Simple Summary
Epithelial–mesenchymal transition (EMT) and its reverse process mesenchymal–epithelial transition (MET) are considered critical events in the cancer progression. These programs are tightly connected with the development of metastasis–the lethal stage of the disease. Both EMT and MET shape the biology of unusually aggressive and heterogeneous triple-negative breast cancer (TNBC). In this review, we summarize the current knowledge of EMT/MET plasticity in the context of TNBC, with a special focus on drivers and mechanisms behind these processes.
Abstract
Triple-negative breast cancer (TNBC) is a subtype of breast carcinoma known for its unusually aggressive behavior and poor clinical outcome. Besides the lack of molecular targets for therapy and profound intratumoral heterogeneity, the relatively quick overt metastatic spread remains a major obstacle in effective clinical management. The metastatic colonization of distant sites by primary tumor cells is affected by the microenvironment, epigenetic state of particular subclones, and numerous other factors. One of the most prominent processes contributing to the intratumoral heterogeneity is an epithelial–mesenchymal transition (EMT), an evolutionarily conserved developmental program frequently hijacked by tumor cells, strengthening their motile and invasive features. In response to various intrinsic and extrinsic stimuli, malignant cells can revert the EMT state through the mesenchymal–epithelial transition (MET), a process that is believed to be critical for the establishment of macrometastasis at secondary sites. Notably, cancer cells rarely undergo complete EMT and rather exist in a continuum of E/M intermediate states, preserving high levels of plasticity, as demonstrated in primary tumors and, ultimately, in circulating tumor cells, representing a simplified element of the metastatic cascade. In this review, we focus on cellular drivers underlying EMT/MET phenotypic plasticity and its detrimental consequences in the context of TNBC cancer.
1. Introduction
Triple-negative breast cancer (TNBC), accounting for approximately 10–15% of all breast carcinomas, associates with an earlier age of onset and frequently manifests inherently aggressive clinical behavior [1,2]. TNBC is characterized by profound heterogeneity and absence of the expression of classical breast cancer markers: estrogen (ER), progesterone (PR), and human epidermal growth factor 2 (HER2) receptors. For that reason, TNBC patients do not benefit from therapies targeting estrogens, progestins and HER2, and conventional chemotherapy and radiotherapy often remain the only treatment option [3,4,5,6]. Based on PAM50 intrinsic subtype classification (Basal-like, HER2-enriched, Luminal A, Luminal B, Normal-like, Claudin-low), most of clinical TNBC samples harbor basal like-signature [7,8]. Further comprehensive molecular studies dissected TNBC diversity and characterized 4 distinct subtypes, each displaying unique disease biology: basal-like 1, basal-like 2, mesenchymal, and luminal androgen receptor type [9,10]. Besides cancer cells, the TNBC tumor mass composes of extracellular matrix and different stromal populations, such as immune and endothelial cells, fibroblasts and others, collectively known as the tumor microenvironment (TME) [11]. TME plays a crucial role in all stages of carcinogenesis, including tumor growth, immune evasion, therapy resistance, and metastatic dissemination [12,13,14,15]. Intratumoral heterogeneity is determined by the phenotypic and molecular diversity within the tumor and can be explained by several concepts, including the differentiation state of the cancer cell of origin, mechanisms of cell plasticity, dynamic genomic alterations, and clonal evolution and selection [16,17,18]. One of the key mechanisms that contributes to phenotypic plasticity and heterogeneity is the epithelial–mesenchymal transition (EMT) and its reverse program known as mesenchymal–epithelial transition (MET). Both of these fundamental programs play specific roles in embryonic development and adult tissue homeostasis. During EMT, cells can undergo dynamic transitions from an epithelial (E) state, characterized by the presence of cell-to-cell junctions, apical-basal polarity, and interactions with basement membrane to a mesenchymal (M) phenotype, identified by fibroblast-like morphology linked with increased migratory and invasive properties [19,20]. Aberrant activation of EMT by cancer cells that enhances their invasive and motile behavior represents a critical event in tumor progression. In this context, EMT was shown to facilitate the spread of cancer cells from primary tumor to circulation, followed by extravasation and dissemination to secondary sites [21,22]. On the other hand, reversion of cells back to epithelial state during MET enables tumor cells to colonize distant organs with higher efficiency and form macrometastasis [23,24]. Apart from their roles in metastatic progression, both of these programs influence therapy resistance, prevent senescence, enhance survival and foster entrance of both normal and neoplastic mammary epithelial cells into stem-cell states [25,26,27,28]. Malignant cells have the capacity to dynamically switch between EMT and MET, which allows them to exist in a wide array of hybrid E/M states, co-expressing both epithelial and mesenchymal markers and partially bearing features of both phenotypes. Some of these highly plastic and aggressive hybrid phenotypic states provide cancer cells with increased fitness and flexibility required for tumor development. These states are believed to be generated in a stepwise manner through the E/M continuum and strongly depend on specific biological context [29,30,31,32,33,34,35]. According to a recent consensus statement, the favored term for the ability of cancer cells to adopt mixed E/M features is epithelial–mesenchymal plasticity (EMP) [20]. While the role and importance of EMT in tumor progression have been widely studied and emphasized in the past, hybrid phenotypes and MET did not get such attention.
According to recent classification, the most commonly used TNBC cell lines in discussed studies possess basal (MDA-MB-468, HCC1806) or mesenchymal phenotype (MDA-MB-231, BT-549, SUM-159 and HS578T) [36]. In addition, few studies employed also non-transformed MCF10A and HMLE cell lines that are immortalized and non-tumorigenic cells per se; however, they can exhibit features similar to TNBC cancer cell lines, including the lack of hormone receptors and HER2 expression [37,38].
Insights into TNBC Heterogeneity
Apart from lack of reliable biomarkers, TNBC tumors exhibit a heterogeneous clinical behavior associated with poor prognosis; therefore, a concerted effort has been undertaken to decipher TNBC heterogeneity. On a single-cell level, five distinct subgroups of malignant cells shared by tumors have been identified, including one clinically relevant subpopulation characterized by multiple signatures of treatment resistance, metastasis, activation of glycosphingolipid metabolism, and associated innate immunity sensing and inflammation [39]. Similarly, in matched primary tumors and micrometastases from basal-like TNBC patient-derived xenografts (PDX) models, a profound transcriptional diversity among tumor cells was observed, as individual tumor clusters associated with genes related to fatty-acid metabolism, proteasome function, extracellular matrix (ECM) modulatory signature, or EMT gene signature [40]. Moreover, scRNA-seq revealed transcriptional reprogramming of tumor cells undergoing chemotherapy pressure of docetaxel and epirubicin [41]. On the protein level, imaging mass cytometry identified tumor populations with high levels of Ki-67, p53, EGFR, and CAIX; basal cytokeratins; and luminal cytokeratins [42]. Besides tumor cells with a basal phenotype characterized by SMA expression, Vimentin, and EMT-like tumor cells, TNBC clinical specimens also consist of fibroblasts, endothelial and immune cells [43]. Immune cells, particularly tumor-infiltrating lymphocytes are often implicated in TNBC prognosis [44,45] and such a complex immune landscape is composed of specific B cell populations, tissue macrophages and T-lymphocytes with distinct transcriptomic signatures including naïve/early costimulatory, exhausted, or cytotoxic [46]. In the context of stromal cells, usually represented in tumors by cancer-associated fibroblasts (CAFs) [47,48], Wu and colleagues identified distinct subsets of myofibroblast-like and inflammatory CAFs along with differentiated and immature perivascular-like stromal cells [15]. Myofibroblastic and inflammatory fibroblasts comprised bulk tumors in the different patient cohort although sc-RNAseq analysis was performed on already presorted CAF subset defined by FAPhi CD29med-hi SMAhi expression [49].
In addition to extensive intratumoral heterogeneity, TNBC patients display a heterogeneity in somatic mutations and copy number aberrations (CNA) [8] and this mutational profile is tightly connected with TNBC subtype [18,50]. For example, TP53 is more frequently mutated in basal-like TNBC, with enriched nonsense and frameshift mutations and along with PIK3CA and PTEN is involved in the early stages of breast cancer development [18,50]. In addition, metastatic TNBC demonstrate enrichment of driver and targetable CNA relative to primary tumors [51] and majority of patients exhibit complex aneuploid rearrangements [52,53,54].
Nevertheless, these studies provide mostly descriptive data of TNBC cellular and genomic heterogeneity, and further mechanistic studies are needed to functionally link particular populations with EMP.
2. Mechanisms Regulating Epithelial–Mesenchymal Plasticity
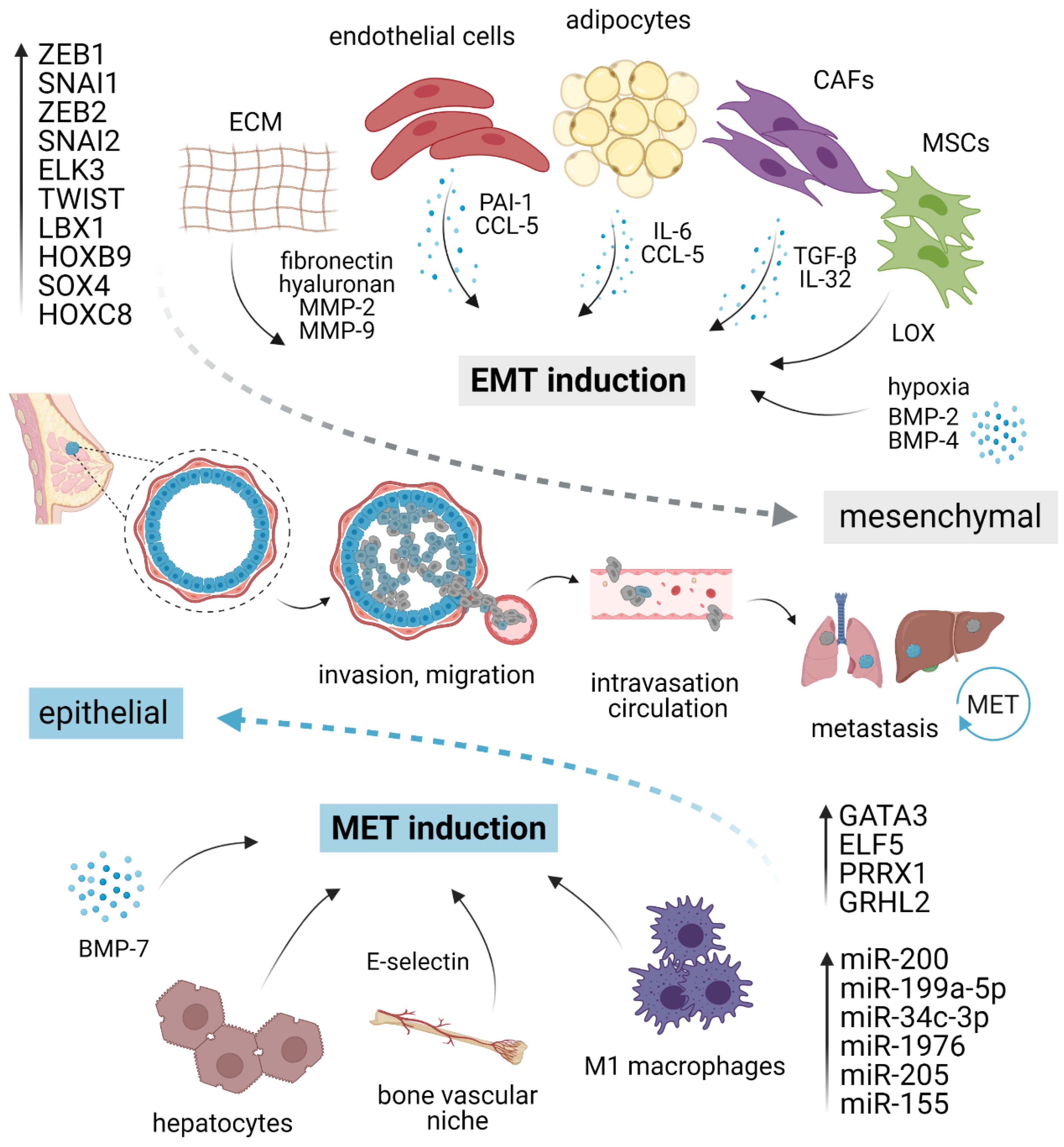
Owing to the transient and dynamic nature of EMT/MET during invasion, dissemination, and metastatic outgrowth, a growing body of evidence suggests that EMP in cancer cells is fueled by the external microenvironmental cues from primary TME or pre-existing metastatic niche, rather than driven by the cell-intrinsic genetic alterations [23]. A complex network of stimuli converges inside the nucleus to up- or down-regulate genes required for epithelial or mesenchymal characteristics, including those involved in migration, invasion, and stemness (Figure 1).
