PARP1: Liaison of Chromatin Remodeling and Transcription
Abstract
:Simple Summary
Abstract
1. Introduction
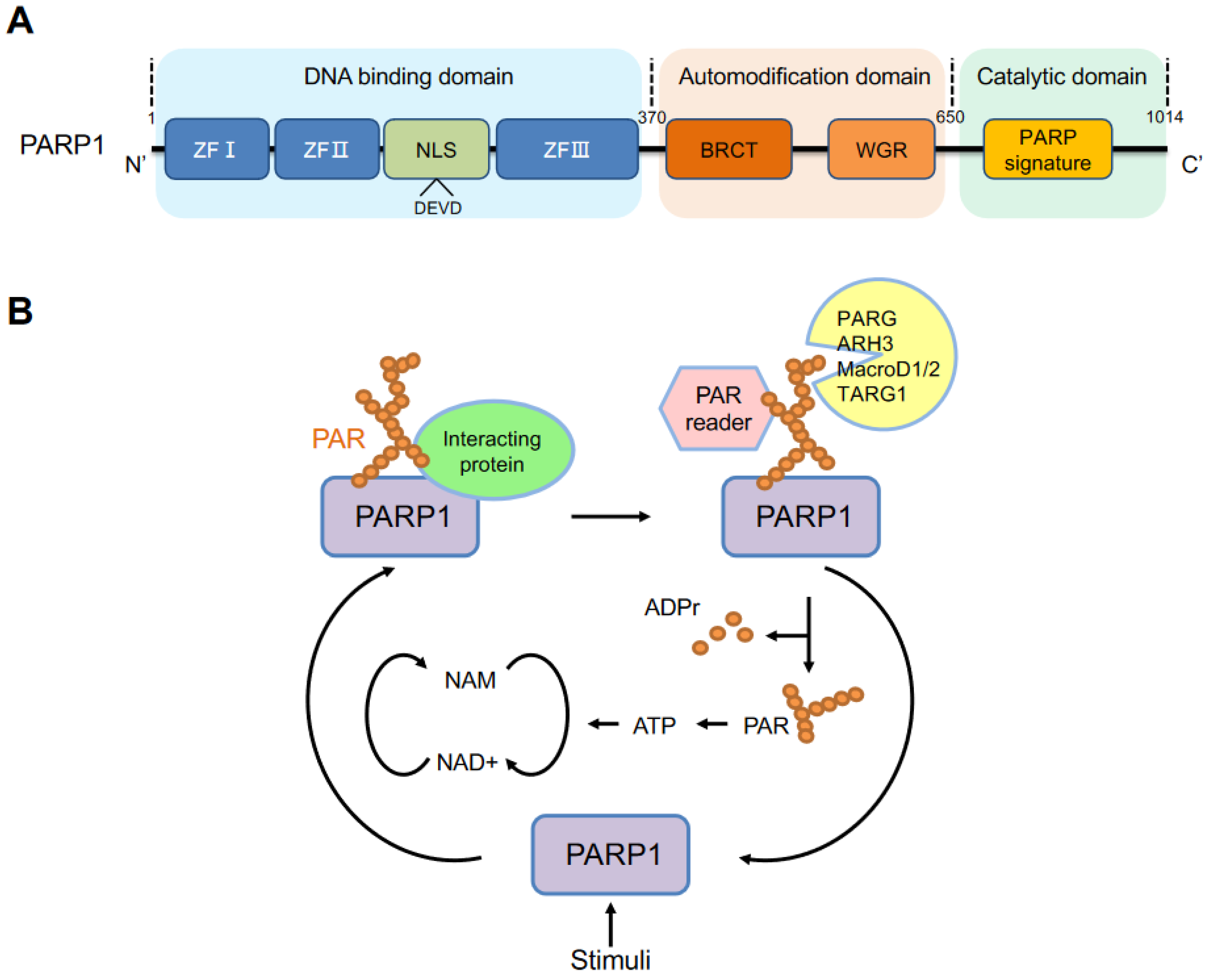
2. PARP1 Structure and Activation
3. PARP1 and Chromatin Remodeling
3.1. Linker Histone H1
3.2. Core Histones and Other Histone Variants
4. Chromatin Remodeling Complexes
5. PARP1 as a Modulator of DNA Methylation
6. PARP1 and PARylation Regulate Gene Transcription
6.1. PARP1 in Inflammatory Response
6.2. Embryonic Development and Cell Differentiation
6.3. Other Cellular Processes
7. Perspectives
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Luscher, B.; Ahel, I.; Altmeyer, M.; Ashworth, A.; Bai, P.; Chang, P.; Cohen, M.; Corda, D.; Dantzer, F.; Daugherty, M.D.; et al. ADP-ribosyltransferases, an update on function and nomenclature. FEBS J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Vyas, S.; Matic, I.; Uchima, L.; Rood, J.; Zaja, R.; Hay, R.T.; Ahel, I.; Chang, P. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat. Commun. 2014, 5, 4426. [Google Scholar] [CrossRef] [PubMed]
- Karlberg, T.; Klepsch, M.; Thorsell, A.G.; Andersson, C.D.; Linusson, A.; Schuler, H. Structural basis for lack of ADP-ribosyltransferase activity in poly(ADP-ribose) polymerase-13/zinc finger antiviral protein. J. Biol. Chem. 2015, 290, 7336–7344. [Google Scholar] [CrossRef] [PubMed]
- Rack, J.G.M.; Palazzo, L.; Ahel, I. (ADP-ribosyl) hydrolases: Structure, function, and biology. Genes Dev. 2020, 34, 263–284. [Google Scholar] [CrossRef]
- Kamaletdinova, T.; Fanaei-Kahrani, Z.; Wang, Z.Q. The Enigmatic Function of PARP1: From PARylation Activity to PAR Readers. Cells 2019, 8, 1625. [Google Scholar] [CrossRef]
- Shieh, W.M.; Ame, J.C.; Wilson, M.V.; Wang, Z.Q.; Koh, D.W.; Jacobson, M.K.; Jacobson, E.L. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Biol. Chem. 1998, 273, 30069–30072. [Google Scholar] [CrossRef]
- Ray Chaudhuri, A.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef]
- Wei, H.; Yu, X. Functions of PARylation in DNA Damage Repair Pathways. Genom. Proteom. Bioinform. 2016, 14, 131–139. [Google Scholar] [CrossRef]
- Posavec Marjanović, M.; Crawford, K.; Ahel, I. PARP, transcription and chromatin modeling. Semin. Cell Dev. Biol. 2017, 63, 102–113. [Google Scholar] [CrossRef]
- Ciccarone, F.; Zampieri, M.; Caiafa, P. PARP1 orchestrates epigenetic events setting up chromatin domains. Semin. Cell Dev. Biol. 2017, 63, 123–134. [Google Scholar] [CrossRef]
- Alkhatib, H.M.; Chen, D.F.; Cherney, B.; Bhatia, K.; Notario, V.; Giri, C.; Stein, G.; Slattery, E.; Roeder, R.G.; Smulson, M.E. Cloning and expression of cDNA for human poly(ADP-ribose). Proc. Natl. Acad. Sci. USA 1987, 84, 1224–1228. [Google Scholar] [CrossRef] [PubMed]
- Uchida, K.; Morita, T.; Sato, T.; Ogura, T.; Yamashita, R.; Noguchi, S.; Suzuki, H.; Nyunoya, H.; Miwa, M.; Sugimura, T. Nucleotide sequence of a full-length cDNA for human fibroblast poly(ADP-ribose) polymerase. Biochem. Biophys. Res. Commun. 1987, 148, 617–622. [Google Scholar] [CrossRef]
- Kurosaki, T.; Ushiro, H.; Mitsuuchi, Y.; Suzuki, S.; Matsuda, M.; Matsuda, Y.; Katunuma, N.; Kangawa, K.; Matsuo, H.; Hirose, T. Primary structure of human poly(ADP-ribose) synthetase as deduced from cDNA sequence. J. Biol. Chem. 1987, 262, 15990–15997. [Google Scholar] [CrossRef]
- Baumgartner, M.; Schneider, R.; Auer, B.; Herzog, H.; Schweiger, M.; Hirsch-Kauffmann, M. Fluorescence in situ mapping of the human nuclear NAD+ ADP-ribosyltransferase gene (ADPRT) and two secondary sites to human chromosomal bands 1q42, 13q34, and 14q24. Cytogenet. Cell Genet. 1992, 61, 172–174. [Google Scholar] [CrossRef] [PubMed]
- Perina, D.; Mikoc, A.; Ahel, J.; Cetkovic, H.; Zaja, R.; Ahel, I. Distribution of protein poly(ADP-ribosyl)ation systems across all domains of life. DNA Repair 2014, 23, 4–16. [Google Scholar] [CrossRef]
- Ikejima, M.; Noguchi, S.; Yamashita, R.; Ogura, T.; Sugimura, T.; Gill, D.M.; Miwa, M. The zinc fingers of human poly(ADP-ribose) polymerase are differentially required for the recognition of DNA breaks and nicks and the consequent enzyme activation. Other structures recognize intact DNA. J. Biol. Chem. 1990, 265, 21907–21913. [Google Scholar] [CrossRef]
