Inhibitor of Growth Factors Regulate Cellular Senescence
Abstract
Simple Summary
Abstract
1. Introduction
2. Cell Line Studies and Experiments in Clinical Samples
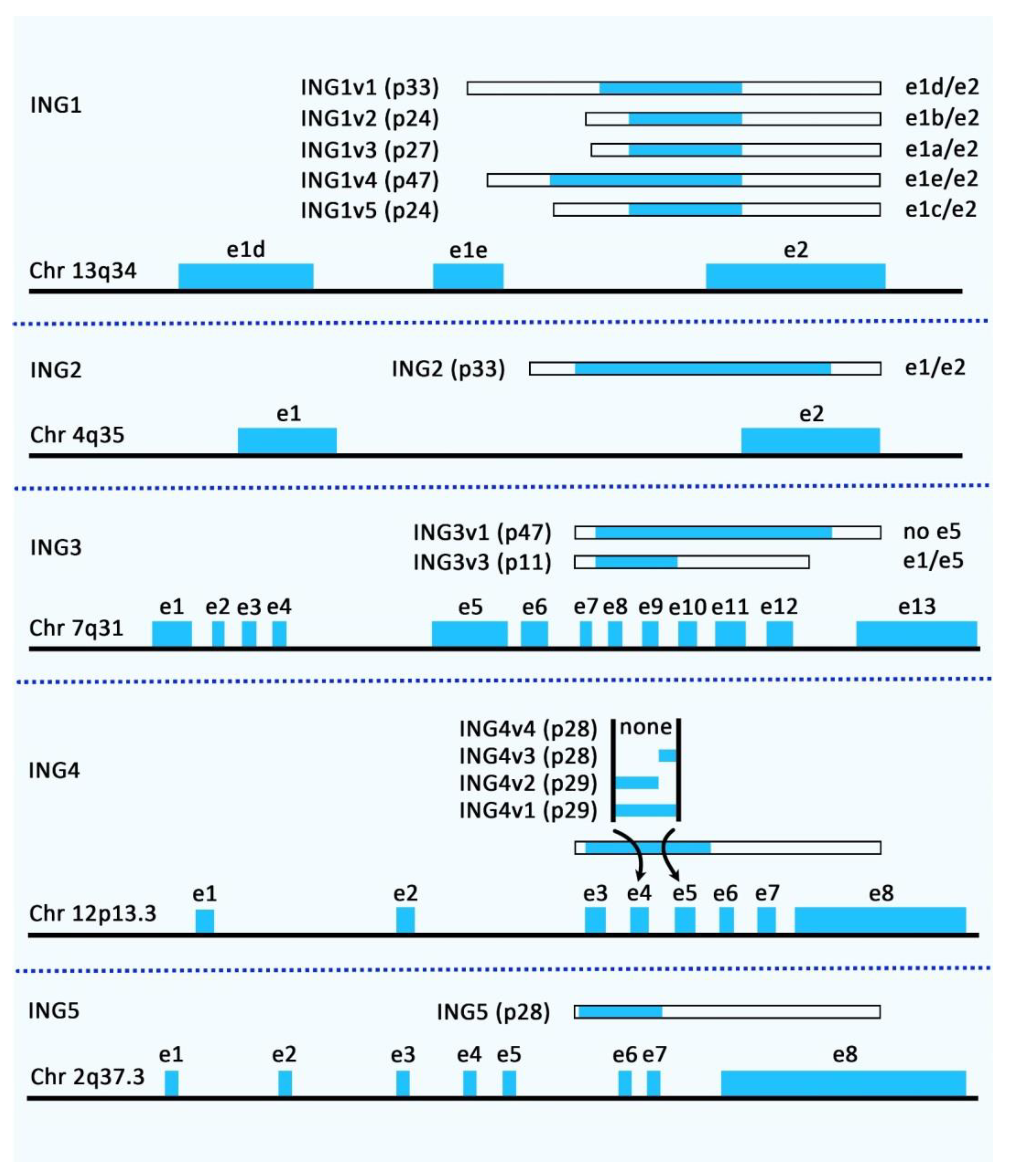
2.1. ING1
2.2. ING2
2.3. ING3
2.4. ING4
2.5. ING5
2.6. Animal Studies
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ludwig, S.; Klitzsch, A.; Baniahmad, A. The ING tumor suppressors in cellular senescence and chromatin. Cell Biosci. 2011, 1, 25. [Google Scholar] [CrossRef] [PubMed]
- Coles, A.H.; Jones, S.N. The ING gene family in the regulation of cell growth and tumorigenesis. J. Cell. Physiol. 2009, 218, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Archambeau, J.; Blondel, A.; Pedeux, R. Focus-ING on DNA Integrity: Implication of ING Proteins in Cell Cycle Regulation and DNA Repair Modulation. Cancers 2019, 12, 58. (In English) [Google Scholar] [CrossRef] [PubMed]
- Doyon, Y.; Cayrou, C.; Ullah, M.; Landry, A.J.; Côté, V.; Selleck, W.; Lane, W.S.; Tan, S.; Yang, X.J.; Côté, J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 2006, 21, 51–64. (In English) [Google Scholar] [CrossRef]
- Campos, E.I.; Chin, M.Y.; Kuo, W.H.; Li, G. Biological functions of the ING family tumor suppressors. Cell. Mol. Life Sci. CMLS 2004, 61, 2597–2613. (In English) [Google Scholar] [CrossRef]
- Soliman, M.A.; Riabowol, K. After a decade of study-ING, a PHD for a versatile family of proteins. Trends Biochem. Sci. 2007, 32, 509–519. (In English) [Google Scholar] [CrossRef]
- Garkavtsev, I.; Kazarov, A.; Gudkov, A.; Riabowol, K. Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat. Genet. 1996, 14, 415–420. (In English) [Google Scholar] [CrossRef]
- Kim, S. HuntING4 new tumor suppressors. Cell Cycle 2005, 4, 516–517. (In English) [Google Scholar] [CrossRef]
- Kim, S.; Chin, K.; Gray, J.W.; Bishop, J.M. A screen for genes that suppress loss of contact inhibition: Identification of ING4 as a candidate tumor suppressor gene in human cancer. Proc. Natl. Acad. Sci. USA 2004, 101, 16251–16256. (In English) [Google Scholar] [CrossRef]
- Unoki, M.; Shen, J.C.; Zheng, Z.M.; Harris, C.C. Novel splice variants of ING4 and their possible roles in the regulation of cell growth and motility. J. Biol. Chem. 2006, 281, 34677–34686. (In English) [Google Scholar] [CrossRef]
- Cheung, K.J., Jr.; Li, G. The tumor suppressor ING1: Structure and function. Exp. Cell Res. 2001, 268, 1–6. (In English) [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Hara, Y.; Riabowol, K. Different HATS of the ING1 gene family. Trends Cell Biol. 2002, 12, 532–538. (In English) [Google Scholar] [CrossRef]
