Intestinal Klebsiella pneumoniae Contributes to Pneumonia by Synthesizing Glutamine in Multiple Myeloma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Samples
2.2. Cell Lines, Antibodies, and Reagents
2.3. Metagenomic Sequencing and Identification of Differential Species
2.4. The 16S Ribosomal DNA qPCR
2.5. The 5TGM1 MM Mouse Model
2.6. Construction and Confirmation of K. pneumonia Containing Mutant glnA
2.7. Intestinal Colonization with K. pneumonia In Vivo
2.8. Immunohistochemistry
2.9. Targeted Metabolomic Assays
2.10. Real-Time Quantitative PCR
2.11. Plate Colony and CCK8 Assay
2.12. Western Blotting
2.13. Statistical Analysis
3. Results
3.1. Subsection K. pneumonia Is Enriched in the Intestine and Is Linked to MM with Pneumonia
3.2. Intestinal K. pneumonia Accelerates Pneumonia in 5TGM1 MM Mice
3.3. Glutamine Is Elevated in PB Plasma of MM Patients with Pneumonia
3.4. Intestinal K. pneumonia Synthesizes Glutamine to Bolster Pneumonia in MM In Vivo
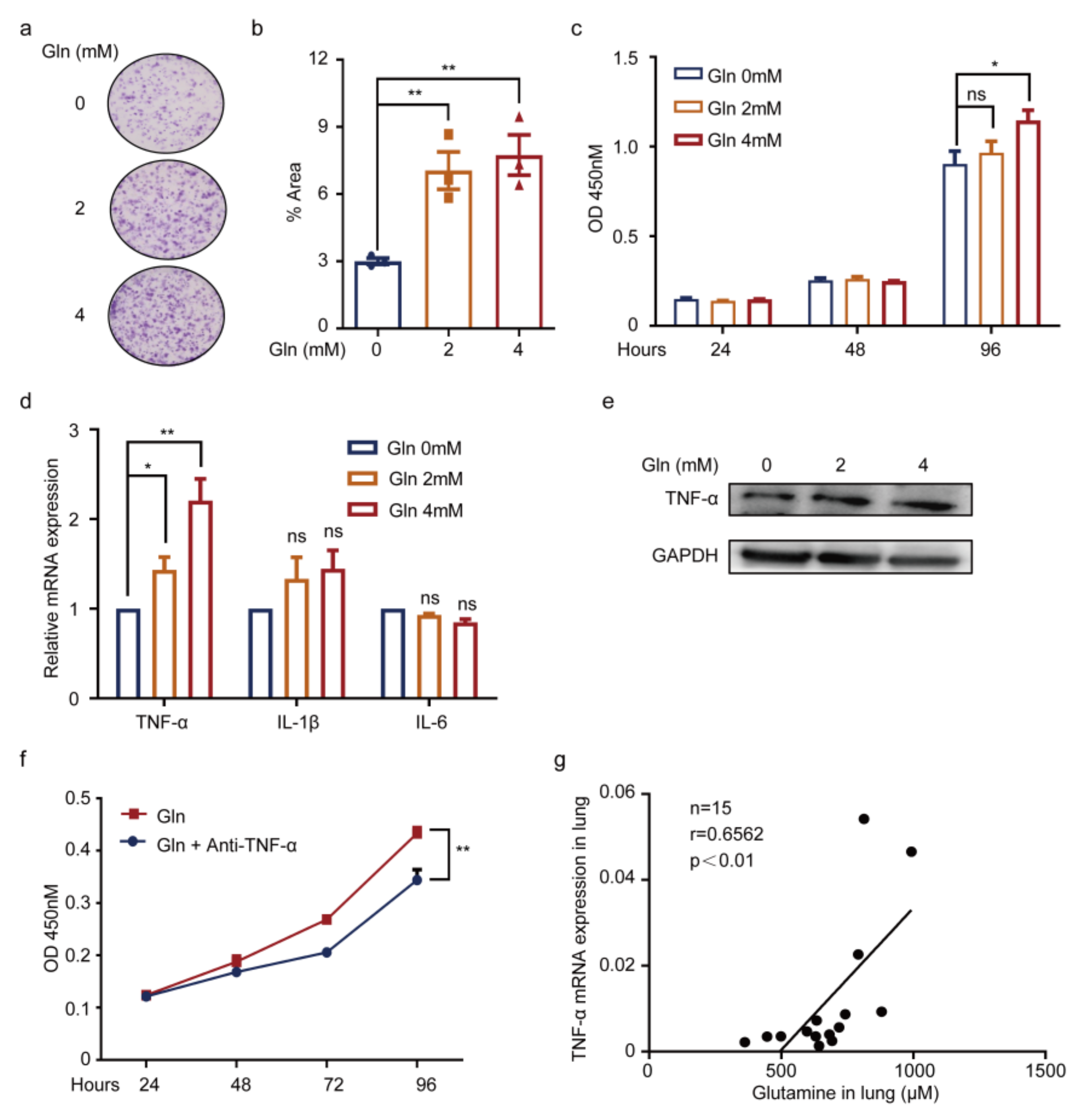
3.5. Glutamine Contributes to Pneumonia by Promoting TNF-α Expression of WI38 Cells
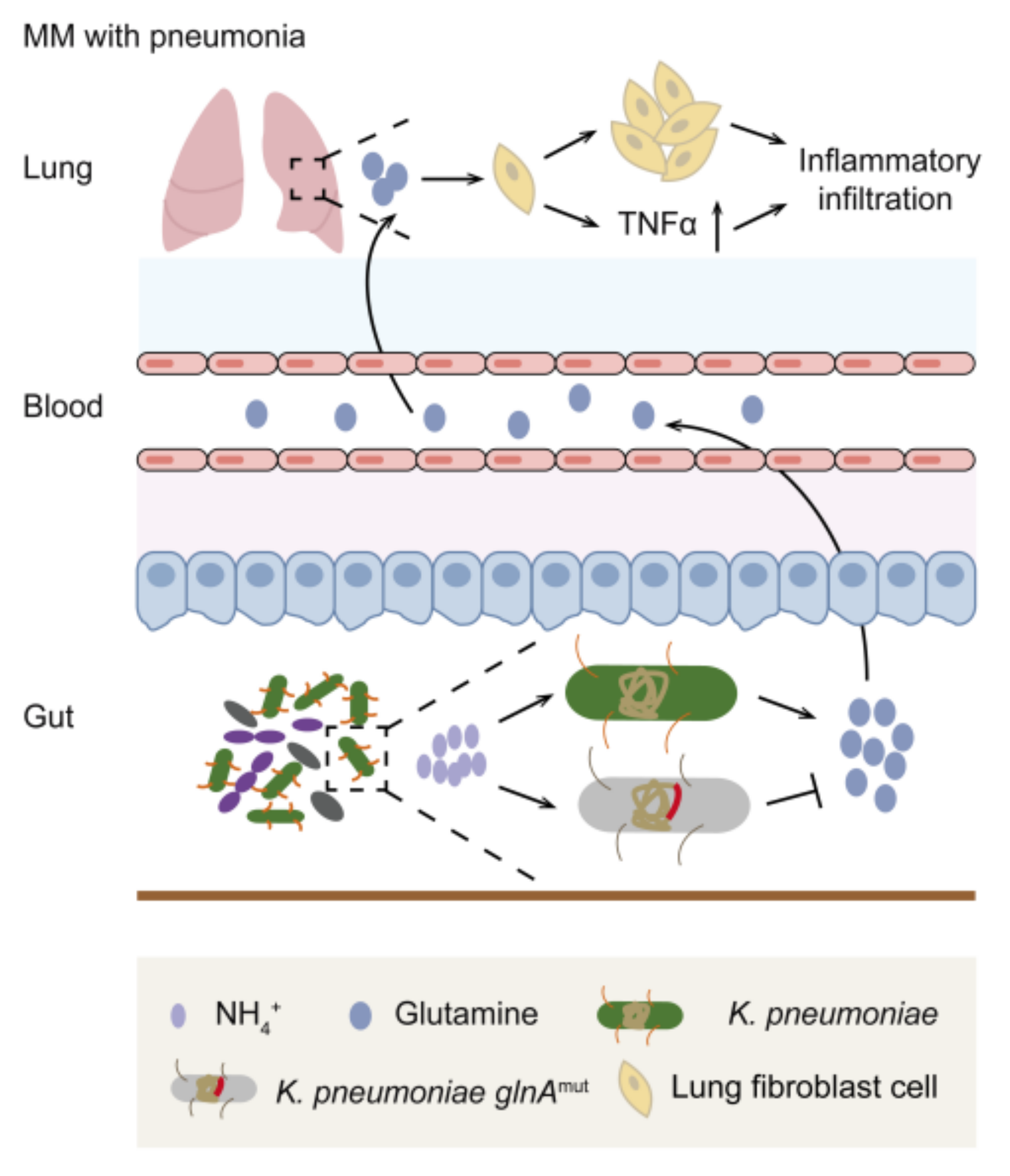
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cowan, A.J.; Green, D.J.; Kwok, M.; Lee, S.; Coffey, D.G.; Holmberg, L.A.; Tuazon, S.; Gopal, A.K.; Libby, E.N. Diagnosis and Management of Multiple Myeloma: A Review. JAMA 2022, 327, 464–477. [Google Scholar] [CrossRef] [PubMed]
- Raje, N.S.; Anaissie, E.; Kumar, S.K.; Lonial, S.; Martin, T.; Gertz, M.A.; Krishnan, A.; Hari, P.; Ludwig, H.; O’Donnell, E.; et al. Consensus guidelines and recommendations for infection prevention in multiple myeloma: A report from the International Myeloma Working Group. Lancet Haematol. 2022, 9, e143–e161. [Google Scholar] [CrossRef]
- Nucci, M.; Anaissie, E. Infections in patients with multiple myeloma in the era of high-dose therapy and novel agents. Clin. Infect. Dis. 2009, 49, 1211–1225. [Google Scholar] [CrossRef] [PubMed]
