c-MYC Protein Stability Is Sustained by MAPKs in Colorectal Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Reagents
2.2. Quantitative Real-Time PCR
2.3. RNA Interference
2.4. Proliferation Assays
2.5. Immunoblot Analysis
2.6. Co-Immunoprecipitation
2.7. In Vitro Pull-Down Assay
2.8. Immunohistochemistry
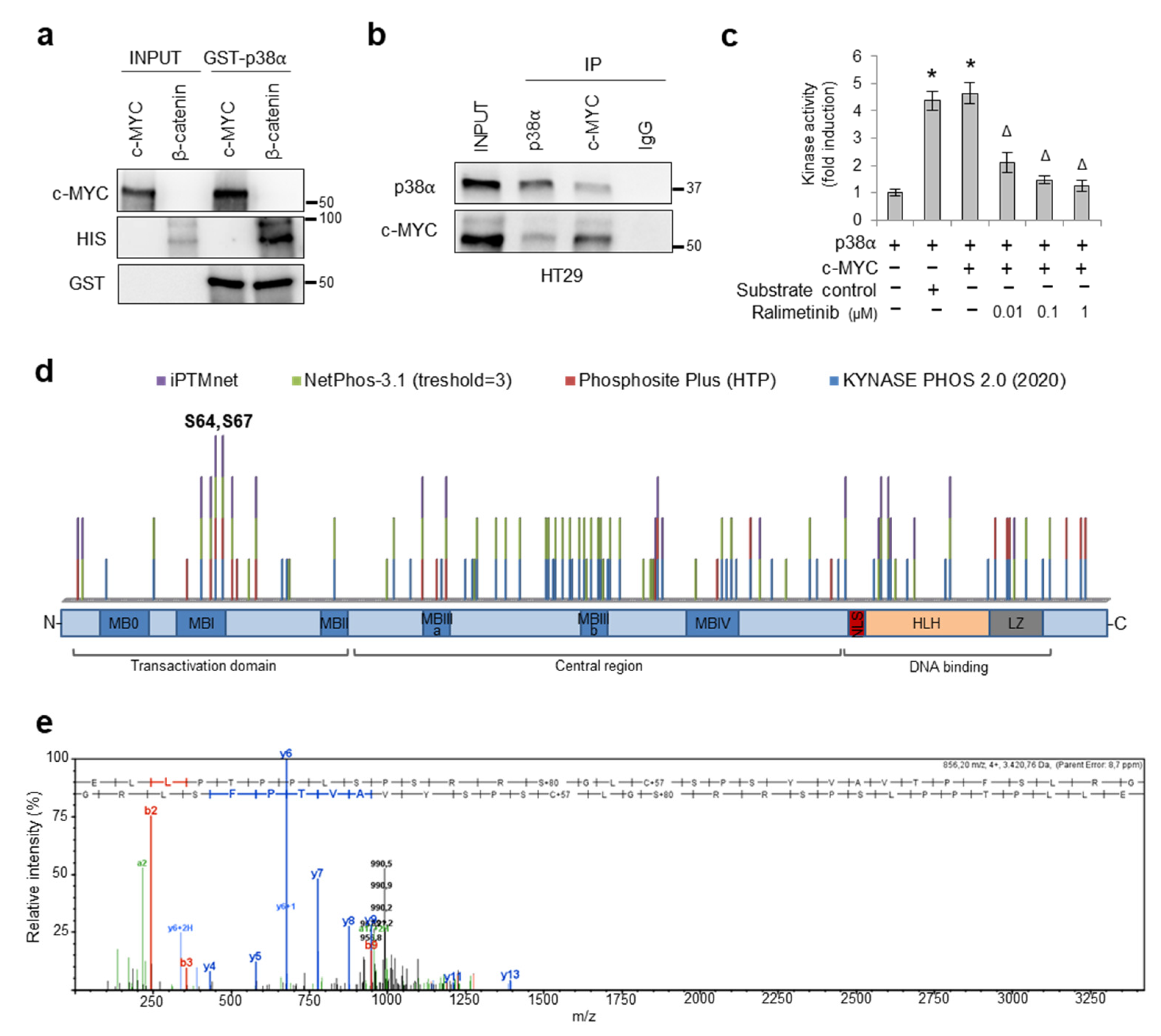
2.9. In Vitro Kinase Assay
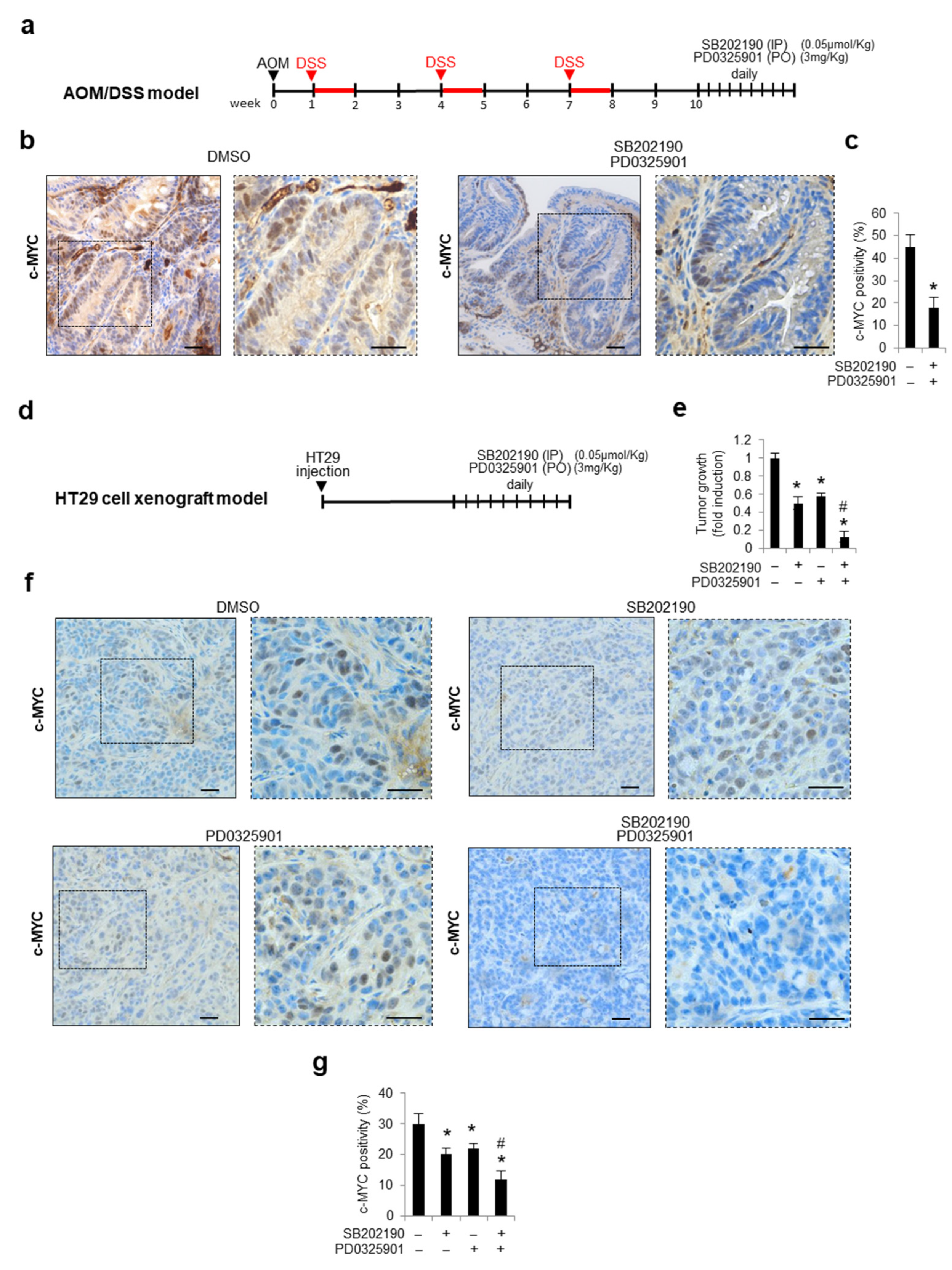
2.10. In Vivo Studies
2.11. Cell Viability Assay
2.12. Ki67 Staining
2.13. Annexin V Staining
2.14. In Silico Prediction Analysis
2.15. Mass Spectrometry Analysis
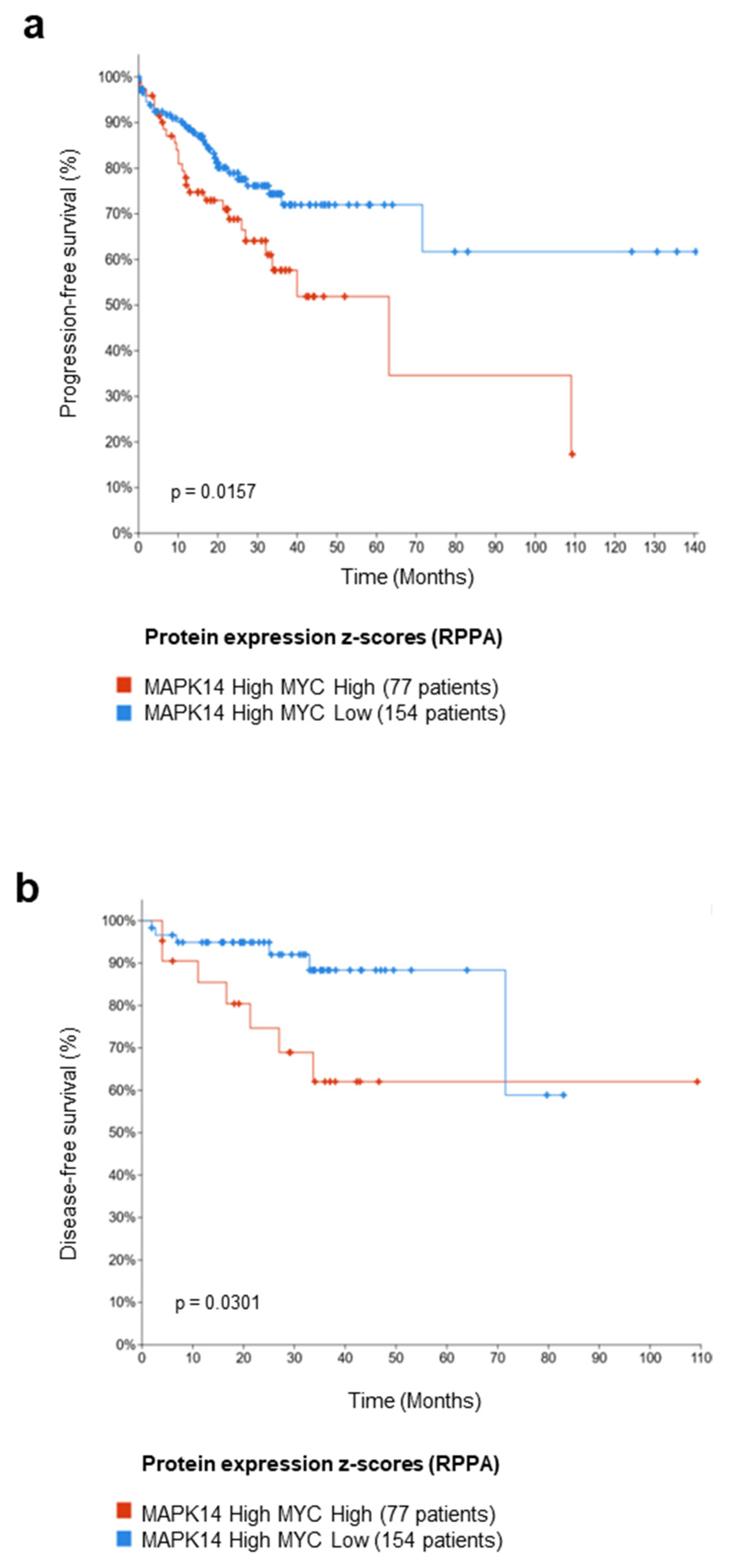
2.16. Dataset Analysis
2.17. Quantification and Statistical Analysis
3. Results
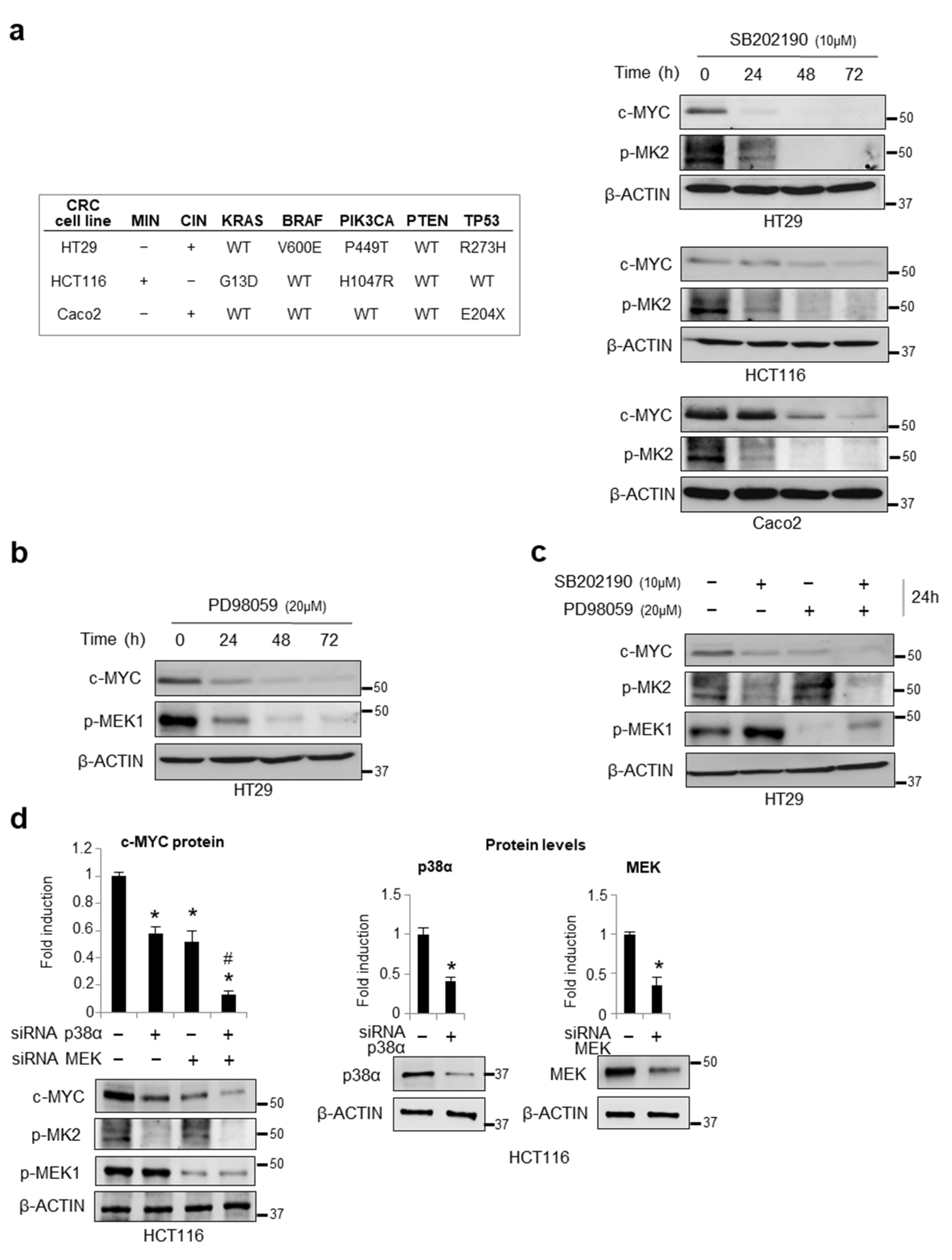
3.1. Chemical Inhibition or Genetic Ablation of the MAPKs p38α and ERK Decreases c-MYC Protein Levels
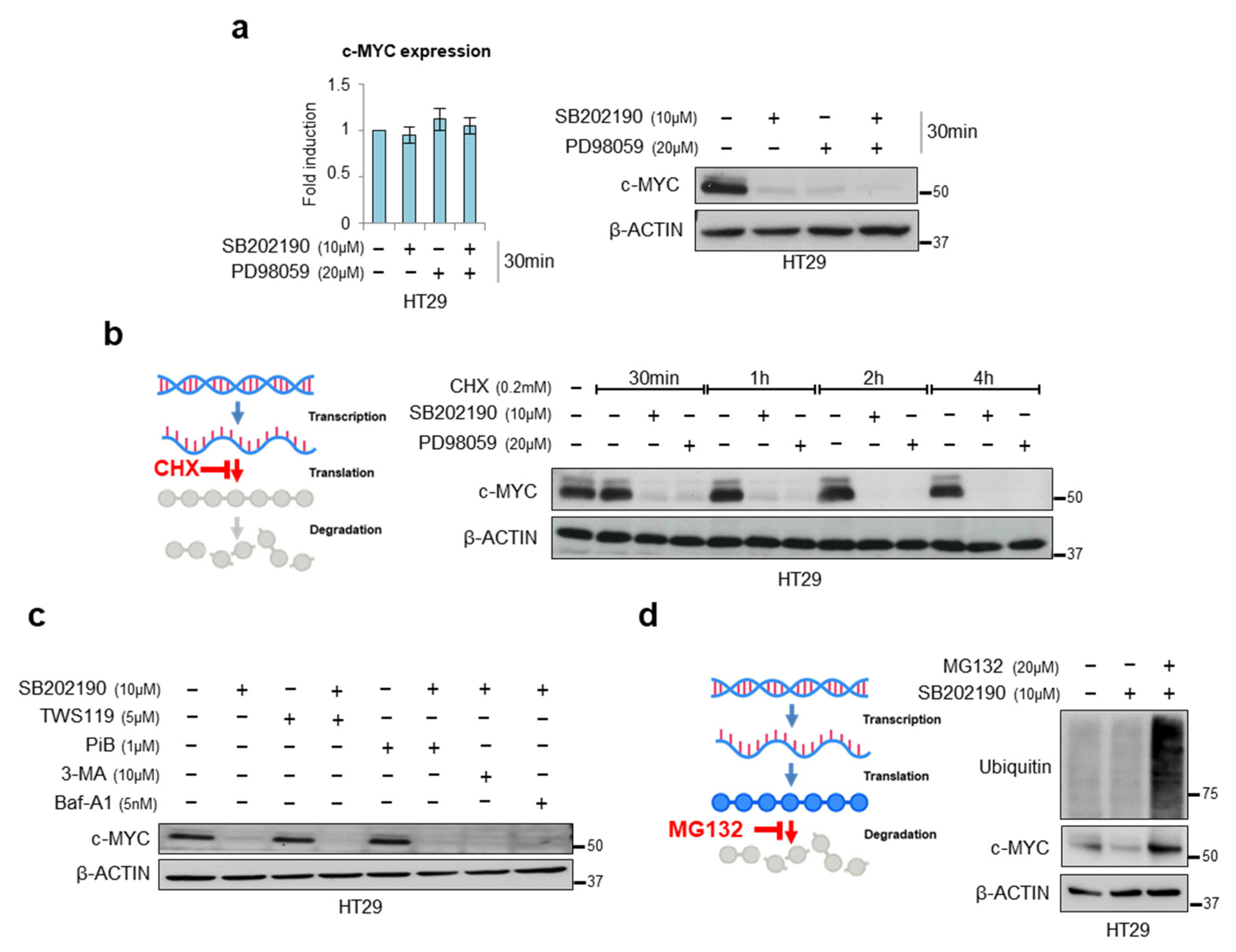
3.2. Characterization of the Mechanism of Action of p38α on c-MYC
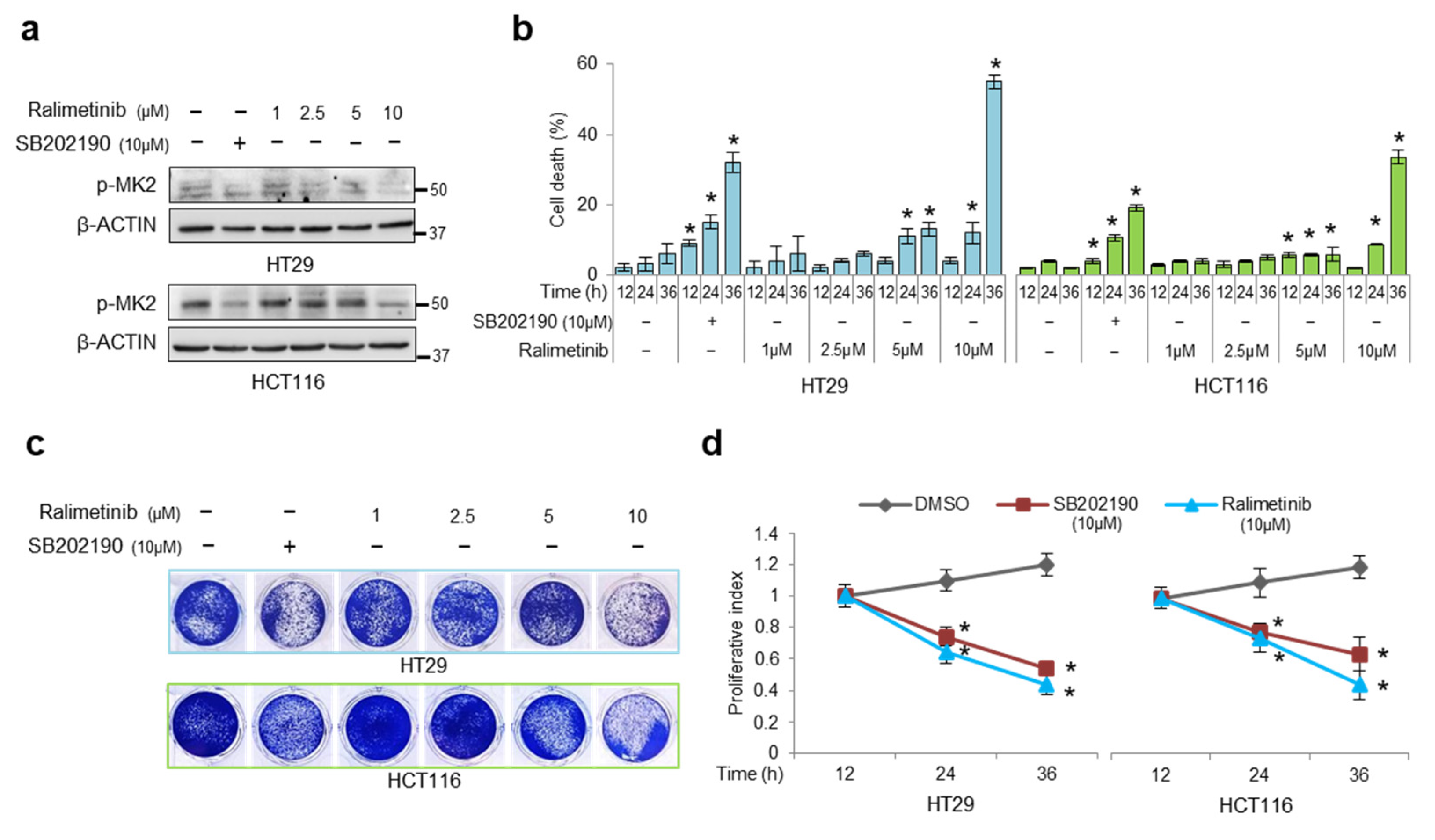
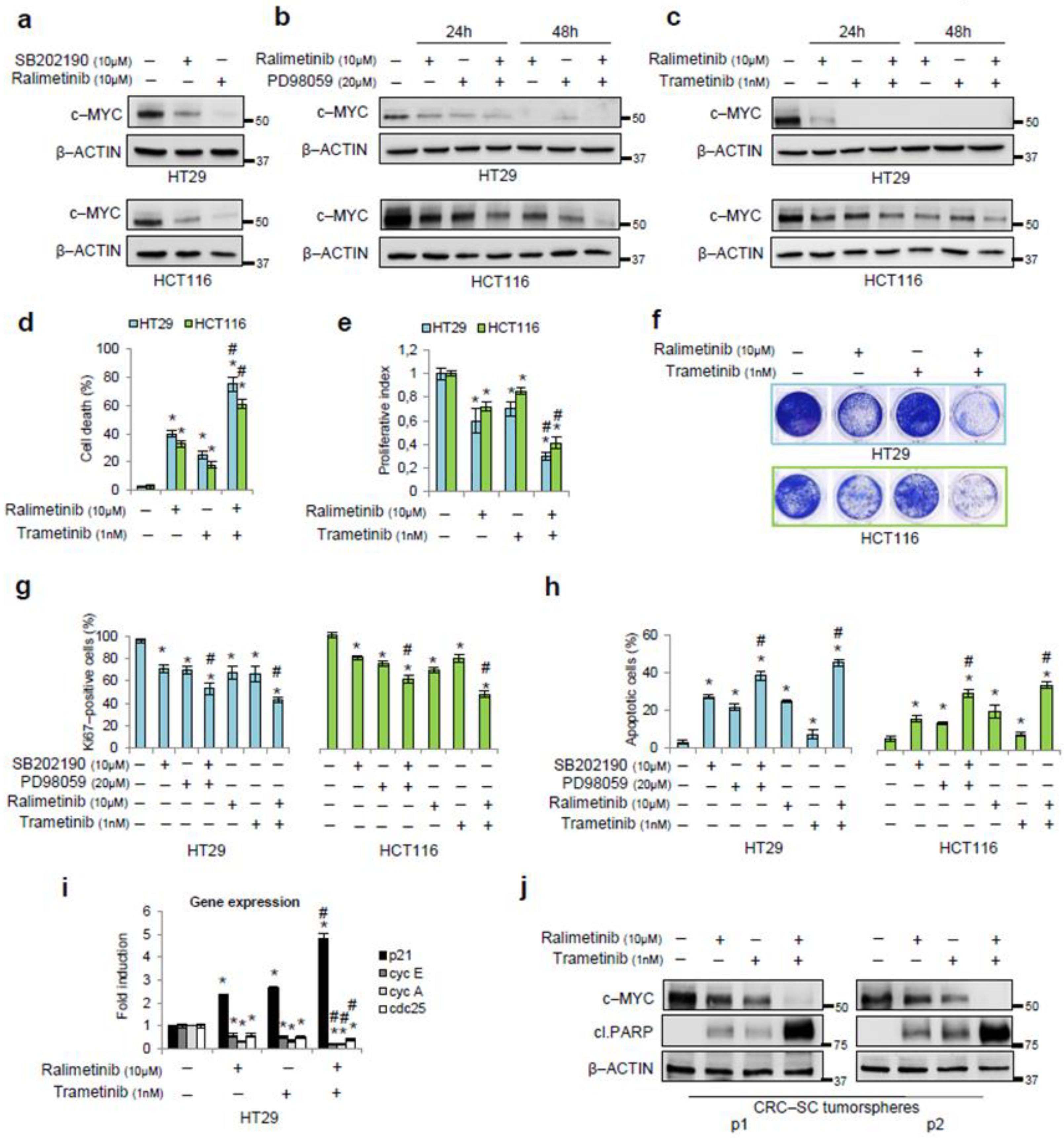
3.3. Pharmacological Targeting of p38α and ERK as a Synthetic Lethality Approach
3.4. p38α Interacts with and Phosphorylates c-MYC
3.5. Combined Inhibition of p38a and MEK1 Affects c-MYC Protein Stability in Preclinical Mouse Models
3.6. Meta-Analysis on a Cohort of Colorectal Tumor Tissues Retrieved from TCGA PanCancer Atlas
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhandari, A.; Woodhouse, M.; Gupta, S. Colorectal Cancer Is a Leading Cause of Cancer Incidence and Mortality among Adults Younger than 50 Years in the USA: A SEER-Based Analysis with Comparison to Other Young-Onset Cancers. J. Investig. Med. 2017, 65, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of Colorectal Cancer: Incidence, Mortality, Survival, and Risk Factors. Przegląd Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Takayama, T.; Miyanishi, K.; Hayashi, T.; Sato, Y.; Niitsu, Y. Colorectal Cancer: Genetics of Development and Metastasis. J. Gastroenterol. 2006, 41, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Noe, O.; Filipiak, L.; Royfman, R.; Campbell, A.; Lin, L.; Hamouda, D.; Stanbery, L.; Nemunaitis, J. Adenomatous Polyposis Coli in Cancer and Therapeutic Implications. Oncol. Rev. 2021, 15, 534. [Google Scholar] [CrossRef]