- Shi, X.; Gozani, O. The fellowships of the INGs. J. Cell. Biochem. 2005, 96, 1127–1136. [Google Scholar] [CrossRef] [PubMed]
- He, G.H.; Helbing, C.C.; Wagner, M.J.; Sensen, C.W.; Riabowol, K. Phylogenetic analysis of the ING family of PHD finger proteins. Mol. Biol. Evol. 2005, 22, 104–116. [Google Scholar] [CrossRef]
- Nouman, G.; Anderson, J.; Lunec, J.; Angus, B. The role of the tumour suppressor p33ING1b in human neoplasia. J. Clin. Pathol. 2003, 56, 491–496. [Google Scholar] [CrossRef][Green Version]
- Ythier, D.; Larrieu, D.; Brambilla, C.; Brambilla, E.; Pedeux, R. The new tumor suppressor genes ING: Genomic structure and status in cancer. Int. J. Cancer 2008, 123, 1483–1490. [Google Scholar] [CrossRef]
- Garkavtsev, I.; Riabowol, K. Extension of the replicative life span of human diploid fibroblasts by inhibition of the p33ING1 candidate tumor suppressor. Mol. Cell. Biol. 1997, 17, 2014–2019. [Google Scholar] [CrossRef]
- Goeman, F.; Thormeyer, D.; Abad, M.; Serrano, M.; Schmidt, O.; Palmero, I.; Baniahmad, A. Growth inhibition by the tumor suppressor p33ING1 in immortalized and primary cells: Involvement of two silencing domains and effect of Ras. Mol. Cell. Biol. 2005, 25, 422–431. [Google Scholar] [CrossRef]
- Soliman, M.A.; Berardi, P.; Pastyryeva, S.; Bonnefin, P.; Feng, X.; Colina, A.; Young, D.; Riabowol, K. ING1a expression increases during replicative senescence and induces a senescent phenotype. Aging Cell 2008, 7, 783–794. [Google Scholar] [CrossRef]
- Rajarajacholan, U.K.; Riabowol, K. Aging with ING: A comparative study of different forms of stress induced premature senescence. Oncotarget 2015, 6, 34118. [Google Scholar] [CrossRef]
- Bertschmann, J.; Thalappilly, S.; Riabowol, K. The ING1a model of rapid cell senescence. Mech. Ageing Dev. 2019, 177, 109–117. (In English) [Google Scholar] [CrossRef] [PubMed]
- Scott, M.; Bonnefin, P.; Vieyra, D.; Boisvert, F.-M.; Young, D.; Bazett-Jones, D.P.; Riabowol, K. UV-induced binding of ING1 to PCNA regulates the induction of apoptosis. J. Cell Sci. 2001, 114, 3455–3462. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Russell, M.; Suzuki, K.; Riabowol, K. Subcellular targeting of p33ING1b by phosphorylation-dependent 14-3-3 binding regulates p21WAF1 expression. Mol. Cell. Biol. 2006, 26, 2947–2954. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Garkavtsev, I.; Grigorian, I.A.; Ossovskaya, V.S.; Chernov, M.V.; Chumakov, P.M.; Gudkov, A.V. The candidate tumour suppressor p33ING1cooperates with p53 in cell growth control. Nature 1998, 391, 295–298. [Google Scholar] [CrossRef]