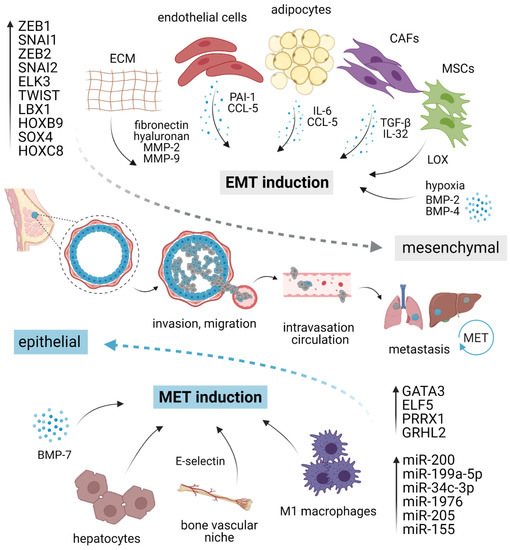
Figure 1.
EMT/MET plasticity in breast cancer. EMT is induced by pro-inflammatory cytokines and molecules secreted by different stromal cells present in the tumor microenvironment, ECM elements, and hypoxia. Different cell types such as macrophages or hepatocytes might activate MET. Each program is executed via a specific set of transcription factors and/or corresponding miRNAs. The activation of EMT and mesenchymal phenotype (gray) grants cancer cells the ability to migrate, invade, intravasate, survive in circulation, and extravasate at distant sites. At the secondary organs, mesenchymal cells revert to an epithelial state (blue) through MET, regaining the ability to form macrometastasis. EMT, epithelial–mesenchymal transition; MET, mesenchymal–epithelial transition; CAFs, cancer-associated fibroblasts; MSCs, mesenchymal stromal cells; ECM, extracellular matrix; MMP, matrix metalloproteinase; LOX, lysyl-oxidase. Created with BioRender.
2.1. Microenvironmental Stimuli
Multiple studies uncovered many microenvironmental factors in promoting EMT/MET machinery, including proinflammatory cytokines secreted by stromal cells, local hypoxic gradients, or specific components of ECM [55]. The vast majority of well-validated EMT inducers are pleiotropic growth factors or cytokines, including EGFs, FGFs, and numerous interleukins, such as IL-6 and IL-8 [55,56]. The most potent EMT inducer is TGF-β (Transforming growth factor-beta), which activates members of the SMAD family of signal transducers. TGF-β can also activate other pathways that are collectively referred to as “non-canonical” TGF-β signaling that complements SMAD action [57,58]. Besides activation of EMT program in epithelial cells, TGF-β promotes expression of cancer stem cells (CSCs) drivers and markers, and together with canonical and non-canonical WNT ligands function in an autocrine manner to maintain mesenchymal state [27,59]. TGF-β can further induce cell invasion through upregulation of Matrix metalloproteinase (MMP)-2 and -9 [60]. In a paracrine manner, TGF-β sculptures local TME and supports immunosuppression [61,62,63,64,65]. Plasma-derived TGF-β-related protein signature consisting of CLIC1, MAPRE1, SERPINA3 genes was predictive of TNBC tumor progression [66]. On the contrary, BMP-7 (Bone morphogenetic protein-7), a TGF-β family member, often acts as a TFG-β antagonist and preserves epithelial phenotype by counteracting SMAD2/3-dependent TGF-β signaling through BMP SMADs 1/5/8. Buijs et al. demonstrated that BMP-7 overexpression in cancer cells or systemic BMP-7 introduction in vivo resulted in enhanced epithelial phenotype and inhibition of bone metastasis development. However, exogenous BMP-7 exposure alone was not sufficient to restore E-cadherin expression. Therefore, it remains unclear whether the decreased growth of bone metastasis is orchestrated by the lack of E-cadherin, as discussed later, or depended on specific bone microenvironment regardless of E-cadherin status [67]. Mechanistically, BMP-7 suppresses cancer cell invasion by inhibiting TGF-β-induced Integrin β3 [68]. These findings were independently confirmed by a different group that further observed EMT attenuation after treatment with nanoparticles targeting Integrin β3 [69]. On the other hand, BMP-2 and BMP-4 were reported as EMT inducers, acting via Retinoblastoma/CD44 and Notch signaling, respectively [70,71].
Although several studies suggested a potential function of resident adipocytes in TNBC plasticity, their role in EMT/MET has not been satisfactorily elucidated yet. For instance, tumor cells exposed to mature adipocytes displayed accelerated invasive properties and enhanced lung metastasis potential [72]. In the experimental settings, human adipose stem cells (ASC) stimulated breast cancer motility and invasion through production of CCL5, as a response to cancer cells [73,74]. Strikingly, CCL5 is detectable in peritumoral adipose tissue of TNBC patients and correlates with lymph node and distant metastases, and poor patient outcome [73]. Sabol and colleagues highlighted the role of obesity in cancer progression [75,76]. Compared to lean donors (BMI < 25), ASC from obese donors (BMI > 30) activated EMT resulting in elevated number of circulating tumor cells (CTC) and increase in metastasis incidence in PDX via leptin signaling but surprisingly did not influence the tumorigenesis of TNBC xenografts [76]. Moreover, adipocyte-derived IL-6 induced EMT through STAT3 signaling [77]. Other stromal cell types capable of inducing EMT include cancer-associated fibroblasts employing paracrine TGF-β [78,79] or IL-32 and Integrin β3–p38 MAPK signaling [80], mesenchymal stromal cells producing lysyl oxidase [81], and endothelial cells via PAI-1 (SERPINE1) and CCL5 signaling [82].
Regarding ECM elements, exogenous fibronectin can accelerate EMT through the activation of Src kinase and ERK/MAP kinase signaling [83]. Alternatively, elevated production of ECM component hyaluronan was sufficient to induce EMT, employing the PI3K/Akt cell survival pathway [84]. Lastly, EMT can occur as a response to a hypoxic environment [85,86], mechanistically observed as a result of hypoxia-induced activation of Notch ligand Jagged2 [87] or governed by Carbonic anhydrase IX [88,89].
The first evidence for direct MET induction associated with E-cadherin restoration in response to microenvironmental cues was provided by Chao et al. In their experimental model, hepatocytes induced the E-cadherin re-expression in otherwise mesenchymal MDA-MB-231 cell line through passive loss of E-cadherin promoter methylation [90,91]. Such E-cadherin re-expression and concomitant quiescent epithelial phenotype also arose after co-cultivation of cancer cells with M1 macrophages [92]. In the bone milieu context, E-selectin from bone vascular niche has been identified as a driver of MET and Wnt activation in cancer cells, promoting bone metastasis [93].
2.2. Tumor Infiltrating Lymphocytes
Various cellular and humoral components of the immune system can constrain or promote the growth of tumors through a dynamic process known as immunoediting. Through various mechanisms of immune-mediated plasticity, including EMT [94], cancer cells change their phenotype to edit their immunogenicity and acquire immunosuppressive traits. The contribution of particular immune cell types on cancer cell plasticity is becoming the focus of mechanistic studies especially due to the rise of immunotherapies. The number of tumor infiltrating lymphocytes (TILs) have a prognostic value in TNBC patients that did not receive adjuvant therapy [95]. Increased incidence of TILs in TNBC can be a consequence of increased mutational burden and neoepitope exposure [96]. In particular, cytotoxic CD8 lymphocytes appear to have an antitumor activity [97]. However, this phenomenon can be suppressed by the CD24-/CD44+ /ALDH+ cancer stem cell population [98]. Conversely, Seo and colleagues showed that TNBC infiltration with CD4+ and CD8+ T cells was positively correlated with the presence of CD44highCD24low tumor cells and Vimentin expression [99]. The cancer stem cell population that underwent beta-catenin-STT3-mediated EMT manifested an accumulation of immunosuppressive molecule PDL1 [100], a target of several immunotherapies. Independent study further showed that mesenchymal breast cancer cells express decreased levels of MHC I and high levels of PDL1, as well as increased number of tumor-associated macrophages in the tumor stroma.
2.3. Transcription Factors
Besides epigenetic stabilization, EMP is largely mediated and maintained by a core set of transcription factors (TFs), most notably SNAI1 (known as Snail), SNAI2 (known as Slug), Twist-related protein 1 (TWIST1), and Zinc-finger E-box-binding homeobox-1 and -2 (ZEB1 and ZEB2, respectively) [19,101]. Expression of these EMT-TFs is often selective, context- and cell type-specific, and transient, and their deregulation in different E/M states imply a high degree of malignant cell plasticity [102]. ZEB1 represents a central switch that induces and determines EMT and is considered and studied as a potential biomarker for poor clinical outcome in triple-negative breast cancer [55,103,104,105,106]. Besides the induction of EMT, activation of ZEB1 by TGF-β can fuel de novo generation of CSCs from non-CSCs populations with poised chromatin within the ZEB1 promoter region and is critical for the maintenance of cells in CSCs-like, CD44hi state [103]. On the other hand, ZEB1 downregulation mediated by PKCα leads to inhibition of mesenchymal phenotype, cell migration, and invasiveness [107]. ZEB1 is known to cooperate with ELK3, a ternary complex factor from the ETS TFs family, to repress E-cadherin by transcriptional activation of ELK3 expression [108]. Suppression of ELK3 through epigenetic activation of GATA3 rendered cancer cells unresponsive to TGF-β and led to their reprogramming into a non-invasive, cuboidal-like, epithelial state, as observed both in vitro and in vivo [109]. In concordance with these findings, a similar effect on epithelial phenotype has been reported for GATA3, a TF considered a master suppressor of metastasis and associated with better prognosis [110]. GATA3 restored E-cadherin expression by binding into GATA-like motifs located within the E-cadherin promoter. Its overexpression reduced the levels of mesenchymal markers Vimentin, N-cadherin, and MMP-9, and resulted in the formation of smaller tumors without overt metastasis [111,112]. Another well-described MET inducer from the ETS family, ELF5, can reactivate epithelial state through transcriptional repression of SNAIL2, a prominent EMT inducer [113]. However, to repress the epithelial branch of EMT program, SNAI2 requires concurrent induction and cooperation with TWIST1 [114]. Similarly, knockdown of SNAI2 alone failed to revert EMT and suppressed the tumor initiation, in contrast to more potent effects associated with loss of SNAI1 [28]. This observation suggests that in TNBC, SNAI2 requires cooperation with other molecules and is insufficient to modulate EMP alone. In other words, SNAI2/Slug seems to induce a weaker EMT than SNAI1/Snail [115,116].
There is growing evidence implying the role of several members of the homeobox (HOX) TF family [117,118,119]. HOXB9 has been reported to promote the mesenchymal state, tumor growth, angiogenesis, and distant metastasis to the lung [120]. Similarly, HOXC8 functions as an EMT and migration inducer through transcriptional activation of MGP (Matrix Gla protein) [121]. Ladybird homeobox 1 (LBX1) displayed an analogous pattern and accelerated EMT through direct upregulation of ZEB1, ZEB2, SNAI1, and TGF-β2. However, MCF10A cells expressing LBX1 alone were not capable of forming tumors. Only cells co-expressing LBX1 and HRASV12 gave rise to tumors, suggesting oncogenic cooperation with LBX1 [122]. On the contrary, HOX member PRRX1 has the capacity to completely reactivate the epithelial state, even in the presence of TWIST1, and the loss of PRRX1 granted cancer cells the ability to colonize lungs [24]. Another widely reported TF involved in EMT/MET is Grainyhead-like 2 (GRHL2), which plays a fundamental role in the maintenance of epithelial phenotype and wound healing. GRHL2 is capable of suppressing TGF-β- and TWIST1-induced or spontaneous EMT occurring in a subpopulation of CD44highCD24low cells within the heterogeneous HMLE cell line [123]. GRHL2 is directly suppressed by ZEB1, which, in turn, represents a direct target for GRHL2-driven repression, suggesting that GRHL2 and ZEB1 form a double-negative regulatory feedback loop [124]. Moreover, GRHL2 expression is lost at the invasive front of primary human breast tumors and cancer cells disseminated into lymph nodes [125].
