; Jaffe, E.S. Hodgkin Lymphoma: An Update on Its Biology with New Insights into Classification. Clin. Lymphoma Myeloma 2009, 9, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Pileri, S.A.; Ascani, S.; Leoncini, L.; Sabattini, E.; Zinzani, P.L.; Piccaluga, P.P.; Pileri, A.; Giunti, M.; Falini, B.; Bolis, G.B.; et al. Hodgkin’s Lymphoma: The Pathologist’s Viewpoint. J. Clin. Pathol. 2002, 55, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Konkay, K.; Paul, T.R.; Uppin, S.G.; Rao, D.R. Hodgkin Lymphoma: A Clinicopathological and Immunophenotypic Study. Indian J. Med. Paediatr. Oncol. 2016, 37, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Steidl, C.; Diepstra, A.; Lee, T.; Chan, F.C.; Farinha, P.; Tan, K.; Telenius, A.; Barclay, L.; Shah, S.P.; Connors, J.M.; et al. Gene Expression Profiling of Microdissected Hodgkin Reed-Sternberg Cells Correlates with Treatment Outcome in Classical Hodgkin Lymphoma. Blood 2012, 120, 3530–3540. [Google Scholar] [CrossRef]
- Tzankov, A.; Matter, M.S.; Dirnhofer, S. Refined Prognostic Role of CD68-Positive Tumor Macrophages in the Context of the Cellular Micromilieu of Classical Hodgkin Lymphoma. Pathobiology 2010, 77, 301–308. [Google Scholar] [CrossRef]
- Deau, B.; Bachy, E.; Ribrag, V.; Delarue, R.; Rubio, M.T.; Bosq, J.; Varet, B.; Brousse, N.; Hermine, O.; Canioni, D. Macrophage, Mast Cell and T Lymphocyte Infiltrations Are Independent Predictive Biomarkers of Primary Refractoriness or Early Relapse in Classical Hodgkin Lymphoma. Leuk. Lymphoma 2013, 54, 41–45. [Google Scholar] [CrossRef]
- Touati, M.; Delage-Corre, M.; Monteil, J.; Abraham, J.; Moreau, S.; Remenieras, L.; Gourin, M.-P.; Dmytruk, N.; Olivrie, A.; Turlure, P.; et al. CD68-Positive Tumor-Associated Macrophages Predict Unfavorable Treatment Outcomes in Classical Hodgkin Lymphoma in Correlation with Interim Fluorodeoxyglucose-Positron Emission Tomography Assessment. Leuk. Lymphoma 2015, 56, 332–341. [Google Scholar] [CrossRef]
- Gupta, S.; Yeh, S.; Chami, R.; Punnett, A.; Chung, C. The Prognostic Impact of Tumour-Associated Macrophages and Reed-Sternberg Cells in Paediatric Hodgkin Lymphoma. Eur. J. Cancer 2013, 49, 3255–3261. [Google Scholar] [CrossRef]
- Cencini, E.; Fabbri, A.; Rigacci, L.; Lazzi, S.; Gini, G.; Cox, M.C.; Mancuso, S.; Abruzzese, E.; Kovalchuk, S.; Goteri, G.; et al. Evaluation of the Prognostic Role of Tumour-Associated Macrophages in Newly Diagnosed Classical Hodgkin Lymphoma and Correlation with Early FDG-PET Assessment. Hematol. Oncol. 2017, 35, 69–78. [Google Scholar] [CrossRef]
- Schreck, S.; Friebel, D.; Buettner, M.; Distel, L.; Grabenbauer, G.; Young, L.S.; Niedobitek, G. Prognostic Impact of Tumour-Infiltrating Th2 and Regulatory T Cells in Classical Hodgkin Lymphoma. Hematol. Oncol. 2009, 27, 31–39. [Google Scholar] [CrossRef]