- Bose, P.; Thakur, S.; Thalappilly, S.; Ahn, B.; Satpathy, S.; Feng, X.; Suzuki, K.; Kim, S.; Riabowol, K. ING1 induces apoptosis through direct effects at the mitochondria. Cell Death Dis. 2013, 4, e788. [Google Scholar] [CrossRef]
- Esmaeili, M.; Pungsrinont, T.; Schaefer, A.; Baniahmad, A. A novel crosstalk between the tumor suppressors ING1 and ING2 regulates androgen receptor signaling. J. Mol. Med. 2016, 94, 1167–1179. [Google Scholar] [CrossRef]
- Esmaeili, M.; Jennek, S.; Ludwig, S.; Klitzsch, A.; Kraft, F.; Melle, C.; Baniahmad, A. The tumor suppressor ING1b is a novel corepressor for the androgen receptor and induces cellular senescence in prostate cancer cells. J. Mol. Cell Biol. 2016, 8, 207–220. [Google Scholar] [CrossRef]
- Rajarajacholan, U.K.; Thalappilly, S.; Riabowol, K. The ING1a tumor suppressor regulates endocytosis to induce cellular senescence via the Rb-E2F pathway. PLoS Biol. 2013, 11, e1001502. [Google Scholar] [CrossRef]
- Bartsch, S.; Mirzakhani, K.; Neubert, L.; Stenzel, A.; Ehsani, M.; Esmaeili, M.; Pungsrinont, T.; Kacal, M.; Rasa, S.M.M.; Kallenbach, J.; et al. Antithetic hTERT Regulation by Androgens in Prostate Cancer Cells: hTERT Inhibition Is Mediated by the ING1 and ING2 Tumor Suppressors. Cancers 2021, 13, 4025. (In English) [Google Scholar] [CrossRef]
- Ricordel, C.; Chaillot, L.; Blondel, A.; Archambeau, J.; Jouan, F.; Mouche, A.; Tiercin, M.; Burel, A.; Lena, H.; Desrues, B.; et al. ING2 tumor suppressive protein translocates into mitochondria and is involved in cellular metabolism homeostasis. Oncogene 2021, 40, 4111–4123. (In English) [Google Scholar] [CrossRef]
- An, J.; Chen, Z.; Ma, Q.; Li, Y.; Shi, F. Liraglutide improves atherosclerosis by regulating long non-coding RNA RMRP/miR-128-1-5P/Gadd45g axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2725–2737. [Google Scholar] [PubMed]
- Han, X.-R.; Bai, X.-Z.; Sun, Y.; Yang, Y. Nuclear ING2 expression is reduced in osteosarcoma. Oncol. Rep. 2014, 32, 1967–1972. [Google Scholar] [CrossRef] [PubMed]
- Goeman, F.; Otto, K.; Kyrylenko, S.; Schmidt, O.; Baniahmad, A. ING2 recruits histone methyltransferase activity with methylation site specificity distinct from histone H3 lysines 4 and 9. Biochim. Et Biophys. Acta 2008, 1783, 1673–1680. (In English) [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shi, X.; Hong, T.; Walter, K.L.; Ewalt, M.; Michishita, E.; Hung, T.; Carney, D.; Peña, P.; Lan, F.; Kaadige, M.R.; et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 2006, 442, 96–99. (In English) [Google Scholar] [CrossRef]