- Augustson, B.M.; Begum, G.; Dunn, J.A.; Barth, N.J.; Davies, F.; Morgan, G.; Behrens, J.; Smith, A.; Child, J.A.; Drayson, M.T. Early mortality after diagnosis of multiple myeloma: Analysis of patients entered onto the United Kingdom Medical Research Council trials between 1980 and 2002—Medical Research Council Adult Leukaemia Working Party. J. Clin. Oncol. 2005, 23, 9219–9226. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.H.; Cho, M.S.; Kim, H.K.; Kim, S.J.; Kim, K.; Cheong, J.W.; Kim, S.J.; Kim, J.S.; Ahn, J.S.; Kim, Y.K.; et al. Risk factors associated with early mortality in patients with multiple myeloma who were treated upfront with a novel agents containing regimen. BMC Cancer 2016, 16, 613. [Google Scholar] [CrossRef] [PubMed]
- Terebelo, H.; Srinivasan, S.; Narang, M.; Abonour, R.; Gasparetto, C.; Toomey, K.; Hardin, J.W.; Larkins, G.; Kitali, A.; Rifkin, R.M.; et al. Recognition of early mortality in multiple myeloma by a prediction matrix. Am. J. Hematol. 2017, 92, 915–923. [Google Scholar] [CrossRef]
- Mai, E.K.; Haas, E.M.; Lucke, S.; Lopprich, M.; Kunz, C.; Pritsch, M.; Knaup-Gregori, P.; Raab, M.S.; Schlenzka, J.; Bertsch, U.; et al. A systematic classification of death causes in multiple myeloma. Blood Cancer J. 2018, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Nucci, M.; Anaissie, E. Infections in patients with multiple myeloma. Semin. Hematol. 2009, 46, 277–288. [Google Scholar] [CrossRef]
- Blimark, C.; Holmberg, E.; Mellqvist, U.H.; Landgren, O.; Bjorkholm, M.; Hultcrantz, M.; Kjellander, C.; Turesson, I.; Kristinsson, S.Y. Multiple myeloma and infections: A population-based study on 9253 multiple myeloma patients. Haematologica 2015, 100, 107–113. [Google Scholar] [CrossRef]
- Schuijt, T.J.; Lankelma, J.M.; Scicluna, B.P.; de Sousa e Melo, F.; Roelofs, J.J.; de Boer, J.D.; Hoogendijk, A.J.; de Beer, R.; de Vos, A.; Belzer, C.; et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 2016, 65, 575–583. [Google Scholar] [CrossRef] [Green Version]
- Shukla, S.D.; Budden, K.F.; Neal, R.; Hansbro, P.M. Microbiome effects on immunity, health and disease in the lung. Clin. Transl. Immunol. 2017, 6, e133. [Google Scholar] [CrossRef]
- Dumas, A.; Bernard, L.; Poquet, Y.; Lugo-Villarino, G.; Neyrolles, O. The role of the lung microbiota and the gut-lung axis in respiratory infectious diseases. Cell Microbiol. 2018, 20, e12966. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, K.; Raundhal, M.; Chen, B.B.; Morse, C.; Tyurina, Y.Y.; Khare, A.; Oriss, T.B.; Huff, R.; Lee, J.S.; St Croix, C.M.; et al. The mito-DAMP cardiolipin blocks IL-10 production causing persistent inflammation during bacterial pneumonia. Nat. Commun. 2017, 8, 13944. [Google Scholar] [CrossRef] [PubMed]