- Gradwohl, G.; Ménissier de Murcia, J.M.; Molinete, M.; Simonin, F.; Koken, M.; Hoeijmakers, J.H.; de Murcia, G. The second zinc-finger domain of poly(ADP-ribose) polymerase determines specificity for single-stranded breaks in DNA. Proc. Natl. Acad. Sci. USA 1990, 87, 2990–2994. [Google Scholar] [CrossRef]
- Langelier, M.F.; Planck, J.L.; Roy, S.; Pascal, J.M. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science 2012, 336, 728–732. [Google Scholar] [CrossRef]
- Spiegel, J.O.; Van Houten, B.; Durrant, J.D. PARP1: Structural insights and pharmacological targets for inhibition. DNA Repair 2021, 103, 103125. [Google Scholar] [CrossRef]
- Rudolph, J.; Muthurajan, U.M.; Palacio, M.; Mahadevan, J.; Roberts, G.; Erbse, A.H.; Dyer, P.N.; Luger, K. The BRCT domain of PARP1 binds intact DNA and mediates intrastrand transfer. Mol. Cell 2021, 81, 4994–5006.e5. [Google Scholar] [CrossRef]
- Palazzo, L.; Leidecker, O.; Prokhorova, E.; Dauben, H.; Matic, I.; Ahel, I. Serine is the major residue for ADP-ribosylation upon DNA damage. Elife 2018, 7, e34334. [Google Scholar] [CrossRef] [PubMed]
- Prokhorova, E.; Zobel, F.; Smith, R.; Zentout, S.; Gibbs-Seymour, I.; Schützenhofer, K.; Peters, A.; Groslambert, J.; Zorzini, V.; Agnew, T.; et al. Serine-linked PARP1 auto-modification controls PARP inhibitor response. Nat. Commun. 2021, 12, 4055. [Google Scholar] [CrossRef] [PubMed]
- Ruf, A.; Mennissier de Murcia, J.; de Murcia, G.; Schulz, G.E. Structure of the catalytic fragment of poly(AD-ribose) polymerase from chicken. Proc. Natl. Acad. Sci. USA 1996, 93, 7481–7485. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Armon, M.; Visochek, L.; Rozensal, D.; Kalal, A.; Geistrikh, I.; Klein, R.; Bendetz-Nezer, S.; Yao, Z.; Seger, R. DNA-Independent PARP-1 Activation by Phosphorylated ERK2 Increases Elk1 Activity: A Link to Histone Acetylation. Mol. Cell 2007, 25, 297–308. [Google Scholar] [CrossRef]
- Hau, A.C.; Grebbin, B.M.; Agoston, Z.; Anders-Maurer, M.; Müller, T.; Groß, A.; Kolb, J.; Langer, J.D.; Döring, C.; Schulte, D. MEIS homeodomain proteins facilitate PARP1/ARTD1-mediated eviction of histone H1. J. Cell Biol. 2017, 216, 2715–2729. [Google Scholar] [CrossRef]
- Thomas, C.J.; Kotova, E.; Andrake, M.; Adolf-Bryfogle, J.; Glaser, R.; Regnard, C.; Tulin, A.V. Kinase-Mediated Changes in Nucleosome Conformation Trigger Chromatin Decondensation via Poly(ADP-Ribosyl)ation. Mol. Cell 2014, 53, 831–842. [Google Scholar] [CrossRef]
- Ummarino, S.; Hausman, C.; Gaggi, G.; Rinaldi, L.; Bassal, M.A.; Zhang, Y.; Seelam, A.J.; Kobayashi, I.S.; Borchiellini, M.; Ebralidze, A.K.; et al. NAD Modulates DNA Methylation and Cell Differentiation. Cells 2021, 10, 2986. [Google Scholar] [CrossRef]
- Brochu, G.; Duchaine, C.; Thibeault, L.; Lagueux, J.; Shah, G.M.; Poirier, G.G. Mode of action of poly (ADP-ribose) glycohydrolase. Biochim. Biophys. Acta 1994, 1219, 342–350. [Google Scholar] [CrossRef]
- Oka, S.; Kato, J.; Moss, J. Identification and Characterization of a Mammalian 39-kDa Poly(ADP-ribose) Glycohydrolase. J. Biol. Chem. 2006, 281, 705–713. [Google Scholar] [CrossRef]
- Jankevicius, G.; Hassler, M.; Golia, B.; Rybin, V.; Zacharias, M.; Timinszky, G.; Ladurner, A.G. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat. Struct. Mol. Biol. 2013, 20, 508–514. [Google Scholar] [CrossRef]
- Sharifi, R.; Morra, R.; Appel, C.D.; Tallis, M.; Chioza, B.; Jankevicius, G.; Simpson, M.A.; Matic, I.; Ozkan, E.; Golia, B.; et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013, 32, 1225–1237. [Google Scholar] [CrossRef] [PubMed]
- Slade, D.; Dunstan, M.S.; Barkauskaite, E.; Weston, R.; Lafite, P.; Dixon, N.; Ahel, M.; Leys, D.; Ahel, I. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature 2011, 477, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Kasamatsu, A.; Oka, S.; Moss, J. The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases. Proc. Natl. Acad. Sci. USA 2006, 103, 16687–16691. [Google Scholar] [CrossRef]
- Peterson, F.C.; Chen, D.; Lytle, B.L.; Rossi, M.N.; Ahel, I.; Denu, J.M.; Volkman, B.F. Orphan macrodomain protein (human C6orf130) is an O-acyl-ADP-ribose deacylase: Solution structure and catalytic properties. J. Biol. Chem. 2011, 286, 35955–35965. [Google Scholar] [CrossRef] [PubMed]