- Peña, P.V.; Davrazou, F.; Shi, X.; Walter, K.L.; Verkhusha, V.V.; Gozani, O.; Zhao, R.; Kutateladze, T.G. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 2006, 442, 100–103. (In English) [Google Scholar] [CrossRef]
- Ohkouchi, C.; Kumamoto, K.; Saito, M.; Ishigame, T.; Suzuki, S.-I.; Takenoshita, S.; Harris, C.C. ING2, a tumor associated gene, enhances PAI-1 and HSPA1A expression with HDAC1 and mSin3A through the PHD domain and C-terminal. Mol. Med. Report. 2017, 16, 7367–7374. [Google Scholar] [CrossRef][Green Version]
- Zhou, L.; Wang, P.; Zhang, J.; Heng, B.C.; Tong, G.Q. ING2 (inhibitor of growth protein-2) plays a crucial role in preimplantation development. Zygote 2016, 24, 89–97. (In English) [Google Scholar] [CrossRef]
- Li, X.; Zhang, Q.; Zhang, M.; Luo, Y.; Fu, Y. Downregulation of nuclear ING3 expression and translocalization to cytoplasm promotes tumorigenesis and progression in head and neck squamous cell carcinoma (HNSCC). Histol. Histopathol. 2019, 35, 681–690. [Google Scholar]
- Nabbi, A.; McClurg, U.L.; Thalappilly, S.; Almami, A.; Mobahat, M.; Bismar, T.A.; Binda, O.; Riabowol, K.T. ING3 promotes prostate cancer growth by activating the androgen receptor. BMC Med. 2017, 15, 103. (In English) [Google Scholar] [CrossRef]
- McClurg, U.L.; Nabbi, A.; Ricordel, C.; Korolchuk, S.; McCracken, S.; Heer, R.; Wilson, L.; Butler, L.M.; Irving-Hooper, B.K.; Pedeux, R. Human ex vivo prostate tissue model system identifies ING3 as an oncoprotein. Br. J. Cancer 2018, 118, 713–726. [Google Scholar] [CrossRef]
- Melekhova, A.; Leeder, M.; Pungsrinont, T.; Schmäche, T.; Kallenbach, J.; Ehsani, M.; Mirzakhani, K.; Rasa, S.M.M.; Neri, F.; Baniahmad, A. A Novel Splice Variant of the Inhibitor of Growth 3 Lacks the Plant Homeodomain and Regulates Epithelial-Mesenchymal Transition in Prostate Cancer Cells. Biomolecules 2021, 11, 1152. (In English) [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.; Soleto, I.; García-Sanz, P.; Moreno-Bueno, G.; Palmero, I. ING4 regulates a secretory phenotype in primary fibroblasts with dual effects on cell proliferation and tumor growth. Oncogene 2014, 33, 1945–1953. (In English) [Google Scholar] [CrossRef] [PubMed]
- Trinh, D.-A.; Shirakawa, R.; Kimura, T.; Sakata, N.; Goto, K.; Horiuchi, H. Inhibitor of Growth 4 (ING4) is a positive regulator of rRNA synthesis. Sci. Rep. 2019, 9, 17235. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.-Q.; Zhao, S.; Yang, L.; Zhao, X.; Zhao, G.-F.; Zhao, S.-P.; Li, Z.-J.; Zheng, H.-C. The nucleocytoplasmic translocation and up-regulation of ING5 protein in breast cancer: A potential target for gene therapy. Oncotarget 2017, 8, 81953. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, S.; Zhao, Z.-J.; He, H.-Y.; Wu, J.-C.; Ding, X.-Q.; Yang, L.; Jia, N.; Li, Z.-J.; Zheng, H.-C. The roles of ING5 in gliomas: A good marker for tumorigenesis and a potential target for gene therapy. Oncotarget 2017, 8, 56558. [Google Scholar] [CrossRef][Green Version]