- Kamper, P.; Bendix, K.; Hamilton-Dutoit, S.; Honoré, B.; Nyengaard, J.R.; d’Amore, F. Tumor-Infiltrating Macrophages Correlate with Adverse Prognosis and Epstein-Barr Virus Status in Classical Hodgkin’s Lymphoma. Haematologica 2011, 96, 269–276. [Google Scholar] [CrossRef]
- Scott, D.W.; Chan, F.C.; Hong, F.; Rogic, S.; Tan, K.L.; Meissner, B.; Ben-Neriah, S.; Boyle, M.; Kridel, R.; Telenius, A.; et al. Gene Expression-Based Model Using Formalin-Fixed Paraffin-Embedded Biopsies Predicts Overall Survival in Advanced-Stage Classical Hodgkin Lymphoma. J. Clin. Oncol. 2013, 31, 692–700. [Google Scholar] [CrossRef]
- Jachimowicz, R.D.; Klapper, W.; Glehr, G.; Müller, H.; Haverkamp, H.; Thorns, C.; Hansmann, M.L.; Möller, P.; Stein, H.; Rehberg, T.; et al. Gene Expression-Based Outcome Prediction in Advanced Stage Classical Hodgkin Lymphoma Treated with BEACOPP. Leukemia 2021, 35, 1–5. [Google Scholar] [CrossRef]
- Sánchez-Aguilera, A.; Montalbán, C.; de la Cueva, P.; Sánchez-Verde, L.; Morente, M.M.; García-Cosío, M.; García-Laraña, J.; Bellas, C.; Provencio, M.; Romagosa, V.; et al. Tumor Microenvironment and Mitotic Checkpoint Are Key Factors in the Outcome of Classic Hodgkin Lymphoma. Blood 2006, 108, 662–668. [Google Scholar] [CrossRef]
- Rancea, M.; Monsef, I.; von Tresckow, B.; Engert, A.; Skoetz, N. High-Dose Chemotherapy Followed by Autologous Stem Cell Transplantation for Patients with Relapsed/Refractory Hodgkin Lymphoma. Cochrane Database Syst. Rev. 2013, 6, CD009411. [Google Scholar] [CrossRef]
- Majhail, N.S.; Weisdorf, D.J.; Defor, T.E.; Miller, J.S.; McGlave, P.B.; Slungaard, A.; Arora, M.; Ramsay, N.K.C.; Orchard, P.J.; MacMillan, M.L.; et al. Long-Term Results of Autologous Stem Cell Transplantation for Primary Refractory or Relapsed Hodgkin’s Lymphoma. Biol. Blood Marrow Transplant. 2006, 12, 1065–1072. [Google Scholar] [CrossRef]
- Moy, R.H.; Younes, A. Immune Checkpoint Inhibition in Hodgkin Lymphoma. Hemasphere 2018, 2, e20. [Google Scholar] [CrossRef]
- Chan, F.C.; Mottok, A.; Gerrie, A.S.; Power, M.; Nijland, M.; Diepstra, A.; van den Berg, A.; Kamper, P.; d’Amore, F.; d’Amore, A.L.; et al. Prognostic Model to Predict Post-Autologous Stem-Cell Transplantation Outcomes in Classical Hodgkin Lymphoma. J. Clin. Oncol. 2017, 35, 3722–3733. [Google Scholar] [CrossRef]
- Schnitter, A.; Kohler, C.W.; Reddemann, K.; Reinke, S.; Thorns, C.; Fend, F.; Federmann, B.; Möller, P.; Szczepanowski, M.; Spang, R.; et al. Therapeutic Targets and Microenvironment in Sequential Biopsies of Classical Hodgkin Lymphoma at Diagnosis and Relapse. J. Hematopathol. 2019, 12, 11–17. [Google Scholar] [CrossRef]
- Steidl, C.; Lee, T.; Shah, S.P.; Farinha, P.; Han, G.; Nayar, T.; Delaney, A.; Jones, S.J.; Iqbal, J.; Weisenburger, D.D.; et al. Tumor-Associated Macrophages and Survival in Classic Hodgkin’s Lymphoma. N. Engl. J. Med. 2010, 362, 875–885. [Google Scholar] [CrossRef]