- Maruta, H.; Okita, N.; Takasawa, R.; Uchiumi, F.; Hatano, T.; Tanuma, S. The involvement of ATP produced via (ADP-Ribose)n in the maintenance of DNA replication apparatus during DNA repair. Biol. Pharm. Bull. 2007, 30, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Morrison, A.J. Chromatin-remodeling links metabolic signaling to gene expression. Mol. Metab. 2020, 38, 100973. [Google Scholar] [CrossRef] [PubMed]
- Bintu, L.; Ishibashi, T.; Dangkulwanich, M.; Wu, Y.Y.; Lubkowska, L.; Kashlev, M.; Bustamante, C. Nucleosomal elements that control the topography of the barrier to transcription. Cell 2012, 151, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Reyes, A.A.; Marcum, R.D.; He, Y. Structure and Function of Chromatin Remodelers. J. Mol. Biol. 2021, 433, 166929. [Google Scholar] [CrossRef]
- Morgan, M.A.J.; Shilatifard, A. Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat. Genet. 2020, 52, 1271–1281. [Google Scholar] [CrossRef]
- Otake, H.; Miwa, M.; Fujimura, S.; Sugimura, T. Binding of ADP-ribose polymer with histone. J. Biochem. 1969, 65, 145–146. [Google Scholar]
- Ueda, K.; Reeder, R.H.; Honjo, T.; Nishizuka, Y.; Hayaishi, O. Poly adenosine diphosphate ribose synthesis associated with chromatin. Biochem. Biophys. Res. Commun. 1968, 31, 379–385. [Google Scholar] [CrossRef]
- Poirier, G.G.; de Murcia, G.; Jongstra-Bilen, J.; Niedergang, C.; Mandel, P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl. Acad. Sci. USA 1982, 79, 3423–3427. [Google Scholar] [CrossRef] [PubMed]
- Huletsky, A.; de Murcia, G.; Muller, S.; Hengartner, M.; Ménard, L.; Lamarre, D.; Poirier, G.G. The effect of poly(ADP-ribosyl)ation on native and H1-depleted chromatin. A role of poly(ADP-ribosyl)ation on core nucleosome structure. J. Biol. Chem. 1989, 264, 8878–8886. [Google Scholar] [CrossRef]
- Tulin, A.; Spradling, A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science 2003, 299, 560–562. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.Y.; Mauro, S.; Gevry, N.; Lis, J.T.; Kraus, W.L. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 2004, 119, 803–814. [Google Scholar] [CrossRef]
- Krishnakumar, R.; Gamble, M.J.; Frizzell, K.M.; Berrocal, J.G.; Kininis, M.; Kraus, W.L. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 2008, 319, 819–821. [Google Scholar] [CrossRef]
- Shan, L.; Li, X.; Liu, L.; Ding, X.; Wang, Q.; Zheng, Y.; Duan, Y.; Xuan, C.; Wang, Y.; Yang, F.; et al. GATA3 cooperates with PARP1 to regulate CCND1 transcription through modulating histone H1 incorporation. Oncogene 2014, 33, 3205–3216. [Google Scholar] [CrossRef]
- Fontán-Lozano, A.; Suárez-Pereira, I.; Horrillo, A.; del-Pozo-Martín, Y.; Hmadcha, A.; Carrión, A.M. Histone H1 poly[ADP]-ribosylation regulates the chromatin alterations required for learning consolidation. J. Neurosci. 2010, 30, 13305–13313. [Google Scholar] [CrossRef]
- Wright, R.H.; Castellano, G.; Bonet, J.; Le Dily, F.; Font-Mateu, J.; Ballaré, C.; Nacht, A.S.; Soronellas, D.; Oliva, B.; Beato, M. CDK2-dependent activation of PARP-1 is required for hormonal gene regulation in breast cancer cells. Genes Dev. 2012, 26, 1972–1983. [Google Scholar] [CrossRef]
- Azad, G.K.; Ito, K.; Sailaja, B.S.; Biran, A.; Nissim-Rafinia, M.; Yamada, Y.; Brown, D.T.; Takizawa, T.; Meshorer, E. PARP1-dependent eviction of the linker histone H1 mediates immediate early gene expression during neuronal activation. J. Cell Biol. 2018, 217, 473–481. [Google Scholar] [CrossRef]
- Kaiser, A.; Kruger, T.; Eiselt, G.; Bechler, J.; Kniemeyer, O.; Huber, O.; Schmidt, M. Identification of PARP-1, Histone H1 and SIRT-1 as New Regulators of Breast Cancer-Related Aromatase Promoter I.3/II. Cells 2020, 9, 427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maluchenko, N.V.; Nilov, D.K.; Pushkarev, S.V.; Kotova, E.Y.; Gerasimova, N.S.; Kirpichnikov, M.P.; Langelier, M.F.; Pascal, J.M.; Akhtar, M.S.; Feofanov, A.V.; et al. Mechanisms of Nucleosome Reorganization by PARP1. Int. J. Mol. Sci. 2021, 22, 12127. [Google Scholar] [CrossRef] [PubMed]
- Bell, N.A.W.; Haynes, P.J.; Brunner, K.; de Oliveira, T.M.; Flocco, M.M.; Hoogenboom, B.W.; Molloy, J.E. Single-molecule measurements reveal that PARP1 condenses DNA by loop stabilization. Sci. Adv. 2021, 7, 33. [Google Scholar] [CrossRef]
- Altmeyer, M.; Messner, S.; Hassa, P.O.; Fey, M.; Hottiger, M.O. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009, 37, 3723–3738. [Google Scholar] [CrossRef]