- Kim, J.C.; Bodmer, W.F. Genomic Landscape of Colorectal Carcinogenesis. J. Cancer Res. Clin. Oncol. 2022, 148, 533–545. [Google Scholar] [CrossRef]
- Piao, S.; Lee, S.-H.; Kim, H.; Yum, S.; Stamos, J.L.; Xu, Y.; Lee, S.-J.; Lee, J.; Oh, S.; Han, J.-K.; et al. Direct Inhibition of GSK3beta by the Phosphorylated Cytoplasmic Domain of LRP6 in Wnt/Beta-Catenin Signaling. PLoS ONE 2008, 3, e4046. [Google Scholar] [CrossRef]
- Wu, G.; Huang, H.; Abreu, J.G.; He, X. Inhibition of GSK3 Phosphorylation of Beta-Catenin via Phosphorylated PPPSPXS Motifs of Wnt Coreceptor LRP6. PLoS ONE 2009, 4, e4926. [Google Scholar] [CrossRef]
- Kim, S.-E.; Huang, H.; Zhao, M.; Zhang, X.; Zhang, A.; Semonov, M.V.; MacDonald, B.T.; Zhang, X.; Garcia Abreu, J.; Peng, L.; et al. Wnt Stabilization of β-Catenin Reveals Principles for Morphogen Receptor-Scaffold Assemblies. Science 2013, 340, 867–870. [Google Scholar] [CrossRef]
- Sansom, O.J.; Meniel, V.S.; Muncan, V.; Phesse, T.J.; Wilkins, J.A.; Reed, K.R.; Vass, J.K.; Athineos, D.; Clevers, H.; Clarke, A.R. Myc Deletion Rescues Apc Deficiency in the Small Intestine. Nature 2007, 446, 676–679. [Google Scholar] [CrossRef]
- Wilkins, J.A.; Sansom, O.J. C-Myc Is a Critical Mediator of the Phenotypes of Apc Loss in the Intestine. Cancer Res. 2008, 68, 4963–4966. [Google Scholar] [CrossRef]
- Sears, R.; Nuckolls, F.; Haura, E.; Taya, Y.; Tamai, K.; Nevins, J.R. Multiple Ras-Dependent Phosphorylation Pathways Regulate Myc Protein Stability. Genes Dev. 2000, 14, 2501–2514. [Google Scholar] [CrossRef] [PubMed]
- Sears, R.C. The Life Cycle of C-Myc: From Synthesis to Degradation. Cell Cycle 2004, 3, 1133–1137. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Hu, L.-L.; Gonzalez-Navajas, J.; Seo, G.S.; Shen, C.; Brick, J.; Herdman, S.; Varki, N.; Corr, M.; Lee, J.; et al. ERK Activation Drives Intestinal Tumorigenesis in Apc(Min/+) Mice. Nat. Med. 2010, 16, 665–670. [Google Scholar] [CrossRef] [PubMed]