The extent of EMT and minimal necessary duration of EMT-TF expression is likely to be TF-dependent, possibly because the dynamic trajectories of EMT in the multi-dimensional landscape may or may not overlap [126]. A specific set of TFs appears to be important for rapid induction of TGF-β-mediated EMT but fails to fully complete and maintain classical EMT. This was demonstrated in a study concerning developmental factor SOX4 [127]. Intriguingly, even the transient TWIST1 expression was sufficient to induce stemness, promote tumor-initiating features, and elicit stably altered, plastic epithelial cell state [128], consistent with observations that a full-blown EMT may be detrimental for stemness and/or metastasis [129,130,131]. Considering the limited number of oncogenic transcription factors deregulated in human cancer, TFs may serve as promising targets in the future. Although TFs have been historically viewed as undruggable, there are currently several TF inhibitors undergoing preclinical and clinical testing for a various cancer types [132].
2.4. MicroRNAs
Recently published data have shown that EMT/MET inducing transcriptional factors often reciprocally interact with microRNAs. MicroRNAs (miRNA) are small, endogenous, non-coding RNAs that regulate gene expression. Their levels are significantly deregulated in human cancers where they might function as either oncogenes or tumor suppressors [133,134]. The miR-200 family is considered to be a “guardian of the epithelial phenotype”, suppressing EMT by directly targeting ZEB1, ZEB2, and other transcriptional repressors of E-cadherin for decay [135,136,137,138]. Reciprocal ZEB/miR-200 feedback loop contributes to EMP and controls central cellular processes [104,139]. Mathematical modeling of this feedback loop can give rise to many interconverting phenotypes—epithelial, mesenchymal, and hybrid E/M [140].
Experimental restoration of miR-200c expression in TNBC cell lines MDA-MB-231, BT549, and SUM159, all of which express miR-200c at relatively low endogenous levels, reversed the EMT gene expression signature, as demonstrated by an increased E-cadherin, decreased ZEB1 and repressed immunosuppressive gene pattern expression [141]. Besides translational repression of ZEB1, miR-200-mediated transcriptional upregulation of E-cadherin was associated with increased acetylation of histone H3 at the E-cadherin promoter [142]. Moreover, the study by Wang et al. demonstrated that an accelerated MET program was orchestrated by an upregulation of miR-200c as a response to tamoxifen exposure that resulted in demethylation of miR-200c promoter [143]. Mechanistically, miR-200 re-expression not only drives epithelial differentiation but also promotes the growth of macrometastasis by directly targeting SEC23A, a regulator of metastasis-suppressive protein secretion, involving Insulin-like growth factor-binding protein 4 (IGFBP4) and Tubulointerstitial nephritis antigen-like 1 (TINAGL1) [144]. Similar results were obtained using isogenic mouse breast cancer models 67NR, 168FARN, 4TO7, and 4T1, that differ in their ability to metastasize when implanted into the mammary fat pad. Only epithelial cell line 4T1, expressing miR-200c and E-cadherin, formed macroscopic metastasis. Mesenchymal cell line 4T07 is capable to disseminate, but forms only micrometastases. Overexpression of miR-200 in 4T07 cell line strikingly augmented MET and facilitated macrometastatic outgrowth [145]. These results collectively point to the possibility that MET induced by the miR-200 family may be crucial for the colonization step, hence necessary for successful completion of the metastatic cascade.
Apart from the miR-200 family, miR-199a-5p overexpression promoted epithelial state, as demonstrated by the reduced cell migration and invasion, as well as increased CDH1 (encoding E-cadherin) and decreased ZEB1 and Twist expression [146]. Additionally, several different miRNAs were identified and described as the regulators of cell plasticity. Increased level of miR-34c-3p resulted in the stimulation of tumor-suppressing features and reversed the EMT through negative modulation MAP3K2 pathway [147]. Similarly, miR-1976 was determined as epithelial “keeper” due to the acceleration of EMT via PIK3CG after its knock-down [148]. miR-205 was, along with miR-200, shown to be significantly reduced in cells undergoing EMT [135]. Finally, miR-155 expression levels inversely correlated with the expression of several EMT markers including SMA, osteonectin, and CD146 in a cohort of TNBC patients. Moreover, miR-155 expression was reported as an independent favorable prognostic factor of distant metastasis-free survival [149].
3. Plasticity in TNBC Progression
Although EMT is widely accepted as a key transcriptional program required for tumor invasion and dissemination, the colonization and outgrowth of recruited cells in the distant organs is believed to require an opposite process, known as MET [23,150]. The MET has been supported mainly by histopathological analyses that revealed morphological resemblance and EMP markers expression between primary tumors and matched metastatic lesions, particularly in E-cadherin expression pattern, which was similar or very often increased in lymph node metastases, relative to the matched primary tumors [151,152,153]. Despite these correlative clinical findings, rigorous functional studies linking MET with metastatic colonization ability are scarce. Major obstacles in investigating MET are associated with the lack of dynamic in vivo models and the sporadic nature of invasion and colonization, making these observations statistically improbable to identify. An important notion is that EMT and MET are not binary processes, and MET does not simply mirror EMT; hence the loss of epithelial features in the cell does not necessarily result in rigid, irreversible mesenchymal phenotype. Cancer cells may not revert to being epithelial if they have crossed a ”tipping point” in terms of induction of EMT [154], and can often reside in a hybrid or partial EMT phenotype endowed with combined epithelial (e.g., cell-to-cell adhesion) and mesenchymal (e.g., motility) features [29,150]. Individual cells with partial EMT phenotype do not fully complete the EMT program and co-express both epithelial markers (such as EpCAM, keratins or E-cadherin) and mesenchymal markers (e.g., Vimentin) along with a differentially expressed set of EMT TFs. Such hybrid cells are presumed to have the highest tumorigenicity and metastatic potential [28,32,33,131,155] and were also observed in breast carcinomas, especially in CTCs exhibiting a spectrum of EMP states [155,156,157,158].
3.1. Circulating Tumor Cells
Recent technological advances allowed isolation and characterization of CTCs, the precursors of metastasis, and their presence in the blood of cancer patients is closely associated with poor prognosis in many tumor types, including TNBC [159,160,161,162]. Individual CTCs or CTC clusters in blood provide a simplified view to metastatic cascade, particularly during the spread to distant sites [163]. Although many CTCs display mesenchymal phenotype, several studies reported that aggressive CTCs might reside in the hybrid E/M phenotype. One of the first studies identified a fraction of rare bi-phenotypic E/M cells in TNBC clinical specimens based on expression of epithelial markers Keratins 5, 7, 8, 18, and 19, EpCAM, CDH1 and mesenchymal markers FN1, CDH2 and SERPINE1/PAI1 [157]. Consistently with TNBC itself being enriched for mesenchymal-like phenotype, CTCs isolated from blood resided predominantly in M, and to a lesser extent also in M/E states. Additionally, in patients with progressive metastatic disease, the proportion of CTCs that shifts to M/E and M states reflects the response to chemotherapy regimen and disease progression, in contrast to proportional E, E/M, E = M, M/E, M states present at the beginning of treatment [157]. Employing a syngeneic 4T1 mouse model, Liu and colleagues associated EMT phenotypes of CTCs and disseminated tumor cells (DTCs) with their ability to form lung metastasis. CTCs bearing hybrid phenotype, characterized predominantly as epithelial (positive for EpCAM, cytokeratin and E-cadherin) with limited mesenchymal traits defined by Vimentin expression represented the most aggressive cells with the strongest lung metastatic capacity when compared to mesenchymal CTCs [155]. In accordance, higher proportions of EpCAM+ cells in the bloodstream of human patients correlated with distant metastases and poor outcome [155]. Similarly, using Vimentin as a mesenchymal marker and cytokeratin as an epithelial marker, CTCs isolated from TNBC PDX models were characterized by mixed E, M, and E/M phenotypes. A unique PDX model rapidly metastasizing to the liver and lungs showed that the great majority of CTCs reflected mixed E/M state [164]. However, a further investigation involving multiple PDX models and human patient cohorts is required to confirm these experimental observations.
3.2. MET and Metastasis
The majority of distant metastases in epithelial carcinomas are well-differentiated, resembling primary tumors. Accumulating data indicate that this re-differentiation is fueled by transient rounds of EMT–MET, which endow cells with properties required to successfully complete the metastatic cascade [23]. In presented experimental studies, MET is usually defined by re-expression of E-cadherin, concomitant reduction in mesenchymal features, and re-differentiated metastatic foci. Of note, E-cadherin is itself required for the growth of primary tumors. Cancer cells with a terminal mesenchymal phenotype characterized by high ZEB1, ZEB2 and SNAI1, exhibited impaired tumor-initiating capacity. In contrast, CD49f+/EpCAM+/E-cadherinhi cells had the highest tumor-initiating capacity in majority of tested TNBC PDXs, when compared to CD49f+/EpCAMlow and CD49flow/ EpCAMlow [165]. Among the first studies inspecting the mechanism of MET, Chao et al. demonstrated that E-cadherin-negative primary tumors originated from MDA-MB-231 cells, gave rise to E-cadherin positive metastatic foci in lungs. Moreover, modeling liver microenvironment by the co-culture of cancer cells with hepatocytes reactivated cancer cell epithelial phenotype and functional E-cadherin re-expression, suggesting the prominent role of the metastatic niche in MET activation [91]. These results have not yet been experimentally reproduced in vivo. Using a similar experimental model, MDA-MB-468, E-cadherin expression sustained in vitro and in vivo proliferation and was indispensable for formation of the lung and lymph node metastasis [166]. Conversely, loss of E-cadherin resulted in increased invasion capacity and dissemination, reduced cell survival and decreased colony formation capacity [167]. Another study linking MET and metastasis took advantage of a dynamic in vivo MDA-MB-468 model, employing Vimentin as an EMP marker. Compared to individually isolated cells or micrometastasis in lungs, lung macrometastasis exhibited more heterogeneous Vimentin and homogeneous E-cadherin expression as observed also in primary tumors. This suggests that MET likely occurred once overt metastases developed [168].
Independently, Dykxhoorn and colleagues demonstrated that only the epithelial, E-cadherin/miR-200/Cytokeratin-expressing subclone of 4T1 cells was capable to form macrometastatic liver and lung metastases, when compared to mesenchymal clone that effectively disseminated, but was only capable of forming micrometastatic lesions [145]. Similarly, re-expression of miR-200 followed by subsequent acquisition of epithelial traits and activation of the metastasis-suppressive program through SEC23A was critical for the formation of macrometastasis in lungs [144]. In the BT-549 model, reactivation of epithelial properties mediated by PRRX1 loss, upregulation of E-cadherin, and loss of Vimentin resulted in tumor-initiating ability and MET, followed by formation of the metastatic foci in lungs. The number of foci was increased in concomitant loss of PRRX1 and TWIST1. Activation of MET was further linked to the acquisition of stem-like properties, hence uncoupling stemness and EMT [24]. Similarly, epithelial features induced by the HLH TF inhibitor ID1, resulting in pulmonary metastasis, were associated with stem-like phenotype and acquired independently of EMT. Remarkably, ID1 was induced by TGF-β only in cells that previously underwent EMT, implying that EMT is a prerequisite for subsequent ID1-induced MET during lung colonization [169]. Tumor initiating abilities of plastic cells were also highlighted in a study by Kröger et al., who observed that cells residing in hybrid E/M state, as determined by SNAI1 and Wnt, formed tumors more effectively than cells in ‘locked’ E or M state. Interestingly, forced transition from hybrid E/M to fully mesenchymal state mediated by ectopic expression of ZEB1 was accompanied by the loss of tumorigenicity, implicating that the existence in the extremes of E/M spectrum is unfavorable for cancer cell aggressiveness [32]. As stressed in previous sections, reversion to the epithelial state is to a large extent orchestrated by microenvironmental tumor-stroma interactions in a context-dependent manner. This idea is supported by a study in a mouse model of TNBC, which demonstrated that the recruitment of bone marrow-derived myeloid progenitors to the pre-metastatic lungs was essential to induce a MET of DTCs and favor the subsequent formation of macrometastases [170].