- Zheng, H.-C.; Zhao, S.; Song, Y.; Ding, X.-Q. The roles of ING5 expression in ovarian carcinogenesis and subsequent progression: A target of gene therapy. Oncotarget 2017, 8, 103449. [Google Scholar] [CrossRef][Green Version]
- Qi, L.; Zhang, Y. Truncation of inhibitor of growth family protein 5 effectively induces senescence, but not apoptosis in human tongue squamous cell carcinoma cell line. Tumor Biol. 2014, 35, 3139–3144. [Google Scholar] [CrossRef]
- Zeremski, M.; Hill, J.E.; Kwek, S.S.; Grigorian, I.A.; Gurova, K.V.; Garkavtsev, I.V.; Diatchenko, L.; Koonin, E.V.; Gudkov, A.V. Structure and regulation of the mouse ing1 gene: Three alternative transcripts encode two PHD finger proteins that have opposite effects on p53 function. J. Biol. Chem. 1999, 274, 32172–32181. [Google Scholar] [CrossRef]
- Kichina, J.; Zeremski, M.; Aris, L.; Gurova, K.; Walker, E.; Franks, R.; Nikitin, A.; Kiyokawa, H.; Gudkov, A. Targeted disruption of the mouse ing1 locus results in reduced body size, hypersensitivity to radiation and elevated incidence of lymphomas. Oncogene 2006, 25, 857–866. [Google Scholar] [CrossRef]
- Saito, M.; Kumamoto, K.; Robles, A.I.; Horikawa, I.; Furusato, B.; Okamura, S.; Goto, A.; Yamashita, T.; Nagashima, M.; Lee, T.-L. Targeted disruption of Ing2 results in defective spermatogenesis and development of soft-tissue sarcomas. PLoS ONE 2010, 5, e15541. [Google Scholar] [CrossRef]
- Fink, D.; Yau, T.; Nabbi, A.; Wagner, B.; Wagner, C.; Hu, S.M.; Lang, V.; Handschuh, S.; Riabowol, K.; Rülicke, T. Loss of Ing3 expression results in growth retardation and embryonic death. Cancers 2019, 12, 80. [Google Scholar] [CrossRef] [PubMed]
- Coles, A.H.; Gannon, H.; Cerny, A.; Kurt-Jones, E.; Jones, S.N. Inhibitor of growth-4 promotes IκB promoter activation to suppress NF-κB signaling and innate immunity. Proc. Natl. Acad. Sci. USA 2010, 107, 11423–11428. [Google Scholar] [CrossRef] [PubMed]
- Dantas, A.; Al Shueili, B.; Yang, Y.; Nabbi, A.; Fink, D.; Riabowol, K. Biological Functions of the ING Proteins. Cancers 2019, 11, 1817. (In English) [Google Scholar] [CrossRef]
- Russell, M.; Berardi, P.; Gong, W.; Riabowol, K. Grow-ING, Age-ING and Die-ING: ING proteins link cancer, senescence and apoptosis. Exp. Cell Res. 2006, 312, 951–961. [Google Scholar] [CrossRef] [PubMed]