- Moser, A.R.; Pitot, H.C.; Dove, W.F. A Dominant Mutation That Predisposes to Multiple Intestinal Neoplasia in the Mouse. Science 1990, 247, 322–324. [Google Scholar] [CrossRef] [PubMed]
- Ros, J.; Baraibar, I.; Sardo, E.; Mulet, N.; Salvà, F.; Argilés, G.; Martini, G.; Ciardiello, D.; Cuadra, J.L.; Tabernero, J.; et al. BRAF, MEK and EGFR Inhibition as Treatment Strategies in BRAF V600E Metastatic Colorectal Cancer. Ther. Adv. Med. Oncol. 2021, 13, 1758835921992974. [Google Scholar] [CrossRef] [PubMed]
- Saliani, M.; Jalal, R.; Javadmanesh, A. Differential Expression Analysis of Genes and Long Non-Coding RNAs Associated with KRAS Mutation in Colorectal Cancer Cells. Sci. Rep. 2022, 12, 7965. [Google Scholar] [CrossRef] [PubMed]
- Raman, M.; Chen, W.; Cobb, M.H. Differential Regulation and Properties of MAPKs. Oncogene 2007, 26, 3100–3112. [Google Scholar] [CrossRef]
- Kyriakis, J.M.; Avruch, J. Mammalian Mitogen-Activated Protein Kinase Signal Transduction Pathways Activated by Stress and Inflammation. Physiol. Rev. 2001, 81, 807–869. [Google Scholar] [CrossRef]
- Kyriakis, J.M.; Avruch, J. Mammalian MAPK Signal Transduction Pathways Activated by Stress and Inflammation: A 10-Year Update. Physiol. Rev. 2012, 92, 689–737. [Google Scholar] [CrossRef]
- Alonso, G.; Ambrosino, C.; Jones, M.; Nebreda, A.R. Differential Activation of P38 Mitogen-Activated Protein Kinase Isoforms Depending on Signal Strength. J. Biol. Chem. 2000, 275, 40641–40648. [Google Scholar] [CrossRef]
- Chiacchiera, F.; Grossi, V.; Cappellari, M.; Peserico, A.; Simonatto, M.; Germani, A.; Russo, S.; Moyer, M.P.; Resta, N.; Murzilli, S.; et al. Blocking P38/ERK Crosstalk Affects Colorectal Cancer Growth by Inducing Apoptosis in Vitro and in Preclinical Mouse Models. Cancer Lett. 2012, 324, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Signorile, M.L.; Grossi, V.; Di Franco, S.; Forte, G.; Disciglio, V.; Fasano, C.; Sanese, P.; De Marco, K.; Susca, F.C.; Mangiapane, L.R.; et al. Pharmacological Targeting of the Novel β-Catenin Chromatin-Associated Kinase P38α in Colorectal Cancer Stem Cell Tumorspheres and Organoids. Cell Death Dis. 2021, 12, 316. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.; Cross, D.; Jänne, P.A. Kinase Drug Discovery 20 Years after Imatinib. Nat. Rev. Drug Discov. 2022. [Google Scholar] [CrossRef] [PubMed]
- Salama, A.K.S.; Li, S.; Macrae, E.R.; Park, J.-I.; Mitchell, E.P.; Zwiebel, J.A.; Chen, H.X.; Gray, R.J.; McShane, L.M.; Rubinstein, L.V.; et al. Dabrafenib and Trametinib in Patients With Tumors With BRAFV600E Mutations: Results of the NCI-MATCH Trial Subprotocol H. J. Clin. Oncol. 2020, 38, 3895–3904. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Williams, T.M.; Robb, R.; Webb, A.; Wei, L.; Chen, W.; Mikhail, S.; Ciombor, K.K.; Cardin, D.B.; Timmers, C.; et al. Phase I Trial of Trametinib with Neoadjuvant Chemoradiation in Patients with Locally Advanced Rectal Cancer. Clin. Cancer Res. 2020, 26, 3117–3125. [Google Scholar] [CrossRef]
- Lin, A.B.; McNeely, S.C.; Beckmann, R.P. Achieving Precision Death with Cell-Cycle Inhibitors That Target DNA Replication and Repair. Clin. Cancer Res. 2017, 23, 3232–3240. [Google Scholar] [CrossRef]
- Biau, J.; Thivat, E.; Chautard, E.; Stefan, D.; Boone, M.; Chauffert, B.; Bourgne, C.; Richard, D.; Molnar, I.; Levesque, S.; et al. Phase 1 Trial of Ralimetinib (LY2228820) with Radiotherapy plus Concomitant Temozolomide in the Treatment of Newly Diagnosed Glioblastoma. Radiother. Oncol. 2021, 154, 227–234. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The CBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the CBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Chiacchiera, F.; Matrone, A.; Ferrari, E.; Ingravallo, G.; Lo Sasso, G.; Murzilli, S.; Petruzzelli, M.; Salvatore, L.; Moschetta, A.; Simone, C. P38alpha Blockade Inhibits Colorectal Cancer Growth in Vivo by Inducing a Switch from HIF1alpha- to FoxO-Dependent Transcription. Cell Death Differ. 2009, 16, 1203–1214. [Google Scholar] [CrossRef]