4. Targeting Plasticity as a Therapeutic Strategy
Intratumoral heterogeneity and EMP as its prerequisite is one of the major challenges in targeting cancer. Both cells with activated EMT and CSC characteristics have been reported to confer drug resistance against multiple conventional therapeutics [29]. Moreover, mesenchymal, basal-like neoplasms tend to be more resistant to neoadjuvant chemotherapy than epithelial, luminal-like tumors [171,172]. The three main strategies have been proposed to combat plasticity: (i) targeting stimuli, which can prevent mesenchymal transitioning; (ii) targeting cells in mesenchymal or hybrid states and therefore prevent MET at secondary niche; (iii) reprogramming mesenchymal cells back to epithelial state [29]. For instance, in the first scenario, chemotherapy-treated TNBC breast cancer displayed enhanced TGF-β signaling and upregulated CSC-related genes. Mechanistically observed in vitro paclitaxel treatment increased IL-8 expression and expanded a CSC population where autocrine TGF-β signaling was active. Pharmacological inhibition using TGF-βR1 kinase inhibitor and clinical candidate galunisertib reduced the paclitaxel-resistant CSC population and prevented re-establishment of tumors after paclitaxel treatment in TNBC xenografts [173]. Moreover, inhibition of TGF-β signaling suppressed paclitaxel-induced EMT, and combined treatment employing TGF-β inhibitor EW-7197 improved the therapeutic effect of paclitaxel by decreasing the lung metastasis and increasing the survival in vivo [174]. Likewise, TGF-β inhibitors also proved their effectiveness in restricting docetaxel-induced EMT, and the combined treatment enhanced cytotoxic efficacy of doxorubicin [175]. The third strategy was demonstrated in study in which overexpression of miR-644a in cancer cells resulted in epithelial state of cells and increased response to doxorubicin mediated by miR-644a/CTBP1/p53 axis [176]. Pattabiraman et al. identified adenylate cyclase activators cholera toxin and forskolin through a compound screening as inducers of CDH1 transcription and MET activators [138]. Cells exposed to these substances reverted to an epithelial state, acquired junctional E-cadherin, became responsive to doxorubicin and paclitaxel treatment, and most importantly lost their tumor initiation capacity [138]. Re-expression of E-cadherin in TNBC has been also associated with decreased sensitivity to protein kinase inhibitor staurosporine and DNA topoisomerase I inhibitor camptothecin [90]. Treatment with eribulin, a non-taxane microtubule inhibitor, induced MET accompanied by decreased in vitro migration, as well as reduced metastatic potential in vivo [177]. Furthermore, a monoclonal antibody that neutralizes IL-8, HuMax-IL8, reverted mesenchymal phenotype in claudin-low TNBC models in vitro and in vivo, and decreased the recruitment of tumor-promoting polymorphonuclear myeloid-derived suppressor cells at the tumor site in xenografts. This effect was further enhanced by combination with docetaxel [178]. Finally, it has been reported that specific downregulation of PD-L1 in claudin-low breast cancer cells showed signs of EMT reversal, implying that patients might benefit from anti-PD-L1 targeted therapy [179]. One other emerging idea is to block plasticity by “fixing cells at a given position on the epithelial–mesenchymal axis” [180]; a potential way to “fix” the cells and block plasticity bi-directionally is to break positive feedback loops in underlying regulatory networks that allow state switching [181]. A proof-of-principle experimental validation of this idea was demonstrated by Celia-Terrassa et al. in a study where breaking the miR-200/ZEB feedback loop reduced metastasis in vivo [182], although further validation of this approach is needed.
Given these data, a combinatorial therapeutic approach leveraging from conventional drugs targeting proliferation and agents modulating cell plasticity might represent promising treatment strategies in the future. Some of the EMT-modulating drugs already tested in clinical trials are summarized in Table 1. However, optimal therapy timing, identification of potential biomarkers, and proper dissection of roles of particular states in tumor evolution are of utmost necessity for future clinical interventions [29].

Table 1.
A list of drugs targeting EMT regulatory components in TNBC clinical trials.
5. Conclusions and Outlook
Recently accumulated data indicate that dynamic reciprocal transition between EMT and MET phenotypes represents the fundamental base of tumor progression in TNBC. Cancer cells comprising the triple-negative breast tumors are often enriched in mesenchymal traits when compared to other breast cancer subtypes, suggesting the presence of high degree of plasticity in TNBC. The EMT and MET programs, aberrantly activated by tumor cells and employed at different stages of cancer progression enable these cancer cells to dynamically change their phenotype, a feature that might be critical for successful metastatic dissemination and overcoming the multiple obstacles on route to a final destination-distant site parenchyma. EMT is considered an early event in breast cancer tumorigenesis, crucial for intravasation; MET, on the other hand, appears to be essential for subsequent re-activation in distant site and macrometastatic outgrowth. Multiple studies also underpinned the involvement of hybrid E/M states in aggressive behavior of tumor cells. However, some of the studies presented here relied solely on traditional molecular markers such as E-cadherin and Vimentin, whereas both EMT and MET programs display complex manifestation. Therefore, studies should identify these processes not only by well-known markers but also relate them to transcription factors and phenotypic features, such as proliferation and invasion. Moreover, enforced manipulations of gene expression usually used in plasticity studies artificially keep cells in fixed states and, therefore, do not recapitulate the whole dynamic EMT/MET spectrum of transient and reversible states. Conclusively, to precisely capture dynamics of the entire spectrum in time-dependent manner, development of more appropriate tools, such as inducible reporter systems or intra-vital imaging, and employment of multi-omics, single-cell based approaches together with integration of multi-scale computational modeling is required to fully reveal the extent of dynamics and implications of EMP in experimentally valid models and clinical specimens.
Author Contributions
B.K., K.S.: conceptualization; B.K.: writing—original draft preparation; J.R., M.K.J.: writing—reviewing and editing; K.S.: supervision, writing—reviewing and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by funds from the Czech Health Research Council (grant nr. 18-08-00245 all rights reserved (KS)) and Czech Science Foundation (grant nr. 20-22984S), and by project “Preclinical Progression of New Organic Compounds with Targeted Biological Activity” (Preclinprogress, CZ.02.1.01/0.0/0.0/16_025/0007381). J.R. was supported by the American Brain Tumor Association Basic Research Fellowship, the Terri Brodeur Breast Cancer Foundation Fellowship, the Druckenmiller Center for Lung Cancer Research, and NCI Cancer Center Support Grant P30 CA008748. M.K.J. was supported by Ramanujan Fellowship awarded by Science and Engineering Research Board (SERB), Department of Science and Technology (DST), Government of India (SB/S2/RJN-049/2018).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
All authors declare no conflict of interest.
References
- Howlader, N.; Altekruse, S.F.; Li, C.I.; Chen, V.W.; Clarke, C.A.; Ries, L.A.; Cronin, K.A. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J. Natl. Cancer Inst. 2014, 106. [Google Scholar] [CrossRef]
- Malorni, L.; Shetty, P.B.; De Angelis, C.; Hilsenbeck, S.; Rimawi, M.F.; Elledge, R.; Osborne, C.K.; De Placido, S.; Arpino, G. Clinical and biologic features of triple-negative breast cancers in a large cohort of patients with long-term follow-up. Breast Cancer Res. Treat. 2012, 136, 795–804. [Google Scholar] [CrossRef]
- Denkert, C.; Liedtke, C.; Tutt, A.; von Minckwitz, G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet 2017, 389, 2430–2442. [Google Scholar] [CrossRef]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef] [PubMed]
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef] [PubMed]
- Kassam, F.; Enright, K.; Dent, R.; Dranitsaris, G.; Myers, J.; Flynn, C.; Fralick, M.; Kumar, R.; Clemons, M. Survival outcomes for patients with metastatic triple-negative breast cancer: Implications for clinical practice and trial design. Clin. Breast Cancer 2009, 9, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Bertucci, F.; Finetti, P.; Cervera, N.; Esterni, B.; Hermitte, F.; Viens, P.; Birnbaum, D. How basal are triple-negative breast cancers? Int. J. Cancer 2008, 123, 236–240. [Google Scholar] [CrossRef]
- Koboldt, D.C.F.; Fulton, R.; McLellan, M.D.; Schmidt, H.; Kalicki-Veizer, J.; McMichael, J.F.; Fulton, L.L.; Dooling, D.J.; Ding, L.; Mardis, E.R.; et al. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Jovanovic, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef]
- Garrido-Castro, A.C.; Lin, N.U.; Polyak, K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov. 2019, 9, 176–198. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Janiszewska, M.; Tabassum, D.P.; Castano, Z.; Cristea, S.; Yamamoto, K.N.; Kingston, N.L.; Murphy, K.C.; Shu, S.; Harper, N.W.; Del Alcazar, C.G.; et al. Subclonal cooperation drives metastasis by modulating local and systemic immune microenvironments. Nat. Cell Biol. 2019, 21, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Keren, L.; Bosse, M.; Marquez, D.; Angoshtari, R.; Jain, S.; Varma, S.; Yang, S.R.; Kurian, A.; Van Valen, D.; West, R.; et al. A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 2018, 174, 1373–1387.e1319. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.S.; Gao, Y.; Welte, T.; Wang, H.; Liu, J.; Janghorban, M.; Sheng, K.; Niu, Y.; Goldstein, A.; Zhao, N.; et al. Immuno-subtyping of breast cancer reveals distinct myeloid cell profiles and immunotherapy resistance mechanisms. Nat. Cell Biol. 2019, 21, 1113–1126. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Z.; Roden, D.L.; Wang, C.; Holliday, H.; Harvey, K.; Cazet, A.S.; Murphy, K.J.; Pereira, B.; Al-Eryani, G.; Bartonicek, N.; et al. Stromal cell diversity associated with immune evasion in human triple-negative breast cancer. EMBO J. 2020, e104063. [Google Scholar] [CrossRef]