- Messner, S.; Altmeyer, M.; Zhao, H.; Pozivil, A.; Roschitzki, B.; Gehrig, P.; Rutishauser, D.; Huang, D.; Caflisch, A.; Hottiger, M.O. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010, 38, 6350–6362. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Zamudio, R.; Ha, H.C. Histone ADP-ribosylation facilitates gene transcription by directly remodeling nucleosomes. Mol. Cell. Biol. 2012, 32, 2490–2502. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Camacho, C.V.; Setlem, R.; Ryu, K.W.; Parameswaran, B.; Gupta, R.K.; Kraus, W.L. Functional Interplay between Histone H2B ADP-Ribosylation and Phosphorylation Controls Adipogenesis. Mol. Cell 2020, 79, 934–949.e14. [Google Scholar] [CrossRef]
- Wang, C.; Xiao, J.; Nowak, K.; Gunasekera, K.; Alippe, Y.; Speckman, S.; Yang, T.; Kress, D.; Abu-Amer, Y.; Hottiger, M.O.; et al. PARP1 Hinders Histone H2B Occupancy at the NFATc1 Promoter to Restrain Osteoclast Differentiation. J. Bone Miner. Res. 2020, 35, 776–788. [Google Scholar] [CrossRef]
- Thomas, C.; Ji, Y.; Wu, C.; Datz, H.; Boyle, C.; MacLeod, B.; Patel, S.; Ampofo, M.; Currie, M.; Harbin, J.; et al. Hit and run versus long-term activation of PARP-1 by its different domains fine-tunes nuclear processes. Proc. Natl. Acad. Sci. USA 2019, 116, 9941–9946. [Google Scholar] [CrossRef]
- Kotova, E.; Lodhi, N.; Jarnik, M.; Pinnola, A.D.; Ji, Y.; Tulin, A.V. Drosophila histone H2A variant (H2Av) controls poly(ADP-ribose) polymerase 1 (PARP1) activation in chromatin. Proc. Natl. Acad. Sci. USA 2011, 108, 6205–6210. [Google Scholar] [CrossRef]
- Nusinow, D.A.; Hernández-Muñoz, I.; Fazzio, T.G.; Shah, G.M.; Kraus, W.L.; Panning, B. Poly(ADP-ribose) Polymerase 1 Is Inhibited by a Histone H2A Variant, MacroH2A, and Contributes to Silencing of the Inactive X Chromosome. J. Biol. Chem. 2007, 282, 12851–12859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnakumar, R.; Kraus, W.L. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell 2010, 39, 736–749. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, A.; Yang, S.H.; Sharrocks, A.D. PARP1 orchestrates variant histone exchange in signal-mediated transcriptional activation. EMBO Rep. 2013, 14, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ruiz, P.D.; Novikov, L.; Casill, A.D.; Park, J.W.; Gamble, M.J. MacroH2A1.1 and PARP-1 cooperate to regulate transcription by promoting CBP-mediated H2B acetylation. Nat. Struct. Mol. Biol. 2014, 21, 981–989. [Google Scholar] [CrossRef]
- Sutcu, H.H.; Matta, E.; Ishchenko, A.A. Role of PARP-catalyzed ADP-ribosylation in the Crosstalk between DNA Strand Breaks and Epigenetic Regulation. J. Mol. Biol. 2019, 432, 1769–1791. [Google Scholar] [CrossRef]
- Gibbs-Seymour, I.; Fontana, P.; Rack, J.G.M.; Ahel, I. HPF1/C4orf27 Is a PARP-1-Interacting Protein that Regulates PARP-1 ADP-Ribosylation Activity. Mol. Cell 2016, 62, 432–442. [Google Scholar] [CrossRef]
- Suskiewicz, M.J.; Zobel, F.; Ogden, T.E.H.; Fontana, P.; Ariza, A.; Yang, J.C.; Zhu, K.; Bracken, L.; Hawthorne, W.J.; Ahel, D.; et al. HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation. Nature 2020, 579, 598–602. [Google Scholar] [CrossRef]
- Sun, F.H.; Zhao, P.; Zhang, N.; Kong, L.L.; Wong, C.C.L.; Yun, C.H. HPF1 remodels the active site of PARP1 to enable the serine ADP-ribosylation of histones. Nat. Commun. 2021, 12, 1028. [Google Scholar] [CrossRef]
- Hendriks, I.A.; Buch-Larsen, S.C.; Prokhorova, E.; Elsborg, J.D.; Rebak, A.; Zhu, K.; Ahel, D.; Lukas, C.; Ahel, I.; Nielsen, M.L. The regulatory landscape of the human HPF1- and ARH3-dependent ADP-ribosylome. Nat. Commun. 2021, 12, 5893. [Google Scholar] [CrossRef]
- LaFargue, C.J.; Dal Molin, G.Z.; Sood, A.K.; Coleman, R.L. Exploring and comparing adverse events between PARP inhibitors. Lancet Oncol. 2019, 20, e15–e28. [Google Scholar] [CrossRef]
- Curtin, N.J.; Szabo, C. Poly(ADP-ribose) polymerase inhibition: Past, present and future. Nat. Rev. Drug Discov. 2020, 19, 711–736. [Google Scholar] [CrossRef] [PubMed]
- Andronikou, C.; Rottenberg, S. Studying PAR-Dependent Chromatin Remodeling to Tackle PARPi Resistance. Trends Mol. Med. 2021, 27, 630–642. [Google Scholar] [CrossRef]