- Herrick, D.J.; Ross, J. The Half-Life of c-Myc MRNA in Growing and Serum-Stimulated Cells: Influence of the Coding and 3’ Untranslated Regions and Role of Ribosome Translocation. Mol. Cell Biol. 1994, 14, 2119–2128. [Google Scholar] [CrossRef]
- Gregory, M.A.; Hann, S.R. C-Myc Proteolysis by the Ubiquitin-Proteasome Pathway: Stabilization of c-Myc in Burkitt’s Lymphoma Cells. Mol. Cell Biol. 2000, 20, 2423–2435. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.; Gonçalves, E.; Mirauta, B.; Ochoa, D.; Stegle, O.; Beltrao, P. Multi-Omics Characterization of Interaction-Mediated Control of Human Protein Abundance Levels. Mol. Cell Proteom. 2019, 18, S114–S125. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ortín, J.E.; Tordera, V.; Chávez, S. Homeostasis in the Central Dogma of Molecular Biology: The Importance of MRNA Instability. RNA Biol. 2019, 16, 1659–1666. [Google Scholar] [CrossRef] [PubMed]
- Mercier, R.; LaPointe, P. The Role of Cellular Proteostasis in Antitumor Immunity. J. Biol. Chem. 2022, 298, 101930. [Google Scholar] [CrossRef]
- Buchanan, B.W.; Lloyd, M.E.; Engle, S.M.; Rubenstein, E.M. Cycloheximide Chase Analysis of Protein Degradation in Saccharomyces Cerevisiae. J. Vis. Exp. 2016, 110, e53975. [Google Scholar] [CrossRef]
- Hann, S.R. Role of Post-Translational Modifications in Regulating c-Myc Proteolysis, Transcriptional Activity and Biological Function. Semin. Cancer Biol. 2006, 16, 288–302. [Google Scholar] [CrossRef]
- Farrell, A.S.; Sears, R.C. MYC Degradation. Cold Spring Harb. Perspect. Med. 2014, 4, a014365. [Google Scholar] [CrossRef]
- Welcker, M.; Clurman, B.E. FBW7 Ubiquitin Ligase: A Tumour Suppressor at the Crossroads of Cell Division, Growth and Differentiation. Nat. Rev. Cancer 2008, 8, 83–93. [Google Scholar] [CrossRef]
- Poole, C.J.; van Riggelen, J. MYC-Master Regulator of the Cancer Epigenome and Transcriptome. Genes 2017, 8, 142. [Google Scholar] [CrossRef]