- Chetaille, B.; Bertucci, F.; Finetti, P.; Esterni, B.; Stamatoullas, A.; Picquenot, J.M.; Copin, M.C.; Morschhauser, F.; Casasnovas, O.; Petrella, T.; et al. Molecular Profiling of Classical Hodgkin Lymphoma Tissues Uncovers Variations in the Tumor Microenvironment and Correlations with EBV Infection and Outcome. Blood 2009, 113, 2765–3775. [Google Scholar] [CrossRef]
- Greaves, P.; Clear, A.; Coutinho, R.; Wilson, A.; Matthews, J.; Owen, A.; Shanyinde, M.; Lister, T.A.; Calaminici, M.; Gribben, J.G. Expression of FOXP3, CD68, and CD20 at Diagnosis in the Microenvironment of Classical Hodgkin Lymphoma Is Predictive of Outcome. J. Clin. Oncol. 2013, 31, 256–262. [Google Scholar] [CrossRef]
- Kanzler, H.; Küppers, R.; Hansmann, M.L.; Rajewsky, K. Hodgkin and Reed-Sternberg Cells in Hodgkin’s Disease Represent the Outgrowth of a Dominant Tumor Clone Derived from (Crippled) Germinal Center B Cells. J. Exp. Med. 1996, 184, 1495–1505. [Google Scholar] [CrossRef]
- Lannutti, B.J.; Meadows, S.A.; Herman, S.E.M.; Kashishian, A.; Steiner, B.; Johnson, A.J.; Byrd, J.C.; Tyner, J.W.; Loriaux, M.M.; Deininger, M.; et al. CAL-101, a P110delta Selective Phosphatidylinositol-3-Kinase Inhibitor for the Treatment of B-Cell Malignancies, Inhibits PI3K Signaling and Cellular Viability. Blood 2011, 117, 591–594. [Google Scholar] [CrossRef]
- Du, J.; Neuenschwander, M.; Yu, Y.; Däbritz, J.H.M.; Neuendorff, N.-R.; Schleich, K.; Bittner, A.; Milanovic, M.; Beuster, G.; Radetzki, S.; et al. Pharmacological Restoration and Therapeutic Targeting of the B-Cell Phenotype in Classical Hodgkin Lymphoma. Blood 2017, 129, 71–81. [Google Scholar] [CrossRef]
- Tzankov, A.; Krugmann, J.; Fend, F.; Fischhofer, M.; Greil, R.; Dirnhofer, S. Prognostic Significance of CD20 Expression in Classical Hodgkin Lymphoma: A Clinicopathological Study of 119 Cases. Clin. Cancer Res. 2003, 9, 1381–1386. [Google Scholar]
- Abuelgasim, K.A.; Shammari, R.A.; Alshieban, S.; Alahmari, B.; Alzahrani, M.; Alhejazi, A.; Alaskar, A.; Damlaj, M. Impact of Cluster of Differentiation 20 Expression and Rituximab Therapy in Classical Hodgkin Lymphoma: Real World Experience. Leuk. Res. Rep. 2021, 15, 100240. [Google Scholar] [CrossRef]
- Sakatani, A.; Igawa, T.; Okatani, T.; Fujihara, M.; Asaoku, H.; Sato, Y.; Yoshino, T. Clinicopathological Significance of CD79a Expression in Classic Hodgkin Lymphoma. J. Clin. Exp. Hematop. 2020, 60, 78–86. [Google Scholar] [CrossRef]
- Teofili, L.; Di Febo, A.L.; Pierconti, F.; Maggiano, N.; Bendandi, M.; Rutella, S.; Cingolani, A.; Di Renzo, N.; Musto, P.; Pileri, S.; et al. Expression of the C-Met Proto-Oncogene and Its Ligand, Hepatocyte Growth Factor, in Hodgkin Disease. Blood 2001, 97, 1063–1069. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, K.; Blumenthal-Barby, F.; Lamprecht, B.; Köchert, K.; Lenze, D.; Hummel, M.; Mathas, S.; Dörken, B.; Janz, M. The IL-15 Cytokine System Provides Growth and Survival Signals in Hodgkin Lymphoma and Enhances the Inflammatory Phenotype of HRS Cells. Leukemia 2015, 29, 1213–1218. [Google Scholar] [CrossRef] [PubMed]