- Thibeault, C.; Suttorp, N.; Opitz, B. The microbiota in pneumonia: From protection to predisposition. Sci. Transl. Med. 2021, 13, eaba0501. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.A.; Coopersmith, C.M. Intestinal crosstalk: A new paradigm for understanding the gut as the “motor” of critical illness. Shock 2007, 28, 384–393. [Google Scholar] [CrossRef]
- Jian, X.; Zhu, Y.; Ouyang, J.; Wang, Y.; Lei, Q.; Xia, J.; Guan, Y.; Zhang, J.; Guo, J.; He, Y.; et al. Alterations of gut microbiome accelerate multiple myeloma progression by increasing the relative abundances of nitrogen-recycling bacteria. Microbiome 2020, 8, 74. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Zhang, J.; Wu, X.; Du, W.; Zhu, Y.; Liu, X.; Liu, Z.; Meng, B.; Guo, J.; Yang, Q.; et al. Blocking glycine utilization inhibits multiple myeloma progression by disrupting glutathione balance. Nat. Commun. 2022, 13, 4007. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Fujimoto, C.; Haruki, Y.; Maeda, T.; Kokeguchi, S.; Petelin, M.; Arai, H.; Tanimoto, I.; Nishimura, F.; Takashiba, S. Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol. Med. Microbiol. 2003, 39, 81–86. [Google Scholar] [CrossRef]
- Tsoi, H.; Chu, E.S.H.; Zhang, X.; Sheng, J.; Nakatsu, G.; Ng, S.C.; Chan, A.W.H.; Chan, F.K.L.; Sung, J.J.Y.; Yu, J. Peptostreptococcus anaerobius Induces Intracellular Cholesterol Biosynthesis in Colon Cells to Induce Proliferation and Causes Dysplasia in Mice. Gastroenterology 2017, 152, 1419–1433.e5. [Google Scholar] [CrossRef]
- Xie, G.; Wang, X.; Wei, R.; Wang, J.; Zhao, A.; Chen, T.; Wang, Y.; Zhang, H.; Xiao, Z.; Liu, X.; et al. Serum metabolite profiles are associated with the presence of advanced liver fibrosis in Chinese patients with chronic hepatitis B viral infection. BMC Med. 2020, 18, 144. [Google Scholar] [CrossRef]