- Timinszky, G.; Till, S.; Hassa, P.O.; Hothorn, M.; Kustatscher, G.; Nijmeijer, B.; Colombelli, J.; Altmeyer, M.; Stelzer, E.H.; Scheffzek, K.; et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009, 16, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, P.D.; Hamilton, G.A.; Park, J.W.; Gamble, M.J. MacroH2A1 Regulation of Poly(ADP-Ribose) Synthesis and Stability Prevents Necrosis and Promotes DNA Repair. Mol. Cell. Biol. 2019, 40, e00230-19. [Google Scholar] [CrossRef] [PubMed]
- Kozlowski, M.; Corujo, D.; Hothorn, M.; Guberovic, I.; Mandemaker, I.K.; Blessing, C.; Sporn, J.; Gutierrez-Triana, A.; Smith, R.; Portmann, T.; et al. MacroH2A histone variants limit chromatin plasticity through two distinct mechanisms. EMBO Rep. 2018, 19, e44445. [Google Scholar] [CrossRef] [PubMed]
- Corujo, D.; Buschbeck, M. Post-Translational Modifications of H2A Histone Variants and Their Role in Cancer. Cancers 2018, 10, 59. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, A.J.; Trivedi, R.D.; Conaway, J.W.; Conaway, R.C. Activation of the SNF2 family ATPase ALC1 by poly(ADP-ribose) in a stable ALC1·PARP1·nucleosome intermediate. J. Biol. Chem. 2012, 287, 43527–43532. [Google Scholar] [CrossRef]
- Ahel, D.; Horejsí, Z.; Wiechens, N.; Polo, S.E.; Garcia-Wilson, E.; Ahel, I.; Flynn, H.; Skehel, M.; West, S.C.; Jackson, S.P.; et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 2009, 325, 1240–1243. [Google Scholar] [CrossRef]
- Mehrotra, P.V.; Ahel, D.; Ryan, D.P.; Weston, R.; Wiechens, N.; Kraehenbuehl, R.; Owen-Hughes, T.; Ahel, I. DNA Repair Factor APLF Is a Histone Chaperone. Mol. Cell 2011, 41, 46–55. [Google Scholar] [CrossRef]
- Moore, S.; Berger, N.D.; Luijsterburg, M.S.; Piett, C.G.; Stanley, F.K.T.; Schrader, C.U.; Fang, S.; Chan, J.A.; Schriemer, D.C.; Nagel, Z.D.; et al. The CHD6 chromatin remodeler is an oxidative DNA damage response factor. Nat. Commun. 2019, 10, 241. [Google Scholar] [CrossRef]
- Fahrer, J.; Popp, O.; Malanga, M.; Beneke, S.; Markovitz, D.M.; Ferrando-May, E.; Burkle, A.; Kappes, F. High-affinity interaction of poly(ADP-ribose) and the human DEK oncoprotein depends upon chain length. Biochemistry 2010, 49, 7119–7130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, A.P.G.; Ryan, D.P.; Galanty, Y.; Low, J.K.K.; Vandevenne, M.; Jackson, S.P.; Mackay, J.P. The N-terminal Region of Chromodomain Helicase DNA-binding Protein 4 (CHD4) Is Essential for Activity and Contains a High Mobility Group (HMG) Box-like-domain That Can Bind Poly(ADP-ribose)*. J. Biol. Chem. 2016, 291, 924–938. [Google Scholar] [CrossRef] [PubMed]
- Chou, D.M.; Adamson, B.; Dephoure, N.E.; Tan, X.; Nottke, A.C.; Hurov, K.E.; Gygi, S.P.; Colaiácovo, M.P.; Elledge, S.J. A chromatin localization screen reveals poly(ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. USA 2010, 107, 18475–18480. [Google Scholar] [CrossRef] [PubMed]
- Hermann, A.; Goyal, R.; Jeltsch, A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 2004, 279, 48350–48359. [Google Scholar] [CrossRef]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef]
- Zardo, G.; D’Erme, M.; Reale, A.; Strom, R.; Perilli, M.; Caiafa, P. Does Poly(ADP-ribosyl)ation Regulate the DNA Methylation Pattern? Biochemistry 1997, 36, 7937–7943. [Google Scholar] [CrossRef]
- Zardo, G.; Caiafa, P. The unmethylated state of CpG islands in mouse fibroblasts depends on the poly(ADP-ribosyl)ation process. J. Biol. Chem. 1998, 273, 16517–16520. [Google Scholar] [CrossRef]
- de Capoa, A.; Febbo, F.R.; Giovannelli, F.; Niveleau, A.; Zardo, G.; Marenzi, S.; Caiafa, P. Reduced levels of poly(ADP-ribosyl)ation result in chromatin compaction and hypermethylation as shown by cell-by-cell computer-assisted quantitative analysis. FASEB J. 1999, 13, 89–93. [Google Scholar] [CrossRef]
- Zampieri, M.; Guastafierro, T.; Calabrese, R.; Ciccarone, F.; Bacalini, M.G.; Reale, A.; Perilli, M.; Passananti, C.; Caiafa, P. ADP-ribose polymers localized on Ctcf-Parp1-Dnmt1 complex prevent methylation of Ctcf target sites. Biochem. J. 2012, 441, 645–652. [Google Scholar] [CrossRef]