- Fiumara, P.; Snell, V.; Li, Y.; Mukhopadhyay, A.; Younes, M.; Gillenwater, A.M.; Cabanillas, F.; Aggarwal, B.B.; Younes, A. Functional Expression of Receptor Activator of Nuclear Factor ΚB in Hodgkin Disease Cell Lines. Blood 2001, 98, 2784–2790. [Google Scholar] [CrossRef] [PubMed]
- Groom, J.R.; Luster, A.D. CXCR3 in T Cell Function. Exp. Cell Res. 2011, 317, 620–631. [Google Scholar] [CrossRef]
- Greaves, P.; Clear, A.; Owen, A.; Iqbal, S.; Lee, A.; Matthews, J.; Wilson, A.; Calaminici, M.; Gribben, J.G. Defining Characteristics of Classical Hodgkin Lymphoma Microenvironment T-Helper Cells. Blood 2013, 122, 2856–2863. [Google Scholar] [CrossRef]
- Brune, M.M.; Juskevicius, D.; Haslbauer, J.; Dirnhofer, S.; Tzankov, A. Genomic Landscape of Hodgkin Lymphoma. Cancers 2021, 13, 682. [Google Scholar] [CrossRef]
- Roemer, M.G.M.; Advani, R.H.; Redd, R.A.; Pinkus, G.S.; Natkunam, Y.; Ligon, A.H.; Connelly, C.F.; Pak, C.J.; Carey, C.D.; Daadi, S.E.; et al. Classical Hodgkin Lymphoma with Reduced Β2M/MHC Class I Expression Is Associated with Inferior Outcome Independent of 9p24.1 Status. Cancer Immunol. Res. 2016, 4, 910–916. [Google Scholar] [CrossRef]
- Kadin, M.E.; Agnarsson, B.A.; Ellingsworth, L.R.; Newcom, S.R. Immunohistochemical Evidence of a Role for Transforming Growth Factor Beta in the Pathogenesis of Nodular Sclerosing Hodgkin’s Disease. Am. J. Pathol. 1990, 136, 1209–1214. [Google Scholar]
- Ansell, S.M.; Lesokhin, A.M.; Borrello, I.; Halwani, A.; Scott, E.C.; Gutierrez, M.; Schuster, S.J.; Millenson, M.M.; Cattry, D.; Freeman, G.J.; et al. PD-1 Blockade with Nivolumab in Relapsed or Refractory Hodgkin’s Lymphoma. N. Engl. J. Med. 2015, 372, 311–319. [Google Scholar] [CrossRef]
- Garcia-Marquez, M.A.; Thelen, M.; Reinke, S.; Keller, D.; Wennhold, K.; Lehmann, J.; Veldman, J.; Borchmann, S.; Rosenwald, A.; Sasse, S.; et al. Reverted Exhaustion Phenotype of Circulating Lymphocytes as Immune Correlate of Anti-PD1 First-Line Treatment in Hodgkin Lymphoma. Leukemia 2022, 36, 760–771. [Google Scholar] [CrossRef]
- Bröckelmann, P.J.; Goergen, H.; Keller, U.; Meissner, J.; Ordemann, R.; Halbsguth, T.V.; Sasse, S.; Sökler, M.; Kerkhoff, A.; Mathas, S.; et al. Efficacy of Nivolumab and AVD in Early-Stage Unfavorable Classic Hodgkin Lymphoma: The Randomized Phase 2 German Hodgkin Study Group NIVAHL Trial. JAMA Oncol. 2020, 6, 872–880. [Google Scholar] [CrossRef]
- Reinke, S.; Bröckelmann, P.J.; Iaccarino, I.; Garcia-Marquez, M.; Borchmann, S.; Jochims, F.; Kotrova, M.; Pal, K.; Brüggemann, M.; Hartmann, E.; et al. Tumor and Microenvironment Response but No Cytotoxic T-Cell Activation in Classic Hodgkin Lymphoma Treated with Anti-PD1. Blood 2020, 136, 2851–2863. [Google Scholar] [CrossRef]
- Oelmann, E.; Stein, H.; Berdel, W.E.; Herbst, H. Expression of Interleukin-1 and Interleukin-1 Receptors Type 1 and Type 2 in Hodgkin Lymphoma. PLoS ONE 2015, 10, e0138747. [Google Scholar] [CrossRef][Green Version]