- Tan, B.; Qiu, Y.; Zou, X.; Chen, T.; Xie, G.; Cheng, Y.; Dong, T.; Zhao, L.; Feng, B.; Hu, X.; et al. Metabonomics identifies serum metabolite markers of colorectal cancer. J. Proteome Res. 2013, 12, 3000–3009. [Google Scholar] [CrossRef]
- Feng, X.; Guo, J.; An, G.; Wu, Y.; Liu, Z.; Meng, B.; He, N.; Zhao, X.; Chen, S.; Zhu, Y.; et al. Genetic Aberrations and Interaction of NEK2 and TP53 Accelerate Aggressiveness of Multiple Myeloma. Adv. Sci. 2022, 9, e2104491. [Google Scholar] [CrossRef]
- Wu, X.; Xia, J.; Zhang, J.; Zhu, Y.; Wu, Y.; Guo, J.; Chen, S.; Lei, Q.; Meng, B.; Kuang, C.; et al. Phosphoglycerate dehydrogenase promotes proliferation and bortezomib resistance through increasing reduced glutathione synthesis in multiple myeloma. Br. J. Haematol. 2020, 190, 52–66. [Google Scholar] [CrossRef] [PubMed]
- Hanada, S.; Pirzadeh, M.; Carver, K.Y.; Deng, J.C. Respiratory Viral Infection-Induced Microbiome Alterations and Secondary Bacterial Pneumonia. Front. Immunol. 2018, 9, 2640. [Google Scholar] [CrossRef]
- Fagundes, C.T.; Amaral, F.A.; Vieira, A.T.; Soares, A.C.; Pinho, V.; Nicoli, J.R.; Vieira, L.Q.; Teixeira, M.M.; Souza, D.G. Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in germfree mice. J. Immunol. 2012, 188, 1411–1420. [Google Scholar] [CrossRef]
- Wu, S.; Jiang, Z.Y.; Sun, Y.F.; Yu, B.; Chen, J.; Dai, C.Q.; Wu, X.L.; Tang, X.L.; Chen, X.Y. Microbiota regulates the TLR7 signaling pathway against respiratory tract influenza A virus infection. Curr. Microbiol. 2013, 67, 414–422. [Google Scholar] [CrossRef]
- Kawahara, T.; Takahashi, T.; Oishi, K.; Tanaka, H.; Masuda, M.; Takahashi, S.; Takano, M.; Kawakami, T.; Fukushima, K.; Kanazawa, H.; et al. Consecutive oral administration of Bifidobacterium longum MM-2 improves the defense system against influenza virus infection by enhancing natural killer cell activity in a murine model. Microbiol. Immunol. 2015, 59, 1–12. [Google Scholar] [CrossRef]
- Jespersen, L.; Tarnow, I.; Eskesen, D.; Morberg, C.M.; Michelsen, B.; Bugel, S.; Dragsted, L.O.; Rijkers, G.T.; Calder, P.C. Effect of Lactobacillus paracasei subsp. paracasei, L. casei 431 on immune response to influenza vaccination and upper respiratory tract infections in healthy adult volunteers: A randomized, double-blind, placebo-controlled, parallel-group study. Am. J. Clin. Nutr. 2015, 101, 1188–1196. [Google Scholar] [CrossRef]