- Patnaik, A.; Haluska, P.; Tolcher, A.W.; Erlichman, C.; Papadopoulos, K.P.; Lensing, J.L.; Beeram, M.; Molina, J.R.; Rasco, D.W.; Arcos, R.R.; et al. A First-in-Human Phase I Study of the Oral P38 MAPK Inhibitor, Ralimetinib (LY2228820 Dimesylate), in Patients with Advanced Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 1095–1102. [Google Scholar] [CrossRef]
- Voon, P.J.; Chen, E.X.; Chen, H.X.; Lockhart, A.C.; Sahebjam, S.; Kelly, K.; Vaishampayan, U.N.; Subbiah, V.; Razak, A.R.; Renouf, D.J.; et al. Phase I Pharmacokinetic Study of Single Agent Trametinib in Patients with Advanced Cancer and Hepatic Dysfunction. J. Exp. Clin. Cancer Res. 2022, 41, 51. [Google Scholar] [CrossRef]
- Bretones, G.; Delgado, M.D.; León, J. Myc and Cell Cycle Control. Biochim. Biophys. Acta 2015, 1849, 506–516. [Google Scholar] [CrossRef]
- Zhang, J.; Song, N.; Zang, D.; Yu, J.; Li, J.; Di, W.; Guo, R.; Zhao, W.; Wang, H. C-Myc Promotes Tumor Proliferation and Anti-apoptosis by Repressing P21 in Rhabdomyosarcomas. Mol. Med. Rep. 2017, 16, 4089–4094. [Google Scholar] [CrossRef]
- Wang, Z.; Kar, S.; Carr, B.I. Cdc25A Protein Phosphatase: A Therapeutic Target for Liver Cancer Therapies. Anticancer Agents Med. Chem. 2008, 8, 863–871. [Google Scholar] [CrossRef]
- Noguchi, K.; Kokubu, A.; Kitanaka, C.; Ichijo, H.; Kuchino, Y. ASK1-Signaling Promotes c-Myc Protein Stability during Apoptosis. Biochem. Biophys. Res. Commun. 2001, 281, 1313–1320. [Google Scholar] [CrossRef]
- Moser, B.; Hochreiter, B.; Basílio, J.; Gleitsmann, V.; Panhuber, A.; Pardo-Garcia, A.; Hoesel, B.; Salzmann, M.; Resch, U.; Noreen, M.; et al. The Inflammatory Kinase IKKα Phosphorylates and Stabilizes C-Myc and Enhances Its Activity. Mol. Cancer 2021, 20, 16. [Google Scholar] [CrossRef]
- Trempolec, N.; Dave-Coll, N.; Nebreda, A.R. SnapShot: p38 MAPK Substrates. Cell 2013, 152, 924–924.e1. [Google Scholar] [CrossRef]