- Koren, S.; Bentires-Alj, M. Breast Tumor Heterogeneity: Source of Fitness, Hurdle for Therapy. Mol. Cell 2015, 60, 537–546. [Google Scholar] [CrossRef]
- Marusyk, A.; Janiszewska, M.; Polyak, K. Intratumor Heterogeneity: The Rosetta Stone of Therapy Resistance. Cancer Cell 2020, 37, 471–484. [Google Scholar] [CrossRef]
- Shah, S.P.; Roth, A.; Goya, R.; Oloumi, A.; Ha, G.; Zhao, Y.; Turashvili, G.; Ding, J.; Tse, K.; Haffari, G.; et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 2012, 486, 395–399. [Google Scholar] [CrossRef]
- Nieto, M.A.; Huang, R.Y.; Jackson, R.A.; Thiery, J.P. Emt: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef]
- Chaffer, C.L.; San Juan, B.P.; Lim, E.; Weinberg, R.A. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016, 35, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef]
- Brabletz, T. To differentiate or not--routes towards metastasis. Nat. Rev. Cancer 2012, 12, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Ocana, O.H.; Corcoles, R.; Fabra, A.; Moreno-Bueno, G.; Acloque, H.; Vega, S.; Barrallo-Gimeno, A.; Cano, A.; Nieto, M.A. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 2012, 22, 709–724. [Google Scholar] [CrossRef] [PubMed]
- Ansieau, S.; Bastid, J.; Doreau, A.; Morel, A.P.; Bouchet, B.P.; Thomas, C.; Fauvet, F.; Puisieux, I.; Doglioni, C.; Piccinin, S.; et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell 2008, 14, 79–89. [Google Scholar] [CrossRef]
- Gal, A.; Sjoblom, T.; Fedorova, L.; Imreh, S.; Beug, H.; Moustakas, A. Sustained TGF beta exposure suppresses Smad and non-Smad signalling in mammary epithelial cells, leading to EMT and inhibition of growth arrest and apoptosis. Oncogene 2008, 27, 1218–1230. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef]
- Ye, X.; Tam, W.L.; Shibue, T.; Kaygusuz, Y.; Reinhardt, F.; Ng Eaton, E.; Weinberg, R.A. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 2015, 525, 256–260. [Google Scholar] [CrossRef]
- Bhatia, S.; Monkman, J.; Blick, T.; Pinto, C.; Waltham, M.; Nagaraj, S.H.; Thompson, E.W. Interrogation of Phenotypic Plasticity between Epithelial and Mesenchymal States in Breast Cancer. J. Clin. Med. 2019, 8, 893. [Google Scholar] [CrossRef] [PubMed]
- Bierie, B.; Pierce, S.E.; Kroeger, C.; Stover, D.G.; Pattabiraman, D.R.; Thiru, P.; Liu Donaher, J.; Reinhardt, F.; Chaffer, C.L.; Keckesova, Z.; et al. Integrin-beta4 identifies cancer stem cell-enriched populations of partially mesenchymal carcinoma cells. Proc. Natl. Acad. Sci. USA 2017, 114, E2337–E2346. [Google Scholar] [CrossRef] [PubMed]
- Jolly, M.K.; Tripathi, S.C.; Jia, D.; Mooney, S.M.; Celiktas, M.; Hanash, S.M.; Mani, S.A.; Pienta, K.J.; Ben-Jacob, E.; Levine, H. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget 2016, 7, 27067–27084. [Google Scholar] [CrossRef]
- Kroger, C.; Afeyan, A.; Mraz, J.; Eaton, E.N.; Reinhardt, F.; Khodor, Y.L.; Thiru, P.; Bierie, B.; Ye, X.; Burge, C.B.; et al. Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc. Natl. Acad. Sci. USA 2019, 116, 7353–7362. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Van Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the tumour transition states occurring during EMT. Nature 2018, 556, 463–468. [Google Scholar] [CrossRef]
- Zhang, J.; Tian, X.J.; Zhang, H.; Teng, Y.; Li, R.; Bai, F.; Elankumaran, S.; Xing, J. TGF-beta-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Sci. Signal 2014, 7, ra91. [Google Scholar] [CrossRef]
- Tripathi, S.; Chakraborty, P.; Levine, H.; Jolly, M.K. A mechanism for epithelial-mesenchymal heterogeneity in a population of cancer cells. PLoS Comput. Biol. 2020, 16, e1007619. [Google Scholar] [CrossRef]
- Espinosa Fernandez, J.R.; Eckhardt, B.L.; Lee, J.; Lim, B.; Pearson, T.; Seitz, R.S.; Hout, D.R.; Schweitzer, B.L.; Nielsen, T.J.; Lawrence, O.R.; et al. Identification of triple-negative breast cancer cell lines classified under the same molecular subtype using different molecular characterization techniques: Implications for translational research. PLoS ONE 2020, 15, e0231953. [Google Scholar] [CrossRef] [PubMed]
- Kao, J.; Salari, K.; Bocanegra, M.; Choi, Y.L.; Girard, L.; Gandhi, J.; Kwei, K.A.; Hernandez-Boussard, T.; Wang, P.; Gazdar, A.F.; et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS ONE 2009, 4, e6146. [Google Scholar] [CrossRef]
- Neve, R.M.; Chin, K.; Fridlyand, J.; Yeh, J.; Baehner, F.L.; Fevr, T.; Clark, L.; Bayani, N.; Coppe, J.P.; Tong, F.; et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006, 10, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Karaayvaz, M.; Cristea, S.; Gillespie, S.M.; Patel, A.P.; Mylvaganam, R.; Luo, C.C.; Specht, M.C.; Bernstein, B.E.; Michor, F.; Ellisen, L.W. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat. Commun. 2018, 9, 3588. [Google Scholar] [CrossRef]
- Davis, R.T.; Blake, K.; Ma, D.; Gabra, M.B.I.; Hernandez, G.A.; Phung, A.T.; Yang, Y.; Maurer, D.; Lefebvre, A.; Alshetaiwi, H.; et al. Transcriptional diversity and bioenergetic shift in human breast cancer metastasis revealed by single-cell RNA sequencing. Nat. Cell Biol. 2020, 22, 310–320. [Google Scholar] [CrossRef]
- Kim, C.; Gao, R.; Sei, E.; Brandt, R.; Hartman, J.; Hatschek, T.; Crosetto, N.; Foukakis, T.; Navin, N.E. Chemoresistance Evolution in Triple-Negative Breast Cancer Delineated by Single-Cell Sequencing. Cell 2018, 173, 879–893.e813. [Google Scholar] [CrossRef]
- Jackson, H.W.; Fischer, J.R.; Zanotelli, V.R.T.; Ali, H.R.; Mechera, R.; Soysal, S.D.; Moch, H.; Muenst, S.; Varga, Z.; Weber, W.P.; et al. The single-cell pathology landscape of breast cancer. Nature 2020, 578, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.; Rapsomaniki, M.A.; Chevrier, S.; Anzeneder, T.; Langwieder, C.; Dykgers, A.; Rees, M.; Ramaswamy, A.; Muenst, S.; Soysal, S.D.; et al. A Single-Cell Atlas of the Tumor and Immune Ecosystem of Human Breast Cancer. Cell 2019, 177, 1330–1345 e1318. [Google Scholar] [CrossRef] [PubMed]
- Andre, F.; Dieci, M.V.; Dubsky, P.; Sotiriou, C.; Curigliano, G.; Denkert, C.; Loi, S. Molecular pathways: Involvement of immune pathways in the therapeutic response and outcome in breast cancer. Clin. Cancer Res. 2013, 19, 28–33. [Google Scholar] [CrossRef] [PubMed]
- He, T.F.; Yost, S.E.; Frankel, P.H.; Dagis, A.; Cao, Y.; Wang, R.; Rosario, A.; Tu, T.Y.; Solomon, S.; Schmolze, D.; et al. Multi-panel immunofluorescence analysis of tumor infiltrating lymphocytes in triple negative breast cancer: Evolution of tumor immune profiles and patient prognosis. PLoS ONE 2020, 15, e0229955. [Google Scholar] [CrossRef]
- Chung, W.; Eum, H.H.; Lee, H.O.; Lee, K.M.; Lee, H.B.; Kim, K.T.; Ryu, H.S.; Kim, S.; Lee, J.E.; Park, Y.H.; et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun. 2017, 8, 15081. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.; Kieffer, Y.; Scholer-Dahirel, A.; Pelon, F.; Bourachot, B.; Cardon, M.; Sirven, P.; Magagna, I.; Fuhrmann, L.; Bernard, C.; et al. Fibroblast Heterogeneity and Immunosuppressive Environment in Human Breast Cancer. Cancer Cell 2018, 33, 463–479e410. [Google Scholar] [CrossRef] [PubMed]
- Tchou, J.; Kossenkov, A.V.; Chang, L.; Satija, C.; Herlyn, M.; Showe, L.C.; Pure, E. Human breast cancer associated fibroblasts exhibit subtype specific gene expression profiles. BMC Med. Genom. 2012, 5, 39. [Google Scholar] [CrossRef]
- Kieffer, Y.; Hocine, H.R.; Gentric, G.; Pelon, F.; Bernard, C.; Bourachot, B.; Lameiras, S.; Albergante, L.; Bonneau, C.; Guyard, A.; et al. Single-Cell Analysis Reveals Fibroblast Clusters Linked to Immunotherapy Resistance in Cancer. Cancer Discov. 2020, 10, 1330–1351. [Google Scholar] [CrossRef]
- Bareche, Y.; Venet, D.; Ignatiadis, M.; Aftimos, P.; Piccart, M.; Rothe, F.; Sotiriou, C. Unravelling triple-negative breast cancer molecular heterogeneity using an integrative multiomic analysis. Ann. Oncol. 2018, 29, 895–902. [Google Scholar] [CrossRef]
- Stover, D.G.; Parsons, H.A.; Ha, G.; Freeman, S.S.; Barry, W.T.; Guo, H.; Choudhury, A.D.; Gydush, G.; Reed, S.C.; Rhoades, J.; et al. Association of Cell-Free DNA Tumor Fraction and Somatic Copy Number Alterations With Survival in Metastatic Triple-Negative Breast Cancer. J. Clin. Oncol. 2018, 36, 543–553. [Google Scholar] [CrossRef]
- Turner, N.; Lambros, M.B.; Horlings, H.M.; Pearson, A.; Sharpe, R.; Natrajan, R.; Geyer, F.C.; van Kouwenhove, M.; Kreike, B.; Mackay, A.; et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene 2010, 29, 2013–2023. [Google Scholar] [CrossRef]
- Hicks, J.; Krasnitz, A.; Lakshmi, B.; Navin, N.E.; Riggs, M.; Leibu, E.; Esposito, D.; Alexander, J.; Troge, J.; Grubor, V.; et al. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res. 2006, 16, 1465–1479. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Davis, A.; McDonald, T.O.; Sei, E.; Shi, X.; Wang, Y.; Tsai, P.C.; Casasent, A.; Waters, J.; Zhang, H.; et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat. Genet. 2016, 48, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.Y.; Fattet, L.; Yang, J. Molecular pathways: Linking tumor microenvironment to epithelial-mesenchymal transition in metastasis. Clin. Cancer Res. 2015, 21, 962–968. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Massague, J. TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 2012, 13, 616–630. [Google Scholar] [CrossRef]
- Scheel, C.; Eaton, E.N.; Li, S.H.; Chaffer, C.L.; Reinhardt, F.; Kah, K.J.; Bell, G.; Guo, W.; Rubin, J.; Richardson, A.L.; et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 2011, 145, 926–940. [Google Scholar] [CrossRef]
- Wiercinska, E.; Naber, H.P.H.; Pardali, E.; van der Pluijm, G.; van Dam, H.; ten Dijke, P. The TGF-beta/Smad pathway induces breast cancer cell invasion through the up-regulation of matrix metalloproteinase 2 and 9 in a spheroid invasion model system. Breast Cancer Res. Treat 2011, 128, 657–666. [Google Scholar] [CrossRef]
- Kang, Y.; He, W.; Tulley, S.; Gupta, G.P.; Serganova, I.; Chen, C.R.; Manova-Todorova, K.; Blasberg, R.; Gerald, W.L.; Massague, J. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proc. Natl. Acad. Sci. USA 2005, 102, 13909–13914. [Google Scholar] [CrossRef]
- Taylor, M.A.; Amin, J.D.; Kirschmann, D.A.; Schiemann, W.P. Lysyl oxidase contributes to mechanotransduction-mediated regulation of transforming growth factor-beta signaling in breast cancer cells. Neoplasia 2011, 13, 406–418. [Google Scholar] [CrossRef]