- Luoto, R.; Ruuskanen, O.; Waris, M.; Kalliomaki, M.; Salminen, S.; Isolauri, E. Prebiotic and probiotic supplementation prevents rhinovirus infections in preterm infants: A randomized, placebo-controlled trial. J. Allergy Clin. Immunol. 2014, 133, 405–413. [Google Scholar] [CrossRef]
- King, S.; Glanville, J.; Sanders, M.E.; Fitzgerald, A.; Varley, D. Effectiveness of probiotics on the duration of illness in healthy children and adults who develop common acute respiratory infectious conditions: A systematic review and meta-analysis. Br. J. Nutr. 2014, 112, 41–54. [Google Scholar] [CrossRef]
- West, N.P.; Horn, P.L.; Pyne, D.B.; Gebski, V.J.; Lahtinen, S.J.; Fricker, P.A.; Cripps, A.W. Probiotic supplementation for respiratory and gastrointestinal illness symptoms in healthy physically active individuals. Clin. Nutr. 2014, 33, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Clarke, T.B.; Davis, K.M.; Lysenko, E.S.; Zhou, A.Y.; Yu, Y.; Weiser, J.N. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 2010, 16, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Anand, S.; Mande, S.S. Diet, Microbiota and Gut-Lung Connection. Front. Microbiol. 2018, 9, 2147. [Google Scholar] [CrossRef]
- Saint-Criq, V.; Lugo-Villarino, G.; Thomas, M. Dysbiosis, malnutrition and enhanced gut-lung axis contribute to age-related respiratory diseases. Ageing Res. Rev. 2021, 66, 101235. [Google Scholar] [CrossRef]
- Budden, K.F.; Gellatly, S.L.; Wood, D.L.; Cooper, M.A.; Morrison, M.; Hugenholtz, P.; Hansbro, P.M. Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 2017, 15, 55–63. [Google Scholar] [CrossRef]
- Galvao, I.; Tavares, L.P.; Correa, R.O.; Fachi, J.L.; Rocha, V.M.; Rungue, M.; Garcia, C.C.; Cassali, G.; Ferreira, C.M.; Martins, F.S.; et al. The Metabolic Sensor GPR43 Receptor Plays a Role in the Control of Klebsiella pneumoniae Infection in the Lung. Front. Immunol. 2018, 9, 142. [Google Scholar] [CrossRef]
- Sencio, V.; Barthelemy, A.; Tavares, L.P.; Machado, M.G.; Soulard, D.; Cuinat, C.; Queiroz-Junior, C.M.; Noordine, M.L.; Salome-Desnoulez, S.; Deryuter, L.; et al. Gut Dysbiosis during Influenza Contributes to Pulmonary Pneumococcal Superinfection through Altered Short-Chain Fatty Acid Production. Cell Rep. 2020, 30, 2934–2947.e6. [Google Scholar] [CrossRef]