- Yu, W.; Ginjala, V.; Pant, V.; Chernukhin, I.; Whitehead, J.; Docquier, F.; Farrar, D.; Tavoosidana, G.; Mukhopadhyay, R.; Kanduri, C.; et al. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat. Genet. 2004, 36, 1105–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guastafierro, T.; Cecchinelli, B.; Zampieri, M.; Reale, A.; Riggio, G.; Sthandier, O.; Zupi, G.; Calabrese, L.; Caiafa, P. CCCTC-binding Factor Activates PARP-1 Affecting DNA Methylation Machinery. J. Biol. Chem. 2008, 283, 21873–21880. [Google Scholar] [CrossRef] [PubMed]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Ciccarone, F.; Klinger, F.G.; Catizone, A.; Calabrese, R.; Zampieri, M.; Bacalini, M.G.; De Felici, M.; Caiafa, P. Poly(ADP-ribosyl)ation acts in the DNA demethylation of mouse primordial germ cells also with DNA damage-independent roles. PLoS ONE 2012, 7, e46927. [Google Scholar] [CrossRef] [PubMed]
- Ciccarone, F.; Valentini, E.; Zampieri, M.; Caiafa, P. 5mC-hydroxylase activity is influenced by the PARylation of TET1 enzyme. Oncotarget 2015, 6, 24333–24347. [Google Scholar] [CrossRef]
- Tolić, A.; Grdović, N.; Dinić, S.; Rajić, J.; Đorđević, M.; Sinadinović, M.; Arambašić Jovanović, J.; Mihailović, M.; Poznanović, G.; Uskoković, A.; et al. Absence of PARP-1 affects Cxcl12 expression by increasing DNA demethylation. J. Cell. Mol. Med. 2019, 23, 2610–2618. [Google Scholar] [CrossRef]
- Kumbhar, R.; Sanchez, A.; Perren, J.; Gong, F.; Corujo, D.; Medina, F.; Devanathan, S.K.; Xhemalce, B.; Matouschek, A.; Buschbeck, M.; et al. Poly(ADP-ribose) binding and macroH2A mediate recruitment and functions of KDM5A at DNA lesions. J. Cell Biol. 2021, 220, e202006149. [Google Scholar] [CrossRef]
- Doege, C.A.; Inoue, K.; Yamashita, T.; Rhee, D.B.; Travis, S.; Fujita, R.; Guarnieri, P.; Bhagat, G.; Vanti, W.B.; Shih, A.; et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 2012, 488, 652–655. [Google Scholar] [CrossRef]
- Zampieri, M.; Bacalini, M.G.; Barchetta, I.; Scalea, S.; Cimini, F.A.; Bertoccini, L.; Tagliatesta, S.; De Matteis, G.; Zardo, G.; Cavallo, M.G.; et al. Increased PARylation impacts the DNA methylation process in type 2 diabetes mellitus. Clin. Epigenetics 2021, 13, 114. [Google Scholar] [CrossRef]
- Osada, T.; Rydén, A.M.; Masutani, M. Poly(ADP-ribosylation) regulates chromatin organization through histone H3 modification and DNA methylation of the first cell cycle of mouse embryos. Biochem. Biophys. Res. Commun. 2013, 434, 15–21. [Google Scholar] [CrossRef]
- Nalabothula, N.; Al-jumaily, T.; Eteleeb, A.M.; Flight, R.M.; Xiaorong, S.; Moseley, H.; Rouchka, E.C.; Fondufe-Mittendorf, Y.N. Genome-Wide Profiling of PARP1 Reveals an Interplay with Gene Regulatory Regions and DNA Methylation. PLoS ONE 2015, 10, e0135410. [Google Scholar] [CrossRef] [PubMed]
- Hassa, P.; Hottiger, M. The functional role of poly(ADP-ribose) polymerase 1 as novel coactivator of NF-κB in inflammatory disorders. Cell Mol. Life Sci. 2002, 59, 1534–1553. [Google Scholar] [CrossRef]
- Ju, B.-G.; Solum, D.; Song, E.J.; Lee, K.-J.; Rose, D.W.; Glass, C.K.; Rosenfeld, M.G. Activating the PARP-1 Sensor Component of the Groucho/TLE1 Corepressor Complex Mediates a CaMKinase IIδ-Dependent Neurogenic Gene Activation Pathway. Cell 2004, 119, 815–829. [Google Scholar] [CrossRef]
- Gao, F.; Kwon, S.W.; Zhao, Y.; Jin, Y. PARP1 Poly(ADP-ribosyl)ates Sox2 to Control Sox2 Protein Levels and FGF4 Expression during Embryonic Stem Cell Differentiation. J. Biol. Chem. 2009, 284, 22263–22273. [Google Scholar] [CrossRef]
- Valdor, R.; Schreiber, V.; Saenz, L.; Martínez, T.; Muñoz-Suano, A.; Dominguez-Villar, M.; Ramírez, P.; Parrilla, P.; Aguado, E.; García-Cózar, F. Regulation of NFAT by poly(ADP-ribose) polymerase activity in T cells. Mol. Immunol. 2008, 45, 1863–1871. [Google Scholar] [CrossRef] [PubMed]
- Choudhuri, S.; Garg, N.J. Trypanosoma cruzi Induces the PARP1/AP-1 Pathway for Upregulation of Metalloproteinases and Transforming Growth Factor β in Macrophages: Role in Cardiac Fibroblast Differentiation and Fibrosis in Chagas Disease. mBio 2020, 11, e01853-20. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Zhang, G. The role of PARP1 in neurodegenerative diseases and aging. FEBS J. 2021, 289, 2013–2024. [Google Scholar] [CrossRef] [PubMed]
- Rosado, M.M.; Bennici, E.; Novelli, F.; Pioli, C. Beyond DNA repair, the immunological role of PARP-1 and its siblings. Immunology 2013, 139, 428–437. [Google Scholar] [CrossRef]
- Welsby, I.; Hutin, D.; Leo, O. Complex roles of members of the ADP-ribosyl transferase super family in immune defences: Looking beyond PARP1. Biochem. Pharmacol. 2012, 84, 11–20. [Google Scholar] [CrossRef]