- Padua, D.; Zhang, X.H.; Wang, Q.; Nadal, C.; Gerald, W.L.; Gomis, R.R.; Massague, J. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell 2008, 133, 66–77. [Google Scholar] [CrossRef]
- Stuber, T.; Monjezi, R.; Wallstabe, L.; Kuhnemundt, J.; Nietzer, S.L.; Dandekar, G.; Wockel, A.; Einsele, H.; Wischhusen, J.; Hudecek, M. Inhibition of TGF-beta-receptor signaling augments the antitumor function of ROR1-specific CAR T-cells against triple-negative breast cancer. J. Immunother Cancer 2020, 8. [Google Scholar] [CrossRef]
- Hanks, B.A.; Holtzhausen, A.; Evans, K.S.; Jamieson, R.; Gimpel, P.; Campbell, O.M.; Hector-Greene, M.; Sun, L.; Tewari, A.; George, A.; et al. Type III TGF-beta receptor downregulation generates an immunotolerant tumor microenvironment. J. Clin. Investig. 2013, 123, 3925–3940. [Google Scholar] [CrossRef]
- Katayama, H.; Tsou, P.; Kobayashi, M.; Capello, M.; Wang, H.; Esteva, F.; Disis, M.L.; Hanash, S. A plasma protein derived TGF beta signature is a prognostic indicator in triple negative breast cancer. NPJ Precis. Oncol. 2019, 3. [Google Scholar] [CrossRef]
- Buijs, J.T.; Henriquez, N.V.; Van Overveld, P.G.M.; Van der Horst, G.; Que, I.; Schwaninger, R.; Rentsch, C.; Ten Dijke, P.; Cleton-Jansen, A.M.; Driouch, K.; et al. Bone morphogenetic protein 7 in the development and treatment of bone Metastases from breast cancer. Cancer Res. 2007, 67, 8742–8751. [Google Scholar] [CrossRef] [PubMed]
- Naber, H.P.; Wiercinska, E.; Pardali, E.; van Laar, T.; Nirmala, E.; Sundqvist, A.; van Dam, H.; van der Horst, G.; van der Pluijm, G.; Heckmann, B.; et al. BMP-7 inhibits TGF-beta-induced invasion of breast cancer cells through inhibition of integrin beta(3) expression. Cell Oncol. 2012, 35, 19–28. [Google Scholar] [CrossRef]
- Parvani, J.G.; Gujrati, M.D.; Mack, M.A.; Schiemann, W.P.; Lu, Z.R. Silencing beta3 Integrin by Targeted ECO/siRNA Nanoparticles Inhibits EMT and Metastasis of Triple-Negative Breast Cancer. Cancer Res. 2015, 75, 2316–2325. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Yu, J.; Park, A.; Dubon, M.J.; Do, J.; Kim, Y.; Nam, D.; Noh, J.; Park, K.S. BMP-4 enhances epithelial mesenchymal transition and cancer stem cell properties of breast cancer cells via Notch signaling. Sci. Rep. 2019, 9, 11724. [Google Scholar] [CrossRef]
- Huang, P.; Chen, A.; He, W.; Li, Z.; Zhang, G.; Liu, Z.; Liu, G.; Liu, X.; He, S.; Xiao, G.; et al. BMP-2 induces EMT and breast cancer stemness through Rb and CD44. Cell Death Discov. 2017, 3, 17039. [Google Scholar] [CrossRef]
- Dirat, B.; Bochet, L.; Dabek, M.; Daviaud, D.; Dauvillier, S.; Majed, B.; Wang, Y.Y.; Meulle, A.; Salles, B.; Le Gonidec, S.; et al. Cancer-associated adipocytes exhibit an activated phenotype and contribute to breast cancer invasion. Cancer Res. 2011, 71, 2455–2465. [Google Scholar] [CrossRef]
- D’Esposito, V.; Liguoro, D.; Ambrosio, M.R.; Collina, F.; Cantile, M.; Spinelli, R.; Raciti, G.A.; Miele, C.; Valentino, R.; Campiglia, P.; et al. Adipose microenvironment promotes triple negative breast cancer cell invasiveness and dissemination by producing CCL5. Oncotarget 2016, 7, 24495–24509. [Google Scholar] [CrossRef] [PubMed]
- Pinilla, S.; Alt, E.; Abdul Khalek, F.J.; Jotzu, C.; Muehlberg, F.; Beckmann, C.; Song, Y.H. Tissue resident stem cells produce CCL5 under the influence of cancer cells and thereby promote breast cancer cell invasion. Cancer Lett. 2009, 284, 80–85. [Google Scholar] [CrossRef]
- Olson, O.C.; Quail, D.F.; Joyce, J.A. Obesity and the tumor microenvironment. Science 2017, 358, 1130–1131. [Google Scholar] [CrossRef]
- Sabol, R.A.; Bowles, A.C.; Cote, A.; Wise, R.; O’Donnell, B.; Matossian, M.D.; Hossain, F.M.; Burks, H.E.; Del Valle, L.; Miele, L.; et al. Leptin produced by obesity-altered adipose stem cells promotes metastasis but not tumorigenesis of triple-negative breast cancer in orthotopic xenograft and patient-derived xenograft models. Breast Cancer Res. 2019, 21, 67. [Google Scholar] [CrossRef]
- Gyamfi, J.; Lee, Y.H.; Eom, M.; Choi, J. Interleukin-6/STAT3 signalling regulates adipocyte induced epithelial-mesenchymal transition in breast cancer cells. Sci. Rep. 2018, 8, 8859. [Google Scholar] [CrossRef]
- Yu, Y.; Xiao, C.H.; Tan, L.D.; Wang, Q.S.; Li, X.Q.; Feng, Y.M. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of breast cancer cells through paracrine TGF-beta signalling. Br. J. Cancer 2014, 110, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Jia, H.H.; Xu, Y.Q.; Zhou, X.; Zhao, X.H.; Wang, Y.F.; Song, X.; Zhu, Z.Y.; Sun, T.; Dou, Y.; et al. Paracrine and epigenetic control of CAF-induced metastasis: The role of HOTAIR stimulated by TGF-ss1 secretion. Mol. Cancer 2018, 17, 5. [Google Scholar] [CrossRef]
- Wen, S.; Hou, Y.; Fu, L.; Xi, L.; Yang, D.; Zhao, M.; Qin, Y.; Sun, K.; Teng, Y.; Liu, M. Cancer-associated fibroblast (CAF)-derived IL32 promotes breast cancer cell invasion and metastasis via integrin beta3-p38 MAPK signalling. Cancer Lett. 2019, 442, 320–332. [Google Scholar] [CrossRef] [PubMed]
- El-Haibi, C.P.; Bell, G.W.; Zhang, J.; Collmann, A.Y.; Wood, D.; Scherber, C.M.; Csizmadia, E.; Mariani, O.; Zhu, C.; Campagne, A.; et al. Critical role for lysyl oxidase in mesenchymal stem cell-driven breast cancer malignancy. Proc. Natl. Acad. Sci. USA 2012, 109, 17460–17465. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, J.; Fang, H.; Tang, L.; Chen, W.; Sun, Q.; Zhang, Q.; Yang, F.; Sun, Z.; Cao, L.; et al. Endothelial cells promote triple-negative breast cancer cell metastasis via PAI-1 and CCL5 signaling. FASEB J. 2018, 32, 276–288. [Google Scholar] [CrossRef]
- Park, J.; Schwarzbauer, J.E. Mammary epithelial cell interactions with fibronectin stimulate epithelial-mesenchymal transition. Oncogene 2014, 33, 1649–1657. [Google Scholar] [CrossRef]
- Zoltan-Jones, A.; Huang, L.; Ghatak, S.; Toole, B.P. Elevated hyaluronan production induces mesenchymal and transformed properties in epithelial cells. J. Biol. Chem. 2003, 278, 45801–45810. [Google Scholar] [CrossRef] [PubMed]
- Louie, E.; Nik, S.; Chen, J.S.; Schmidt, M.; Song, B.; Pacson, C.; Chen, X.F.; Park, S.; Ju, J.; Chen, E.I. Identification of a stem-like cell population by exposing metastatic breast cancer cell lines to repetitive cycles of hypoxia and reoxygenation. Breast Cancer Res. 2010, 12, R94. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, K.; Nordenskjold, B.; Landberg, G. Hypoxia, Snail and incomplete epithelial-mesenchymal transition in breast cancer. Br. J. Cancer 2009, 101, 1769–1781. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Okuda, H.; Watabe, M.; Kobayashi, A.; Pai, S.K.; Liu, W.; Pandey, P.R.; Fukuda, K.; Hirota, S.; Sugai, T.; et al. Hypoxia-induced Jagged2 promotes breast cancer metastasis and self-renewal of cancer stem-like cells. Oncogene 2011, 30, 4075–4086. [Google Scholar] [CrossRef]
- Lock, F.E.; McDonald, P.C.; Lou, Y.; Serrano, I.; Chafe, S.C.; Ostlund, C.; Aparicio, S.; Winum, J.Y.; Supuran, C.T.; Dedhar, S. Targeting carbonic anhydrase IX depletes breast cancer stem cells within the hypoxic niche. Oncogene 2013, 32, 5210–5219. [Google Scholar] [CrossRef]
- Ciccone, V.; Filippelli, A.; Angeli, A.; Supuran, C.T.; Morbidelli, L. Pharmacological Inhibition of CA-IX Impairs Tumor Cell Proliferation, Migration and Invasiveness. Int. J. Mol. Sci. 2020, 21, 2983. [Google Scholar] [CrossRef]
- Chao, Y.; Wu, Q.; Shepard, C.; Wells, A. Hepatocyte induced re-expression of E-cadherin in breast and prostate cancer cells increases chemoresistance. Clin. Exp. Metastasis 2012, 29, 39–50. [Google Scholar] [CrossRef]
- Chao, Y.L.; Shepard, C.R.; Wells, A. Breast carcinoma cells re-express E-cadherin during mesenchymal to epithelial reverting transition. Mol. Cancer 2010, 9, 179. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Ma, B.; Shao, H.; Clark, A.M.; Wells, A. Macrophage phenotypic subtypes diametrically regulate epithelial-mesenchymal plasticity in breast cancer cells. BMC Cancer 2016, 16, 419. [Google Scholar] [CrossRef] [PubMed]
- Esposito, M.; Mondal, N.; Greco, T.M.; Wei, Y.; Spadazzi, C.; Lin, S.C.; Zheng, H.; Cheung, C.; Magnani, J.L.; Lin, S.H.; et al. Bone vascular niche E-selectin induces mesenchymal-epithelial transition and Wnt activation in cancer cells to promote bone metastasis. Nat. Cell Biol. 2019, 21, 627–639. [Google Scholar] [CrossRef]
- Knutson, K.L.; Lu, H.; Stone, B.; Reiman, J.M.; Behrens, M.D.; Prosperi, C.M.; Gad, E.A.; Smorlesi, A.; Disis, M.L. Immunoediting of cancers may lead to epithelial to mesenchymal transition. J. Immunol. 2006, 177, 1526–1533. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Jonas, S.F.; Bataillon, G.; Criscitiello, C.; Salgado, R.; Loi, S.; Viale, G.; Lee, H.J.; Dieci, M.V.; Kim, S.B.; et al. Prognostic value of tumor-infiltrating lymphocytes in patients with early-stage triple-negative breast cancers (TNBC) who did not receive adjuvant chemotherapy. Ann. Oncol. 2019, 30, 1941–1949. [Google Scholar] [CrossRef]
- Disis, M.L.; Stanton, S.E. Triple-negative breast cancer: Immune modulation as the new treatment paradigm. Am. Soc. Clin. Oncol. Educ. Book 2015, e25–e30. [Google Scholar] [CrossRef]
- Mahmoud, S.M.; Paish, E.C.; Powe, D.G.; Macmillan, R.D.; Grainge, M.J.; Lee, A.H.; Ellis, I.O.; Green, A.R. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J. Clin. Oncol. 2011, 29, 1949–1955. [Google Scholar] [CrossRef]
- Akalay, I.; Janji, B.; Hasmim, M.; Noman, M.Z.; Andre, F.; De Cremoux, P.; Bertheau, P.; Badoual, C.; Vielh, P.; Larsen, A.K.; et al. Epithelial-to-mesenchymal transition and autophagy induction in breast carcinoma promote escape from T-cell-mediated lysis. Cancer Res. 2013, 73, 2418–2427. [Google Scholar] [CrossRef]
- Seo, A.N.; Lee, H.J.; Kim, E.J.; Kim, H.J.; Jang, M.H.; Lee, H.E.; Kim, Y.J.; Kim, J.H.; Park, S.Y. Tumour-infiltrating CD8+ lymphocytes as an independent predictive factor for pathological complete response to primary systemic therapy in breast cancer. Br. J. Cancer 2013, 109, 2705–2713. [Google Scholar] [CrossRef]
- Hsu, J.M.; Xia, W.; Hsu, Y.H.; Chan, L.C.; Yu, W.H.; Cha, J.H.; Chen, C.T.; Liao, H.W.; Kuo, C.W.; Khoo, K.H.; et al. STT3-dependent PD-L1 accumulation on cancer stem cells promotes immune evasion. Nat. Commun. 2018, 9, 1908. [Google Scholar] [CrossRef] [PubMed]
- Taube, J.H.; Herschkowitz, J.I.; Komurov, K.; Zhou, A.Y.; Gupta, S.; Yang, J.; Hartwell, K.; Onder, T.T.; Gupta, P.B.; Evans, K.W.; et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl. Acad. Sci. USA 2010, 107, 15449–15454. [Google Scholar] [CrossRef] [PubMed]