- Bolzoni, M.; Chiu, M.; Accardi, F.; Vescovini, R.; Airoldi, I.; Storti, P.; Todoerti, K.; Agnelli, L.; Missale, G.; Andreoli, R.; et al. Dependence on glutamine uptake and glutamine addiction characterize myeloma cells: A new attractive target. Blood 2016, 128, 667–679. [Google Scholar] [CrossRef]
- Li, M.; Chiang, Y.L.; Lyssiotis, C.A.; Teater, M.R.; Hong, J.Y.; Shen, H.; Wang, L.; Hu, J.; Jing, H.; Chen, Z.; et al. Non-oncogene Addiction to SIRT3 Plays a Critical Role in Lymphomagenesis. Cancer Cell 2019, 35, 916–931.e9. [Google Scholar] [CrossRef]




| Patients’ Characteristics | High K. pneumonia (n = 10; n/N × 100%) | Low K. pneumonia (n = 19; n/N × 100%) | p-Value |
|---|---|---|---|
| Sex | 0.6999 | ||
| Male | 6/10 (60.00) | 9/19 (47.37) | |
| Female | 4/10 (40.00) | 10/19 (52.63) | |
| Age (Year) | 59.8 ± 9.19 | 59.53 ± 11.73 | 0.9495 |
| Subtype of immunoglobulin | 1 | ||
| IgA | 1/10 (10.00) | 3/19 (15.79) | |
| IgG | 6/10 (60.00) | 12/19 (63.16) | |
| Subtype of immunoglobulin | 0.2701 | ||
| κ | 6/10 (60.00) | 7/19 (36.84) | |
| λ | 4/10 (40.00) | 12/19 (63.16) | |
| DS stage | 0.6942 | ||
| I + II | 3/10 (30.00) | 8/19 (42.11) | |
| III | 7/10 (70.00) | 11/19 (57.89) | |
| ISS stage | 1 | ||
| I | 1/10 (10.00) | 2/19 (10.53) | |
| II + III | 9/10 (90.00) | 17/19 (89.47) | |
| Pneumonia | 0.0418 * | ||
| Yes | 7/10 (70.00) | 6/19 (31.58) | |
| No | 2/10 (20.00) | 13/19 (68.42) | |
| LDH (U/L) | 197 ± 55.19 | 191.32 ± 43.2 | 0.7619 |
| Immunoparesis | 1 | ||
| Yes | 10/10 (100.00) | 18/19 (94.74) | |
| No | 0/10 (0) | 1/19 (5.26) | |
| Plasma cells in bone marrow (%) | 19.96 ± 16.11 | 30.36 ± 24.96 | 0.2713 |
| Ca+ (mmol/L) | 2.37 ± 0.48 | 2.18 ± 0.28 | 0.1792 |
| Creatinine (μmol/L) | 141.4 ± 105.4 | 111.02 ± 74.6 | 0.3747 |
| Patients’ Characteristics | High Glutamine (n = 57; n/N × 100%) | Low Glutamine (n = 58; n/N × 100%) | p-Value |
|---|---|---|---|
| Sex | 0.2234 | ||
| Male | 33/57 (57.89) | 27/58 (46.55) | |
| Female | 24/57 (42.11) | 31/58 (53.44) | |
| Age (Year) | 59.7 ± 10.04 | 56.38 ± 10.84 | |
| Subtype of immunoglobulin | 0.4002 | ||