- Oliver, F.J.; Menissier-de Murcia, J.; Nacci, C.; Decker, P.; Andriantsitohaina, R.; Muller, S.; de la Rubia, G.; Stoclet, J.C.; de Murcia, G. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly(ADP-ribose) polymerase-1 deficient mice. Embo J. 1999, 18, 4446–4454. [Google Scholar] [CrossRef]
- Pacher, P.; Szabó, C. Role of poly(ADP-ribose) polymerase 1 (PARP-1) in cardiovascular diseases: The therapeutic potential of PARP inhibitors. Cardiovasc. Drug Rev. 2007, 25, 235–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassa, P.O.; Covic, M.; Hasan, S.; Imhof, R.; Hottiger, M.O. The Enzymatic and DNA Binding Activity of PARP-1 Are Not Required for NF-κB Coactivator Function. J. Biol. Chem. 2001, 276, 45588–45597. [Google Scholar] [CrossRef] [PubMed]
- Hassa, P.O.; Buerki, C.; Lombardi, C.; Imhof, R.; Hottiger, M.O. Transcriptional coactivation of nuclear factor-kappaB-dependent gene expression by p300 is regulated by poly(ADP)-ribose polymerase-1. J. Biol. Chem. 2003, 278, 45145–45153. [Google Scholar] [CrossRef]
- Hassa, P.O.; Haenni, S.S.; Buerki, C.; Meier, N.I.; Lane, W.S.; Owen, H.; Gersbach, M.; Imhof, R.; Hottiger, M.O. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J. Biol. Chem. 2005, 280, 40450–40464. [Google Scholar] [CrossRef]
- Kameoka, M.; Ota, K.; Tetsuka, T.; Tanaka, Y.; Itaya, A.; Okamoto, T.; Yoshihara, K. Evidence for regulation of NF-kappaB by poly(ADP-ribose) polymerase. Biochem. J. 2000, 346, 641–649. [Google Scholar] [CrossRef]
- Liu, L.; Ke, Y.; Jiang, X.; He, F.; Pan, L.; Xu, L.; Zeng, X.; Ba, X. Lipopolysaccharide activates ERK-PARP-1-RelA pathway and promotes nuclear factor-κB transcription in murine macrophages. Hum. Immunol. 2012, 73, 439–447. [Google Scholar] [CrossRef]
- Chang, W.-J.; Alvarez-Gonzalez, R. The Sequence-specific DNA Binding of NF-κB Is Reversibly Regulated by the Automodification Reaction of Poly(ADP-ribose) Polymerase 1. J. Biol. Chem. 2001, 276, 47664–47670. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, H.; Nagaso, H.; Kakui, N.; Ishikawa, M.; Hiranuma, T.; Hoshiko, S. Critical role of the automodification of poly(ADP-ribose) polymerase-1 in nuclear factor-kappaB-dependent gene expression in primary cultured mouse glial cells. J. Biol. Chem. 2004, 279, 42774–42786. [Google Scholar] [CrossRef] [PubMed]
- Zerfaoui, M.; Errami, Y.; Naura, A.S.; Suzuki, Y.; Kim, H.; Ju, J.; Liu, T.; Hans, C.P.; Kim, J.G.; Abd Elmageed, Z.Y.; et al. Poly(ADP-ribose) polymerase-1 is a determining factor in Crm1-mediated nuclear export and retention of p65 NF-kappa B upon TLR4 stimulation. J. Immunol. 2010, 185, 1894–1902. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Bai, L.; Zhou, W.; Yang, Y.; Zhang, J.; Li, L.; Jiang, M.; Mi, Y.; Li, T.T.; Zhang, X.; et al. PARP-1-regulated TNF-α expression in the dorsal root ganglia and spinal dorsal horn contributes to the pathogenesis of neuropathic pain in rats. Brain Behav. Immun. 2020, 88, 482–496. [Google Scholar] [CrossRef]
- Petrilli, V.; Herceg, Z.; Hassa, P.O.; Patel, N.S.; Di Paola, R.; Cortes, U.; Dugo, L.; Filipe, H.M.; Thiemermann, C.; Hottiger, M.O.; et al. Noncleavable poly(ADP-ribose) polymerase-1 regulates the inflammation response in mice. J. Clin. Investig. 2004, 114, 1072–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohio, A.A.; Sattout, A.; Wang, R.; Wang, K.; Sah, R.K.; Guo, X.; Zeng, X.; Ke, Y.; Boldogh, I.; Ba, X. c-Abl-Mediated Tyrosine Phosphorylation of PARP1 Is Crucial for Expression of Proinflammatory Genes. J. Immunol. 2019, 203, 1521–1531. [Google Scholar] [CrossRef] [PubMed]
- Zingarelli, B.; Hake, P.W.; Burroughs, T.J.; Piraino, G.; O’Connor, M.; Denenberg, A. Activator protein-1 signalling pathway and apoptosis are modulated by poly(ADP-ribose) polymerase-1 in experimental colitis. Immunology 2004, 113, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Olabisi, O.A.; Soto-Nieves, N.; Nieves, E.; Yang, T.T.C.; Yang, X.; Yu, R.Y.L.; Suk, H.Y.; Macian, F.; Chow, C.-W. Regulation of Transcription Factor NFAT by ADP-Ribosylation. Mol. Cell. Biol. 2008, 28, 2860–2871. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Chen, X.; Xu, X.; Qian, Y.; Liang, G.; Yao, F.; Yao, Z.; Wu, H.; Zhang, J.; He, Q.; et al. PARP1 Suppresses the Transcription of PD-L1 by Poly(ADP-Ribosyl)ating STAT3. Cancer Immunol. Res. 2019, 7, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Xia, W.; Yamaguchi, H.; Wei, Y.; Chen, M.K.; Hsu, J.M.; Hsu, J.L.; Yu, W.H.; Du, Y.; Lee, H.H.; et al. PARP Inhibitor Upregulates PD-L1 Expression and Enhances Cancer-Associated Immunosuppression. Clin. Cancer Res. 2017, 23, 3711–3720. [Google Scholar] [CrossRef] [PubMed]