| ING Member | Specific Function | Redundant Function | Related Pathways | Reference |
|---|---|---|---|---|
| ING1 | Down-regulation of cyclin B1 and possibly cyclin E, prevention of cell transformation, and induction of apoptosis in cooperation with c-Myc | Regulation of G1/S and G2/M cell cycle transition, regulation of the p53 pathway, chromatin remodeling, and regulation of nucleotide excision repair (NER) in response to UV exposure | NF-κB, ARFMDM2-p53 | [1,2] |
| ING2 | Regulation of senescence, regulation of Fas expression, and regulation of apoptosis in response to UV exposure | Regulation of the p53 pathway, regulation of NER, chromatin remodeling, and regulation of the cell cycle and apoptosis | NF-κB, TGFβ | |
| ING3 | - | Regulation of the cell cycle and apoptosis, chromatin remodeling and neural development | - | |
| ING4 | Inhibition of colony formation, reduction of the percentage of cells in S phase, up-regulation of Bax expression, angiogenesis, and cell migration | Regulation of the p53 pathway and chromatin remodeling | NF-κB, HIF-1α | |
| ING5 | Reduction of colony-forming efficiency, inhibition of S-phase, and induction of p21 | Regulation of the p53 pathway, chromatin remodeling, and regulation of the cell cycle and apoptosis | - |
| Disease/Cellular Mechanism | ING Type and Signaling Pathways | Cell Line | Function | Reference |
|---|---|---|---|---|
| Aging | ING1a and Rb pathway | Hs68 fibroblast cells, and EA.hy926 and HaCaT cells | ↑↑ ING1a: ↑ senescence in the absence of activating p53-mediated DNA damage signaling by activating the Rb pathway | [20] |
| Breast cancer | ING5 | MDA-MB-231 and MCF-7 cells | ↑↑ ING5: ↓ cell viability, glycolysis, and mitochondrial respiration, ↑ apoptosis, S arrest, autophagy, or senescence, and ↑ chemoresistance to Cisplatin, MG132, paclitaxel, and SAHA | [44] |
| Glioma | ING5 | U87 | ↑↑ ING5: ↓ proliferation, energy metabolism, migration, and invasion, and ↑ G2/M arrest, apoptosis, dedifferentiation, and senescence | [45] |
| Head and neck squamous cell carcinoma | ING3, p300, p21, and p53 | HNSCC cells | ↑↑ ING3: ↑ p53-mediated cell-cycle arrest, senescence, and/or apoptosis via interacting with p300 and p21 | [38] |
| Induction of apoptosis | ING1 and BAX | SKBR3; MDA-MB468, BT474, and T47D lines; HCT116, and HEK293 | The degree of mitochondrial translocation is correlated with the ability of ING1 to induce apoptosis. Binding and activation of BAX by ING1 are needed for the induction of apoptosis. | [25] |
| Osteosarcoma | ING2 | HOS cells | ↑↑ ING2: ↑ apoptosis, G1 phase arrest, and senescence | [32] |