- Scheeren, F.A.; Diehl, S.A.; Smit, L.A.; Beaumont, T.; Naspetti, M.; Bende, R.J.; Blom, B.; Karube, K.; Ohshima, K.; van Noesel, C.J.M.; et al. IL-21 Is Expressed in Hodgkin Lymphoma and Activates STAT5: Evidence That Activated STAT5 Is Required for Hodgkin Lymphomagenesis. Blood 2008, 111, 4706–4715. [Google Scholar] [CrossRef]

| All cHL Patients N = 42 | r/r cHL Patients n = 30 | non-r/r cHL Patients n = 12 | |
|---|---|---|---|
| Age, median (range), year | 30.5 (15–83) | 30 (15–83) | 31 (23–58) |
| Histological subtypes, n (%) | |||
| Nodular sclerosis | 24 (57.1) | 17 (56.6) | 7 (58.3) |
| Mixed-cellularity | 15 (35.7) | 11 (36.7) | 4 (33.4) |
| Lymphocyte-rich | 2 (4.8) | 2 (6.7) | 0 |
| Lymphocyte-depleted | 1 (2.4) | 0 | 1 (8.3) |
| EBV status, n (%) | |||
| EBV positive | 8 (19.0) | 5 (16.7) | 3 (25.0) |
| EBV negative Missing | 29 (69.0) 5 (12.0) | 22 (73.3) 3 (10.0) | 7 (58.3) 2 (16.7) |
| Ann Arbor Stage, n (%) | |||
| I-II | 9/41 (22) | 9/29 (31) | 0 |
| III-IV | 32/41 (78) | 20/29 (69) | 12 (100) |
| Treatment, n (%) | |||
| BEACOPP | 11 (26.2) | 6 (20.0) | 5 (41.7) |
| ABVD | 24 (57.1) | 17 (56.6) | 7 (58.3) |
| Other chemotherapy * | 5 (11.9) | 5 (16.7) | 0 |
| Missing | 2 (4.8) | 2 (6.7) | 0 |
| Treatment response, n (%) | |||
| Complete remission | 35 (83.3) | 23 (76.7) | 12 (100) |
| Progression | 6 (14.3) | 6 (20) | 0 |
| Missing | 1 (2.4) | 1 (3.3) | 0 |
| Death, n (%) | 8 (19) | 8 (26.7) | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lamaison, C.; Ferrant, J.; Gravelle, P.; Traverse-Glehen, A.; Ghesquières, H.; Tosolini, M.; Rossi, C.; Ysebaert, L.; Brousset, P.; Laurent, C.; et al. Histological Subtypes Drive Distinct Prognostic Immune Signatures in Classical Hodgkin Lymphoma. Cancers 2022, 14, 4893. https://doi.org/10.3390/cancers14194893
Lamaison C, Ferrant J, Gravelle P, Traverse-Glehen A, Ghesquières H, Tosolini M, Rossi C, Ysebaert L, Brousset P, Laurent C, et al. Histological Subtypes Drive Distinct Prognostic Immune Signatures in Classical Hodgkin Lymphoma. Cancers. 2022; 14(19):4893. https://doi.org/10.3390/cancers14194893
Chicago/Turabian StyleLamaison, Claire, Juliette Ferrant, Pauline Gravelle, Alexandra Traverse-Glehen, Hervé Ghesquières, Marie Tosolini, Cédric Rossi, Loic Ysebaert, Pierre Brousset, Camille Laurent, and et al. 2022. "Histological Subtypes Drive Distinct Prognostic Immune Signatures in Classical Hodgkin Lymphoma" Cancers 14, no. 19: 4893. https://doi.org/10.3390/cancers14194893
APA StyleLamaison, C., Ferrant, J., Gravelle, P., Traverse-Glehen, A., Ghesquières, H., Tosolini, M., Rossi, C., Ysebaert, L., Brousset, P., Laurent, C., & Syrykh, C. (2022). Histological Subtypes Drive Distinct Prognostic Immune Signatures in Classical Hodgkin Lymphoma. Cancers, 14(19), 4893. https://doi.org/10.3390/cancers14194893