- De Robertis, M.; Massi, E.; Poeta, M.L.; Carotti, S.; Morini, S.; Cecchetelli, L.; Signori, E.; Fazio, V.M. The AOM/DSS Murine Model for the Study of Colon Carcinogenesis: From Pathways to Diagnosis and Therapy Studies. J. Carcinog. 2011, 10, 9. [Google Scholar] [CrossRef]
- Kudchadkar, S.; Ahmed, S.; Mukherjee, T.; Sagar, J. Current Guidelines in the Surgical Management of Hereditary Colorectal Cancers. World J. Gastrointest. Oncol. 2022, 14, 833–841. [Google Scholar] [CrossRef]
- Fang, J.Y.; Richardson, B.C. The MAPK Signalling Pathways and Colorectal Cancer. Lancet Oncol. 2005, 6, 322–327. [Google Scholar] [CrossRef]
- Thompson, N.; Lyons, J. Recent Progress in Targeting the Raf/MEK/ERK Pathway with Inhibitors in Cancer Drug Discovery. Curr. Opin. Pharmacol. 2005, 5, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Junttila, M.R.; Ala-Aho, R.; Jokilehto, T.; Peltonen, J.; Kallajoki, M.; Grenman, R.; Jaakkola, P.; Westermarck, J.; Kähäri, V.-M. P38alpha and P38delta Mitogen-Activated Protein Kinase Isoforms Regulate Invasion and Growth of Head and Neck Squamous Carcinoma Cells. Oncogene 2007, 26, 5267–5279. [Google Scholar] [CrossRef] [PubMed]
- Allevato, M.; Bolotin, E.; Grossman, M.; Mane-Padros, D.; Sladek, F.M.; Martinez, E. Sequence-Specific DNA Binding by MYC/MAX to Low-Affinity Non-E-Box Motifs. PLoS ONE 2017, 12, e0180147. [Google Scholar] [CrossRef]
- Welcker, M.; Orian, A.; Jin, J.; Grim, J.E.; Grim, J.A.; Harper, J.W.; Eisenman, R.N.; Clurman, B.E. The Fbw7 Tumor Suppressor Regulates Glycogen Synthase Kinase 3 Phosphorylation-Dependent c-Myc Protein Degradation. Proc. Natl. Acad. Sci. USA 2004, 101, 9085–9090. [Google Scholar] [CrossRef]
- Cash, T.; Fox, E.; Liu, X.; Minard, C.G.; Reid, J.M.; Scheck, A.C.; Weigel, B.J.; Wetmore, C. A Phase 1 Study of Prexasertib (LY2606368), a CHK1/2 Inhibitor, in Pediatric Patients with Recurrent or Refractory Solid Tumors, Including CNS Tumors: A Report from the Children’s Oncology Group Pediatric Early Phase Clinical Trials Network (ADVL1515). Pediatr. Blood Cancer 2021, 68, e29065. [Google Scholar] [CrossRef]