- Stemmler, M.P.; Eccles, R.L.; Brabletz, S.; Brabletz, T. Non-redundant functions of EMT transcription factors. Nat. Cell Biol. 2019, 21, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Chaffer, C.L.; Marjanovic, N.D.; Lee, T.; Bell, G.; Kleer, C.G.; Reinhardt, F.; D’Alessio, A.C.; Young, R.A.; Weinberg, R.A. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 2013, 154, 61–74. [Google Scholar] [CrossRef]
- Drapela, S.; Bouchal, J.; Jolly, M.K.; Culig, Z.; Soucek, K. ZEB1: A Critical Regulator of Cell Plasticity, DNA Damage Response, and Therapy Resistance. Front. Mol. Biosci. 2020, 7, 36. [Google Scholar] [CrossRef]
- Karihtala, P.; Auvinen, P.; Kauppila, S.; Haapasaari, K.M.; Jukkola-Vuorinen, A.; Soini, Y. Vimentin, zeb1 and Sip1 are up-regulated in triple-negative and basal-like breast cancers: Association with an aggressive tumour phenotype. Breast Cancer Res. Treat 2013, 138, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.T.; Zhong, H.T.; Li, G.W.; Shen, J.X.; Ye, Q.Q.; Zhang, M.L.; Liu, J. Oncogenic functions of the EMT-related transcription factor ZEB1 in breast cancer. J. Transl. Med. 2020, 18, 51. [Google Scholar] [CrossRef]
- Llorens, M.C.; Rossi, F.A.; Garcia, I.A.; Cooke, M.; Abba, M.C.; Lopez-Haber, C.; Barrio-Real, L.; Vaglienti, M.V.; Rossi, M.; Bocco, J.L.; et al. PKCalpha Modulates Epithelial-to-Mesenchymal Transition and Invasiveness of Breast Cancer Cells Through ZEB1. Front. Oncol. 2019, 9, 1323. [Google Scholar] [CrossRef]
- Cho, H.J.; Oh, N.; Park, J.H.; Kim, K.S.; Kim, H.K.; Lee, E.; Hwang, S.; Kim, S.J.; Park, K.S. ZEB1 Collaborates with ELK3 to Repress E-Cadherin Expression in Triple-Negative Breast Cancer Cells. Mol. Cancer Res. 2019, 17, 2257–2266. [Google Scholar] [CrossRef]
- Kong, S.Y.; Kim, K.S.; Kim, J.; Kim, M.K.; Lee, K.H.; Lee, J.Y.; Oh, N.; Park, J.I.; Park, J.H.; Heo, S.H.; et al. The ELK3-GATA3 axis orchestrates invasion and metastasis of breast cancer cells in vitro and in vivo. Oncotarget 2016, 7, 65137–65146. [Google Scholar] [CrossRef] [PubMed]
- Dydensborg, A.B.; Rose, A.A.; Wilson, B.J.; Grote, D.; Paquet, M.; Giguere, V.; Siegel, P.M.; Bouchard, M. GATA3 inhibits breast cancer growth and pulmonary breast cancer metastasis. Oncogene 2009, 28, 2634–2642. [Google Scholar] [CrossRef]
- Chu, I.M.; Lai, W.C.; Aprelikova, O.; El Touny, L.H.; Kouros-Mehr, H.; Green, J.E. Expression of GATA3 in MDA-MB-231 triple-negative breast cancer cells induces a growth inhibitory response to TGFss. PLoS ONE 2013, 8, e61125. [Google Scholar] [CrossRef]
- Yan, W.; Cao, Q.J.; Arenas, R.B.; Bentley, B.; Shao, R. GATA3 inhibits breast cancer metastasis through the reversal of epithelial-mesenchymal transition. J. Biol. Chem. 2010, 285, 14042–14051. [Google Scholar] [CrossRef]
- Chakrabarti, R.; Hwang, J.; Andres Blanco, M.; Wei, Y.; Lukacisin, M.; Romano, R.A.; Smalley, K.; Liu, S.; Yang, Q.; Ibrahim, T.; et al. Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2. Nat. Cell Biol. 2012, 14, 1212–1222. [Google Scholar] [CrossRef]
- Casas, E.; Kim, J.; Bendesky, A.; Ohno-Machado, L.; Wolfe, C.J.; Yang, J. Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer Res. 2011, 71, 245–254. [Google Scholar] [CrossRef]
- Gras, B.; Jacqueroud, L.; Wierinckx, A.; Lamblot, C.; Fauvet, F.; Lachuer, J.; Puisieux, A.; Ansieau, S. Snail family members unequally trigger EMT and thereby differ in their ability to promote the neoplastic transformation of mammary epithelial cells. PLoS ONE 2014, 9, e92254. [Google Scholar] [CrossRef] [PubMed]
- Subbalakshmi, A.R.; Sahoo, S.; Biswas, K.; Jolly, M.K. A Computational Systems Biology Approach Identifies SLUG as a Mediator of Partial Epithelial-Mesenchymal Transition (EMT). Cells Tissues Organs 2021, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Cantile, M.; Pettinato, G.; Procino, A.; Feliciello, I.; Cindolo, L.; Cillo, C. In vivo expression of the whole HOX gene network in human breast cancer. Eur. J. Cancer 2003, 39, 257–264. [Google Scholar] [CrossRef]
- De Bessa Garcia, S.A.; Araujo, M.; Pereira, T.; Mouta, J.; Freitas, R. HOX genes function in Breast Cancer development. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188358. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Huang, Q.; Wei, G.H. The Role of HOX Transcription Factors in Cancer Predisposition and Progression. Cancers 2019, 11, 528. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, T.; Takahashi, F.; Chiba, N.; Brachtel, E.; Takahashi, M.; Godin-Heymann, N.; Gross, K.W.; Vivanco, M.; Wijendran, V.; Shioda, T.; et al. HOXB9, a gene overexpressed in breast cancer, promotes tumorigenicity and lung metastasis. Proc. Natl. Acad. Sci. USA 2010, 107, 1100–1105. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Zou, J.; Zhang, M.; Zhang, J.; Xu, S.; Zhu, S.; Yang, M.; Li, D.; Wang, Y.; Shi, J.; et al. Upregulation of MGP by HOXC8 promotes the proliferation, migration, and EMT processes of triple-negative breast cancer. Mol. Carcinog. 2019, 58, 1863–1875. [Google Scholar] [CrossRef]
- Yu, M.; Smolen, G.A.; Zhang, J.; Wittner, B.; Schott, B.J.; Brachtel, E.; Ramaswamy, S.; Maheswaran, S.; Haber, D.A. A developmentally regulated inducer of EMT, LBX1, contributes to breast cancer progression. Genes Dev. 2009, 23, 1737–1742. [Google Scholar] [CrossRef]
- Cieply, B.; Riley, P.t.; Pifer, P.M.; Widmeyer, J.; Addison, J.B.; Ivanov, A.V.; Denvir, J.; Frisch, S.M. Suppression of the epithelial-mesenchymal transition by Grainyhead-like-2. Cancer Res. 2012, 72, 2440–2453. [Google Scholar] [CrossRef]
- Mooney, S.M.; Talebian, V.; Jolly, M.K.; Jia, D.; Gromala, M.; Levine, H.; McConkey, B.J. The GRHL2/ZEB Feedback Loop-A Key Axis in the Regulation of EMT in Breast Cancer. J. Cell Biochem. 2017, 118, 2559–2570. [Google Scholar] [CrossRef]
- Werner, S.; Frey, S.; Riethdorf, S.; Schulze, C.; Alawi, M.; Kling, L.; Vafaizadeh, V.; Sauter, G.; Terracciano, L.; Schumacher, U.; et al. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. J. Biol. Chem. 2013, 288, 22993–23008. [Google Scholar] [CrossRef]
- Jolly, M.K.; Somarelli, J.A.; Sheth, M.; Biddle, A.; Tripathi, S.C.; Armstrong, A.J.; Hanash, S.M.; Bapat, S.A.; Rangarajan, A.; Levine, H. Hybrid epithelial/mesenchymal phenotypes promote metastasis and therapy resistance across carcinomas. Pharmacol. Ther. 2019, 194, 161–184. [Google Scholar] [CrossRef]
- Vervoort, S.J.; Lourenco, A.R.; van Boxtel, R.; Coffer, P.J. SOX4 mediates TGF-beta-induced expression of mesenchymal markers during mammary cell epithelial to mesenchymal transition. PLoS ONE 2013, 8, e53238. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.M.; Panzilius, E.; Bartsch, H.S.; Irmler, M.; Beckers, J.; Kari, V.; Linnemann, J.R.; Dragoi, D.; Hirschi, B.; Kloos, U.J.; et al. Stem-cell-like properties and epithelial plasticity arise as stable traits after transient Twist1 activation. Cell Rep. 2015, 10, 131–139. [Google Scholar] [CrossRef]
- Grosse-Wilde, A.; Fouquier d’Herouel, A.; McIntosh, E.; Ertaylan, G.; Skupin, A.; Kuestner, R.E.; del Sol, A.; Walters, K.A.; Huang, S. Stemness of the hybrid Epithelial/Mesenchymal State in Breast Cancer and Its Association with Poor Survival. PLoS ONE 2015, 10, e0126522. [Google Scholar] [CrossRef] [PubMed]
- Grosse-Wilde, A.; Kuestner, R.E.; Skelton, S.M.; MacIntosh, E.; d′Herouel, A.F.; Ertaylan, G.; Del Sol, A.; Skupin, A.; Huang, S. Loss of inter-cellular cooperation by complete epithelial-mesenchymal transition supports favorable outcomes in basal breast cancer patients. Oncotarget 2018, 9, 20018–20033. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Mauri, F.; Song, Y.; de Cock, F.; Meeusen, B.; Swedlund, B.; Impens, F.; Van Haver, D.; Opitz, M.; Thery, M.; et al. Fat1 deletion promotes hybrid EMT state, tumour stemness and metastasis. Nature 2021, 589, 448–455. [Google Scholar] [CrossRef]
- Bushweller, J.H. Targeting transcription factors in cancer—From undruggable to reality. Nat. Rev. Cancer 2019, 19, 611–624. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, A.; Scott, J.G.; Harris, A.L.; Buffa, F.M. Pan-cancer characterisation of microRNA across cancer hallmarks reveals microRNA-mediated downregulation of tumour suppressors. Nat. Commun. 2018, 9, 5228. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Hurteau, G.J.; Carlson, J.A.; Spivack, S.D.; Brock, G.J. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007, 67, 7972–7976. [Google Scholar] [CrossRef] [PubMed]
- Korpal, M.; Lee, E.S.; Hu, G.; Kang, Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 2008, 283, 14910–14914. [Google Scholar] [CrossRef]
- Pattabiraman, D.R.; Bierie, B.; Kober, K.I.; Thiru, P.; Krall, J.A.; Zill, C.; Reinhardt, F.; Tam, W.L.; Weinberg, R.A. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science 2016, 351, aad3680. [Google Scholar] [CrossRef]
- Brabletz, S.; Brabletz, T. The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO Rep. 2010, 11, 670–677. [Google Scholar] [CrossRef]
- Jolly, M.K.; Celia-Terrassa, T. Dynamics of Phenotypic Heterogeneity Associated with EMT and Stemness during Cancer Progression. J. Clin. Med. 2019, 8, 1542. [Google Scholar] [CrossRef]
- Rogers, T.J.; Christenson, J.L.; Greene, L.I.; O’Neill, K.I.; Williams, M.M.; Gordon, M.A.; Nemkov, T.; D’Alessandro, A.; Degala, G.D.; Shin, J.; et al. Reversal of Triple-Negative Breast Cancer EMT by miR-200c Decreases Tryptophan Catabolism and a Program of Immunosuppression. Mol. Cancer Res. 2019, 17, 30–41. [Google Scholar] [CrossRef]
- Tryndyak, V.P.; Beland, F.A.; Pogribny, I.P. E-cadherin transcriptional down-regulation by epigenetic and microRNA-200 family alterations is related to mesenchymal and drug-resistant phenotypes in human breast cancer cells. Int. J. Cancer 2010, 126, 2575–2583. [Google Scholar] [CrossRef]
- Wang, Q.; Cheng, Y.; Wang, Y.; Fan, Y.; Li, C.; Zhang, Y.; Dong, Q.; Ma, Y.; Teng, Y.E.; Qu, X.; et al. Tamoxifen reverses epithelial-mesenchymal transition by demethylating miR-200c in triple-negative breast cancer cells. BMC Cancer 2017, 17, 492. [Google Scholar] [CrossRef] [PubMed]
- Korpal, M.; Ell, B.J.; Buffa, F.M.; Ibrahim, T.; Blanco, M.A.; Celia-Terrassa, T.; Mercatali, L.; Khan, Z.; Goodarzi, H.; Hua, Y.; et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med. 2011, 17, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Dykxhoorn, D.M.; Wu, Y.; Xie, H.; Yu, F.; Lal, A.; Petrocca, F.; Martinvalet, D.; Song, E.; Lim, B.; Lieberman, J. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS ONE 2009, 4, e7181. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Shin, V.Y.; Siu, M.T.; Ho, J.C.; Cheuk, I.; Kwong, A. miR-199a-5p confers tumor-suppressive role in triple-negative breast cancer. BMC Cancer 2016, 16, 887. [Google Scholar] [CrossRef]
