RAD50 Loss of Function Variants in the Zinc Hook Domain Associated with Higher Risk of Familial Esophageal Squamous Cell Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. WES, Target Capture Sequencing, and Bioinformatics Analysis
2.2. Statistical Analysis
2.3. Cell Culture
2.4. RAD50 Constructs, Lentiviral Preparation, and Transduction
2.5. Western Blotting
2.6. Ionizing Radiation and Immunofluorescence Staining
2.7. Cell Viability and Colony Formation Assays
3. Results
3.1. WES Analysis Prioritizes RAD50 as Top Candidate Cancer Predisposition Gene (CPG) for Familial ESCC
3.2. Two RAD50 LOF Variants at the Zinc Hook Domain Associate with Increased Risk of Familial ESCC
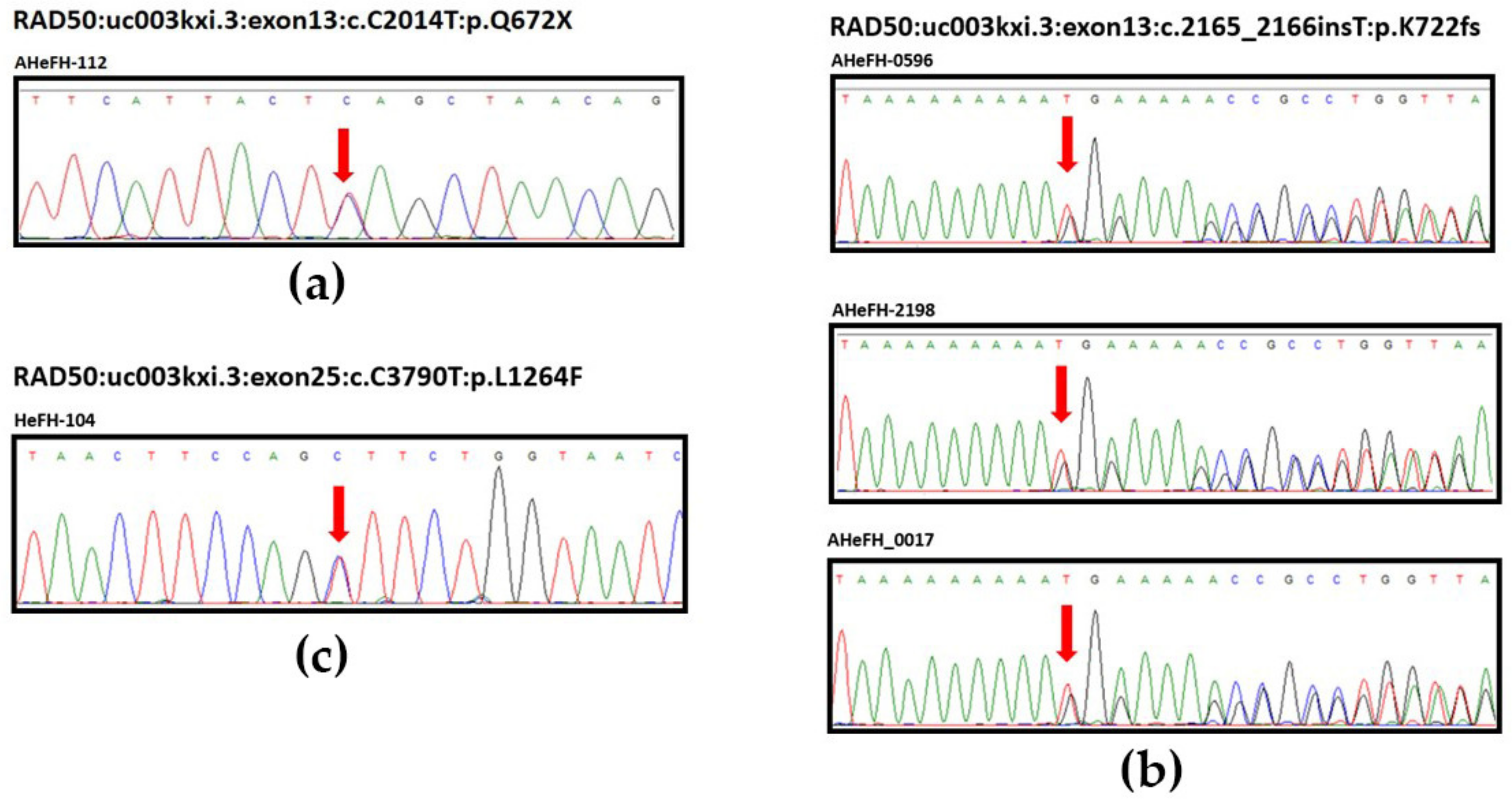
3.3. Sanger Sequencing Validation of RAD50 Germline Variants
3.4. RAD50 Is Indispensable for the Survival of ESCC Cells
3.5. Dominant Negative Overexpression of RAD50Q672X Mutant Delays the Repair of IR-Induced DSBs
3.6. Dominant Negative Overexpression of RAD50 Mutants Sensitize Cells to Formaldehyde and CHK1 Inhibitor AZD7762 Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Abnet, C.C.; Arnold, M.; Wei, W.Q. Epidemiology of Esophageal Squamous Cell Carcinoma. Gastroenterology 2018, 154, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.D.; Zhou, F.Y.; Li, X.M.; Sun, L.D.; Song, X.; Jin, Y.; Li, J.M.; Kong, G.Q.; Qi, H.; Cui, J.; et al. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat. Genet. 2010, 42, 759–763. [Google Scholar] [CrossRef]
- Wu, C.; Wang, Z.; Song, X.; Feng, X.S.; Abnet, C.C.; He, J.; Hu, N.; Zuo, X.B.; Tan, W.; Zhan, Q.; et al. Joint analysis of three genome-wide association studies of esophageal squamous cell carcinoma in Chinese populations. Nat. Genet. 2014, 46, 1001–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, R.; Kamatani, Y.; Takahashi, A.; Usami, M.; Hosono, N.; Kawaguchi, T.; Tsunoda, T.; Kamatani, N.; Kubo, M.; Nakamura, Y.; et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology 2009, 137, 1768–1775. [Google Scholar] [CrossRef] [PubMed]