| Ovarian cancer | ING5 | ES-2, H08910, H08910-PM, OVCAR3, SKOV3/DDP, A2780, and A2780/T | ↑↑ ING5: ↓ cell viability and migration, invasion, and ↑ apoptosis, cell cycle arrest, senescence, and autophagy | [46] |
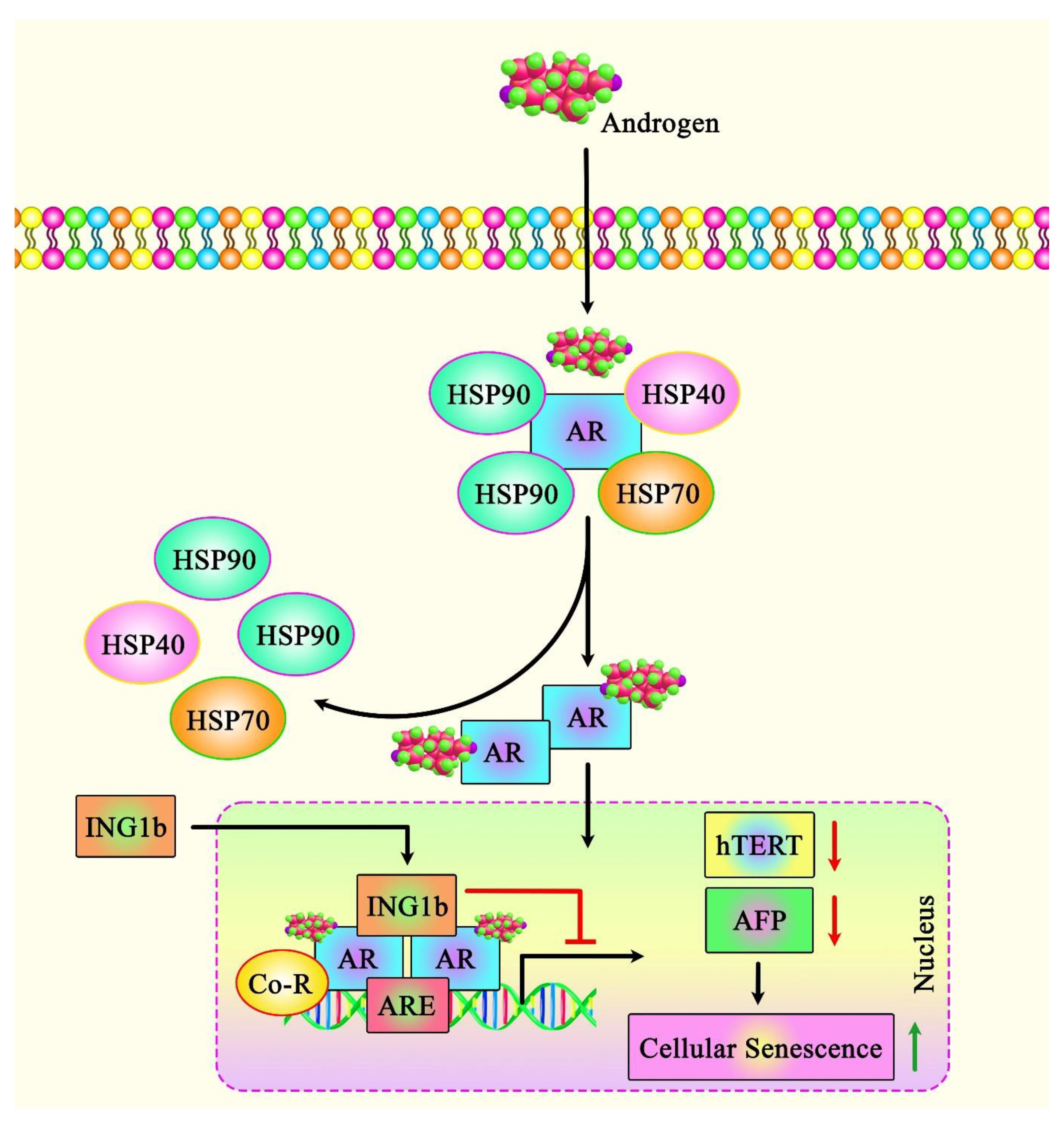
| Prostate cancer | ING1b, ING2, and AR signaling | LNCaP PCa cells | ∆ ING1b: ↓ growth and migration, ↑ induction of cellular senescence and the cell cycle inhibitor p16 INK4a ↑↑ ING2: causes ↑ growth arrest, and induces cellular senescence by interacting with AR and inhibiting AR transcriptional activation | [26] |
| ING1b and AR signaling | ING1b knockout (KO) mouse embryonic fibroblasts (MEFs), PC3 cells, PC3-AR cell line, C4-2 cells, LNCaP, and NIH3T3 S2-6 | ↑↑ ING1b: ↑ cellular senescence, and ↓ growth and migration via inhibiting AR-mediated transactivation | [27] | |
| Regulation of endocytosis | ING1a, Rb-E2F pathway, and ITSN2 | Hs68 and WI38 fibroblast cells | ↑↑ ING1a: ↑ cellular senescence to regulate endocytosis via the Rb-E2F pathway and overexpression of ITSN2 | [28] |
| Tongue squamous cell carcinoma | ING5, two truncated fragments of ING5 (aa 1-184 and aa 107-226), cyclin E, and CDK2 | HSC-3 | ↑↑ ING5: ↓ proliferation and ↑ apoptosis in HSC-3 cells Two truncated fragments of ING5 (aa 1-184 and aa 107-226): ↑ cellular senescence and inhibition of cyclin E and CDK2 expression. | [47] |
| Tumor Type | Animal Models | Results | Reference |
|---|---|---|---|
| Breast cancer | Balb/c nude mice | ↑↑ ING5: ↓ tumor volume and weight | [44] |
| Glioma | Female Balb/c nude mice | ↑↑ ING5: ↓ tumor volume and weight | [45] |
| Ovarian cancer | Female Balb/c nude mice | ↑↑ ING5: ↓ proliferation, and ↑ apoptosis and autophagy | [46] |
| Prostate cancer | Ing1 knockout (KO) mice | ∆ ING1: ↓ endogenous AR target genes | [26] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghafouri-Fard, S.; Taheri, M.; Baniahmad, A. Inhibitor of Growth Factors Regulate Cellular Senescence. Cancers 2022, 14, 3107. https://doi.org/10.3390/cancers14133107
Ghafouri-Fard S, Taheri M, Baniahmad A. Inhibitor of Growth Factors Regulate Cellular Senescence. Cancers. 2022; 14(13):3107. https://doi.org/10.3390/cancers14133107
Chicago/Turabian StyleGhafouri-Fard, Soudeh, Mohammad Taheri, and Aria Baniahmad. 2022. "Inhibitor of Growth Factors Regulate Cellular Senescence" Cancers 14, no. 13: 3107. https://doi.org/10.3390/cancers14133107
APA StyleGhafouri-Fard, S., Taheri, M., & Baniahmad, A. (2022). Inhibitor of Growth Factors Regulate Cellular Senescence. Cancers, 14(13), 3107. https://doi.org/10.3390/cancers14133107