- Bendell, J.C.; Bischoff, H.G.; Hwang, J.; Reinhardt, H.C.; Zander, T.; Wang, X.; Hynes, S.; Pitou, C.; Campbell, R.; Iversen, P.; et al. A Phase 1 Dose-Escalation Study of Checkpoint Kinase 1 (CHK1) Inhibitor Prexasertib in Combination with P38 Mitogen-Activated Protein Kinase (P38 MAPK) Inhibitor Ralimetinib in Patients with Advanced or Metastatic Cancer. Investig. New Drugs 2020, 38, 1145–1155. [Google Scholar] [CrossRef]
- Kudo, M.; Morimoto, M.; Moriguchi, M.; Izumi, N.; Takayama, T.; Yoshiji, H.; Hino, K.; Oikawa, T.; Chiba, T.; Motomura, K.; et al. A Randomized, Double-Blind, Placebo-Controlled, Phase 3 Study of Tivantinib in Japanese Patients with MET-High Hepatocellular Carcinoma. Cancer Sci. 2020, 111, 3759–3769. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lepore Signorile, M.; Grossi, V.; Fasano, C.; Forte, G.; Disciglio, V.; Sanese, P.; De Marco, K.; La Rocca, F.; Armentano, R.; Valentini, A.M.; et al. c-MYC Protein Stability Is Sustained by MAPKs in Colorectal Cancer. Cancers 2022, 14, 4840. https://doi.org/10.3390/cancers14194840
Lepore Signorile M, Grossi V, Fasano C, Forte G, Disciglio V, Sanese P, De Marco K, La Rocca F, Armentano R, Valentini AM, et al. c-MYC Protein Stability Is Sustained by MAPKs in Colorectal Cancer. Cancers. 2022; 14(19):4840. https://doi.org/10.3390/cancers14194840
Chicago/Turabian StyleLepore Signorile, Martina, Valentina Grossi, Candida Fasano, Giovanna Forte, Vittoria Disciglio, Paola Sanese, Katia De Marco, Francesca La Rocca, Raffaele Armentano, Anna Maria Valentini, and et al. 2022. "c-MYC Protein Stability Is Sustained by MAPKs in Colorectal Cancer" Cancers 14, no. 19: 4840. https://doi.org/10.3390/cancers14194840
APA StyleLepore Signorile, M., Grossi, V., Fasano, C., Forte, G., Disciglio, V., Sanese, P., De Marco, K., La Rocca, F., Armentano, R., Valentini, A. M., Giannelli, G., & Simone, C. (2022). c-MYC Protein Stability Is Sustained by MAPKs in Colorectal Cancer. Cancers, 14(19), 4840. https://doi.org/10.3390/cancers14194840