- Menissier de Murcia, J.; Ricoul, M.; Tartier, L.; Niedergang, C.; Huber, A.; Dantzer, F.; Schreiber, V.; Ame, J.C.; Dierich, A.; LeMeur, M.; et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. Embo J. 2003, 22, 2255–2263. [Google Scholar] [CrossRef]
- Liu, Z.; Kraus, W.L. Catalytic-independent functions of PARP-1 determine Sox2 pioneer activity at intractable genomic loci. Mol. Cell 2017, 65, 589–603.e9. [Google Scholar] [CrossRef]
- Lönn, P.; van der Heide, L.P.; Dahl, M.; Hellman, U.; Heldin, C.-H.; Moustakas, A. PARP-1 Attenuates Smad-Mediated Transcription. Mol. Cell 2010, 40, 521–532. [Google Scholar] [CrossRef]
- Luo, X.; Ryu, K.W.; Kim, D.S.; Nandu, T.; Medina, C.J.; Gupte, R.; Gibson, B.A.; Soccio, R.E.; Yu, Y.; Gupta, R.K.; et al. PARP-1 Controls the Adipogenic Transcriptional Program by PARylating C/EBPβ and Modulating Its Transcriptional Activity. Mol. Cell 2017, 65, 260–271. [Google Scholar] [CrossRef]
- Simbulan-Rosenthal, C.M.; Rosenthal, D.S.; Luo, R.; Samara, R.; Espinoza, L.A.; Hassa, P.O.; Hottiger, M.O.; Smulson, M.E. PARP-1 binds E2F-1 independently of its DNA binding and catalytic domains, and acts as a novel coactivator of E2F-1-mediated transcription during re-entry of quiescent cells into S phase. Oncogene 2003, 22, 8460–8471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakamaki, J.; Daitoku, H.; Yoshimochi, K.; Miwa, M.; Fukamizu, A. Regulation of FOXO1-mediated transcription and cell proliferation by PARP-1. Biochem. Biophys. Res. Commun. 2009, 382, 497–502. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, Y.; Wang, L.; Luo, X.; Huang, K.; Wang, C.; Du, M.; Liu, F.; Luo, T.; Huang, D.; et al. Poly(ADP-ribose) Polymerase 1 Is a Key Regulator of Estrogen Receptor α-dependent Gene Transcription. J. Biol. Chem. 2013, 288, 11348–11357. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Wang, X.-J.; Tian, W.; Jaramillo, M.C.; Lau, A.; Zhang, D.D. Poly(ADP-ribose) polymerase-1 modulates Nrf2-dependent transcription. Free Radic. Biol. Med. 2014, 67, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Katoh, Y.; Ikura, T.; Hoshikawa, Y.; Tashiro, S.; Ito, T.; Ohta, M.; Kera, Y.; Noda, T.; Igarashi, K. Methionine adenosyltransferase II serves as a transcriptional corepressor of Maf oncoprotein. Mol. Cell 2011, 41, 554–566. [Google Scholar] [CrossRef]
- Peng, H.; Zhu, Q.S.; Zhong, S.; Levy, D. Transcription of the Human Microsomal Epoxide Hydrolase Gene (EPHX1) Is Regulated by PARP-1 and Histone H1.2. Association with Sodium-Dependent Bile Acid Transport. PLoS ONE 2015, 10, e0125318. [Google Scholar] [CrossRef]
- Cardnell, R.J.; Feng, Y.; Mukherjee, S.; Diao, L.; Tong, P.; Stewart, C.A.; Masrorpour, F.; Fan, Y.; Nilsson, M.; Shen, Y.; et al. Activation of the PI3K/mTOR Pathway following PARP Inhibition in Small Cell Lung Cancer. PLoS ONE 2016, 11, e0152584. [Google Scholar] [CrossRef]
- Vikas, P.; Borcherding, N.; Chennamadhavuni, A.; Garje, R. Therapeutic Potential of Combining PARP Inhibitor and Immunotherapy in Solid Tumors. Front. Oncol. 2020, 10, 570. [Google Scholar] [CrossRef]
- Lord, C.J.; Ashworth, A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zong, W.; Gong, Y.; Sun, W.; Li, T.; Wang, Z.-Q. PARP1: Liaison of Chromatin Remodeling and Transcription. Cancers 2022, 14, 4162. https://doi.org/10.3390/cancers14174162
Zong W, Gong Y, Sun W, Li T, Wang Z-Q. PARP1: Liaison of Chromatin Remodeling and Transcription. Cancers. 2022; 14(17):4162. https://doi.org/10.3390/cancers14174162
Chicago/Turabian StyleZong, Wen, Yamin Gong, Wenli Sun, Tangliang Li, and Zhao-Qi Wang. 2022. "PARP1: Liaison of Chromatin Remodeling and Transcription" Cancers 14, no. 17: 4162. https://doi.org/10.3390/cancers14174162
APA StyleZong, W., Gong, Y., Sun, W., Li, T., & Wang, Z.-Q. (2022). PARP1: Liaison of Chromatin Remodeling and Transcription. Cancers, 14(17), 4162. https://doi.org/10.3390/cancers14174162