| IgA | 14/57 (24.56) | 10/58 (17.24) | |
| IgG | 29/57 (50.88) | 33/58 (56.90) | |
| IgD | 3/57 (5.26) | 6/58 (10.34) | |
| Subtype of immunoglobulin | 0.5803 | ||
| κ | 28/57 (49.12) | 32/58 (55.17) | |
| λ | 28/57 (49.12) | 26/58 (44.83) | |
| DS stage | 0.1136 | ||
| I | 5/57 (8.77) | 1/58 (1.72) | |
| II + III | 52/57 (91.23) | 57/58 (98.28) | |
| ISS stage | 0.0534 | ||
| I | 13/57 (22.81) | 10/58 (17.24) | |
| II | 19/57 (33.33) | 10/58 (17.24) | |
| III | 25/57 (43.86) | 38/58 (65.52) | |
| pneumonia | 0.0093 ** | ||
| Yes | 27/57 (47.36) | 14/58 (24.14) | |
| No | 30/57 (52.63) | 44/58 (75.86) | |
| LDH(U/L) | 158.25 ± 65.67 | 220.79 ± 239.49 | 0.0598 |
| Immunoparesis | 0.7051 | ||
| Yes | 50/57 (87.72) | 53/58 (91.38) | |
| No | 6/57 (10.53) | 5/58 (8.62) | |
| Plasma cells in bone marrow (%) | 31.73 ± 23.36 | 34.48 ± 27.34 | 0.6782 |
| Ca+ (mmol/L) | 2.37 ± 0.39 | 2.28 ± 0.31 | 0.1685 |
| Creatinine (μmol/L) | 115.14 ± 128.56 | 113.71 ± 92.67 | 0.9455 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Yang, Q.; Zhu, Y.; Jian, X.; Guo, J.; Zhang, J.; Kuang, C.; Feng, X.; An, G.; Qiu, L.; et al. Intestinal Klebsiella pneumoniae Contributes to Pneumonia by Synthesizing Glutamine in Multiple Myeloma. Cancers 2022, 14, 4188. https://doi.org/10.3390/cancers14174188
Wang Y, Yang Q, Zhu Y, Jian X, Guo J, Zhang J, Kuang C, Feng X, An G, Qiu L, et al. Intestinal Klebsiella pneumoniae Contributes to Pneumonia by Synthesizing Glutamine in Multiple Myeloma. Cancers. 2022; 14(17):4188. https://doi.org/10.3390/cancers14174188
Chicago/Turabian StyleWang, Yihui, Qin Yang, Yinghong Zhu, Xingxing Jian, Jiaojiao Guo, Jingyu Zhang, Chunmei Kuang, Xiangling Feng, Gang An, Lugui Qiu, and et al. 2022. "Intestinal Klebsiella pneumoniae Contributes to Pneumonia by Synthesizing Glutamine in Multiple Myeloma" Cancers 14, no. 17: 4188. https://doi.org/10.3390/cancers14174188
APA StyleWang, Y., Yang, Q., Zhu, Y., Jian, X., Guo, J., Zhang, J., Kuang, C., Feng, X., An, G., Qiu, L., Li, G., He, Y., & Zhou, W. (2022). Intestinal Klebsiella pneumoniae Contributes to Pneumonia by Synthesizing Glutamine in Multiple Myeloma. Cancers, 14(17), 4188. https://doi.org/10.3390/cancers14174188