- Wu, J.; Li, W.Z.; Huang, M.L.; Wei, H.L.; Wang, T.; Fan, J.; Li, N.L.; Ling, R. Regulation of cancerous progression and epithelial-mesenchymal transition by miR-34c-3p via modulation of MAP3K2 signaling in triple-negative breast cancer cells. Biochem. Biophys. Res. Commun. 2017, 483, 10–16. [Google Scholar] [CrossRef]
- Wang, J.; Li, M.; Han, X.; Wang, H.; Wang, X.; Ma, G.; Xia, T.; Wang, S. MiR-1976 knockdown promotes epithelial-mesenchymal transition and cancer stem cell properties inducing triple-negative breast cancer metastasis. Cell Death Dis. 2020, 11, 500. [Google Scholar] [CrossRef]
- Jang, M.H.; Kim, H.J.; Gwak, J.M.; Chung, Y.R.; Park, S.Y. Prognostic value of microRNA-9 and microRNA-155 expression in triple-negative breast cancer. Hum. Pathol. 2017, 68, 69–78. [Google Scholar] [CrossRef]
- Jolly, M.K.; Ware, K.E.; Gilja, S.; Somarelli, J.A.; Levine, H. EMT and MET: Necessary or permissive for metastasis? Mol. Oncol. 2017, 11, 755–769. [Google Scholar] [CrossRef]
- Bukholm, I.K.; Nesland, J.M.; Borresen-Dale, A.L. Re-expression of E-cadherin, alpha-catenin and beta-catenin, but not of gamma-catenin, in metastatic tissue from breast cancer patients. J. Pathol. 2000, 190, 15–19. [Google Scholar] [CrossRef]
- Kowalski, P.J.; Rubin, M.A.; Kleer, C.G. E-cadherin expression in primary carcinomas of the breast and its distant metastases. Breast Cancer Res. 2003, 5, R217–R222. [Google Scholar] [CrossRef]
- Park, D.; Karesen, R.; Axcrona, U.; Noren, T.; Sauer, T. Expression pattern of adhesion molecules (E-cadherin, alpha-, beta-, gamma-catenin and claudin-7), their influence on survival in primary breast carcinoma, and their corresponding axillary lymph node metastasis. APMIS Acta Pathol. Microbiol. et Immunol. Scand. 2007, 115, 52–65. [Google Scholar] [CrossRef]
- Jia, W.; Deshmukh, A.; Mani, S.A.; Jolly, M.K.; Levine, H. A possible role for epigenetic feedback regulation in the dynamics of the epithelial-mesenchymal transition (EMT). Phys. Biol. 2019, 16, 066004. [Google Scholar] [CrossRef]
- Liu, X.; Li, J.; Cadilha, B.L.; Markota, A.; Voigt, C.; Huang, Z.; Lin, P.P.; Wang, D.D.; Dai, J.; Kranz, G.; et al. Epithelial-type systemic breast carcinoma cells with a restricted mesenchymal transition are a major source of metastasis. Sci. Adv. 2019, 5, eaav4275. [Google Scholar] [CrossRef]
- Khoo, B.L.; Lee, S.C.; Kumar, P.; Tan, T.Z.; Warkiani, M.E.; Ow, S.G.; Nandi, S.; Lim, C.T.; Thiery, J.P. Short-term expansion of breast circulating cancer cells predicts response to anti-cancer therapy. Oncotarget 2015, 6, 15578–15593. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Bocci, F.; Mandal, S.; Tejaswi, T.; Jolly, M.K. Investigating epithelial-mesenchymal heterogeneity of tumors and circulating tumor cells with transcriptomic analysis and biophysical modeling. Comput. Syst. Oncol. 2021, e1015. [Google Scholar] [CrossRef]
- Radovich, M.; Jiang, G.; Hancock, B.A.; Chitambar, C.; Nanda, R.; Falkson, C.; Lynce, F.C.; Gallagher, C.; Isaacs, C.; Blaya, M.; et al. Association of Circulating Tumor DNA and Circulating Tumor Cells After Neoadjuvant Chemotherapy With Disease Recurrence in Patients With Triple-Negative Breast Cancer: Preplanned Secondary Analysis of the BRE12-158 Randomized Clinical Trial. JAMA Oncol. 2020, 6, 1410–1415. [Google Scholar] [CrossRef] [PubMed]
- Gwark, S.C.; Kim, J.; Lee, C.H.; Kim, Y.H.; Kim, M.S.; Hong, S.W.; Choi, M.Y.; Jeon, B.H.; Kwon, N.J.; Kim, K.Y.; et al. Analysis of the serial circulating tumor cell count during neoadjuvant chemotherapy in breast cancer patients. Sci. Rep. 2020, 10, 17466. [Google Scholar] [CrossRef] [PubMed]
- Peeters, D.J.; van Dam, P.J.; Van den Eynden, G.G.; Rutten, A.; Wuyts, H.; Pouillon, L.; Peeters, M.; Pauwels, P.; Van Laere, S.J.; van Dam, P.A.; et al. Detection and prognostic significance of circulating tumour cells in patients with metastatic breast cancer according to immunohistochemical subtypes. Br. J. Cancer 2014, 110, 375–383. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Xenidis, N.; Perraki, M.; Apostolaki, S.; Politaki, E.; Kafousi, M.; Stathopoulos, E.N.; Stathopoulou, A.; Lianidou, E.; Chlouverakis, G.; et al. Different prognostic value of cytokeratin-19 mRNA positive circulating tumor cells according to estrogen receptor and HER2 status in early-stage breast cancer. J. Clin. Oncol. 2007, 25, 5194–5202. [Google Scholar] [CrossRef]
- Micalizzi, D.S.; Haber, D.A.; Maheswaran, S. Cancer metastasis through the prism of epithelial-to-mesenchymal transition in circulating tumor cells. Mol. Oncol. 2017, 11, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Ramani, V.C.; Lemaire, C.A.; Triboulet, M.; Casey, K.M.; Heirich, K.; Renier, C.; Vilches-Moure, J.G.; Gupta, R.; Razmara, A.M.; Zhang, H.; et al. Investigating circulating tumor cells and distant metastases in patient-derived orthotopic xenograft models of triple-negative breast cancer. Breast Cancer Res. 2019, 21, 98. [Google Scholar] [CrossRef] [PubMed]
- Sikandar, S.S.; Kuo, A.H.; Kalisky, T.; Cai, S.; Zabala, M.; Hsieh, R.W.; Lobo, N.A.; Scheeren, F.A.; Sim, S.; Qian, D.; et al. Role of epithelial to mesenchymal transition associated genes in mammary gland regeneration and breast tumorigenesis. Nat. Commun. 2017, 8, 1669. [Google Scholar] [CrossRef] [PubMed]
- Hugo, H.J.; Gunasinghe, N.; Hollier, B.G.; Tanaka, T.; Blick, T.; Toh, A.; Hill, P.; Gilles, C.; Waltham, M.; Thompson, E.W. Epithelial requirement for in vitro proliferation and xenograft growth and metastasis of MDA-MB-468 human breast cancer cells: Oncogenic rather than tumor-suppressive role of E-cadherin. Breast Cancer Res. 2017, 19, 86. [Google Scholar] [CrossRef] [PubMed]
- Padmanaban, V.; Krol, I.; Suhail, Y.; Szczerba, B.M.; Aceto, N.; Bader, J.S.; Ewald, A.J. E-cadherin is required for metastasis in multiple models of breast cancer. Nature 2019, 573, 439–444. [Google Scholar] [CrossRef]
- Bonnomet, A.; Syne, L.; Brysse, A.; Feyereisen, E.; Thompson, E.W.; Noel, A.; Foidart, J.M.; Birembaut, P.; Polette, M.; Gilles, C. A dynamic in vivo model of epithelial-to-mesenchymal transitions in circulating tumor cells and metastases of breast cancer. Oncogene 2012, 31, 3741–3753. [Google Scholar] [CrossRef]
- Stankic, M.; Pavlovic, S.; Chin, Y.; Brogi, E.; Padua, D.; Norton, L.; Massague, J.; Benezra, R. TGF-beta-Id1 signaling opposes Twist1 and promotes metastatic colonization via a mesenchymal-to-epithelial transition. Cell Rep. 2013, 5, 1228–1242. [Google Scholar] [CrossRef]
- Gao, D.; Joshi, N.; Choi, H.; Ryu, S.; Hahn, M.; Catena, R.; Sadik, H.; Argani, P.; Wagner, P.; Vahdat, L.T.; et al. Myeloid progenitor cells in the premetastatic lung promote metastases by inducing mesenchymal to epithelial transition. Cancer Res. 2012, 72, 1384–1394. [Google Scholar] [CrossRef]
- Carey, L.A.; Dees, E.C.; Sawyer, L.; Gatti, L.; Moore, D.T.; Collichio, F.; Ollila, D.W.; Sartor, C.I.; Graham, M.L.; Perou, C.M. The triple negative paradox: Primary tumor chemosensitivity of breast cancer subtypes. Clin. Cancer Res. 2007, 13, 2329–2334. [Google Scholar] [CrossRef] [PubMed]
- Liedtke, C.; Mazouni, C.; Hess, K.R.; Andre, F.; Tordai, A.; Mejia, J.A.; Symmans, W.F.; Gonzalez-Angulo, A.M.; Hennessy, B.; Green, M.; et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 2008, 26, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Bhola, N.E.; Balko, J.M.; Dugger, T.C.; Kuba, M.G.; Sanchez, V.; Sanders, M.; Stanford, J.; Cook, R.S.; Arteaga, C.L. TGF-beta inhibition enhances chemotherapy action against triple-negative breast cancer. J. Clin. Investig. 2013, 123, 1348–1358. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Kim, M.J.; Park, S.A.; Kim, J.S.; Min, K.N.; Kim, D.K.; Lim, W.; Nam, J.S.; Sheen, Y.Y. Combinatorial TGF-beta attenuation with paclitaxel inhibits the epithelial-to-mesenchymal transition and breast cancer stem-like cells. Oncotarget 2015, 6, 37526–37543. [Google Scholar] [CrossRef]
- Bandyopadhyay, A.; Wang, L.; Agyin, J.; Tang, Y.; Lin, S.; Yeh, I.T.; De, K.; Sun, L.Z. Doxorubicin in combination with a small TGFbeta inhibitor: A potential novel therapy for metastatic breast cancer in mouse models. PLoS ONE 2010, 5, e10365. [Google Scholar] [CrossRef] [PubMed]
- Raza, U.; Saatci, O.; Uhlmann, S.; Ansari, S.A.; Eyupoglu, E.; Yurdusev, E.; Mutlu, M.; Ersan, P.G.; Altundag, M.K.; Zhang, J.D.; et al. The miR-644a/CTBP1/p53 axis suppresses drug resistance by simultaneous inhibition of cell survival and epithelial-mesenchymal transition in breast cancer. Oncotarget 2016, 7, 49859–49877. [Google Scholar] [CrossRef]
- Yoshida, T.; Ozawa, Y.; Kimura, T.; Sato, Y.; Kuznetsov, G.; Xu, S.; Uesugi, M.; Agoulnik, S.; Taylor, N.; Funahashi, Y.; et al. Eribulin mesilate suppresses experimental metastasis of breast cancer cells by reversing phenotype from epithelial-mesenchymal transition (EMT) to mesenchymal-epithelial transition (MET) states. Br. J. Cancer 2014, 110, 1497–1505. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, C.; McCampbell, K.K.; David, J.M.; Palena, C. Neutralization of IL-8 decreases tumor PMN-MDSCs and reduces mesenchymalization of claudin-low triple-negative breast cancer. JCI Insight 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Alsuliman, A.; Colak, D.; Al-Harazi, O.; Fitwi, H.; Tulbah, A.; Al-Tweigeri, T.; Al-Alwan, M.; Ghebeh, H. Bidirectional crosstalk between PD-L1 expression and epithelial to mesenchymal transition: Significance in claudin-low breast cancer cells. Mol. Cancer 2015, 14, 149. [Google Scholar] [CrossRef]
- Williams, E.D.; Gao, D.C.; Redfern, N.; Thompson, E.W. Controversies around epithelial-mesenchymal plasticity in cancer metastasis. Nat. Rev. Cancer 2019, 19, 716–732. [Google Scholar] [CrossRef]
- Hari, K.; Sabuwala, B.; Subramani, B.V.; La Porta, C.A.M.; Zapperi, S.; Font-Clos, F.; Jolly, M.K. Identifying inhibitors of epithelial-mesenchymal plasticity using a network topology-based approach. NPJ Syst. Biol. Appl. 2020, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Celia-Terrassa, T.; Bastian, C.; Liu, D.D.; Ell, B.; Aiello, N.M.; Wei, Y.; Zamalloa, J.; Blanco, A.M.; Hang, X.; Kunisky, D.; et al. Author Correction: Hysteresis control of epithelial-mesenchymal transition dynamics conveys a distinct program with enhanced metastatic ability. Nat. Commun. 2019, 10, 527. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).