- Gupta, B.; Kumar, N. Worldwide incidence, mortality and time trends for cancer of the oesophagus. Eur. J. Cancer Prev. 2017, 26, 107–118. [Google Scholar] [CrossRef]
- Dawsey, S.M.; Wang, G.Q.; Weinstein, W.M.; Lewin, K.J.; Liu, F.S.; Wiggett, S.; Nieberg, R.K.; Li, J.Y.; Taylor, P.R. Squamous dysplasia and early esophageal cancer in the Linxian region of China: Distinctive endoscopic lesions. Gastroenterology 1993, 105, 1333–1340. [Google Scholar] [CrossRef]
- Wei, W.Q.; Chen, Z.F.; He, Y.T.; Feng, H.; Hou, J.; Lin, D.M.; Li, X.Q.; Guo, C.L.; Li, S.S.; Wang, G.Q.; et al. Long-Term Follow-Up of a Community Assignment, One-Time Endoscopic Screening Study of Esophageal Cancer in China. J. Clin. Oncol. 2015, 33, 1951–1957. [Google Scholar] [CrossRef]
- He, Z.; Liu, Z.; Liu, M.; Guo, C.; Xu, R.; Li, F.; Liu, A.; Yang, H.; Shen, L.; Wu, Q.; et al. Efficacy of endoscopic screening for esophageal cancer in China (ESECC): Design and preliminary results of a population-based randomised controlled trial. Gut 2019, 68, 198–206. [Google Scholar] [CrossRef]
- Yokoyama, A.; Ohmori, T.; Makuuchi, H.; Maruyama, K.; Okuyama, K.; Takahashi, H.; Yokoyama, T.; Yoshino, K.; Hayashida, M.; Ishii, H. Successful screening for early esophageal cancer in alcoholics using endoscopy and mucosa iodine staining. Cancer 1995, 76, 928–934. [Google Scholar] [CrossRef]
- Hashimoto, C.L.; Iriya, K.; Baba, E.R.; Navarro-Rodriguez, T.; Zerbini, M.C.; Eisig, J.N.; Barbuti, R.; Chinzon, D.; Moraes-Filho, J.P. Lugol’s dye spray chromoendoscopy establishes early diagnosis of esophageal cancer in patients with primary head and neck cancer. Am. J. Gastroenterol. 2005, 100, 275–282. [Google Scholar] [CrossRef]
- Dubuc, J.; Legoux, J.; Winnock, M.; Seyrig, J.; Barbier, J.; Barrioz, T.; Laugier, R.; Boulay, G.; Grasset, D.; Sautereau, D.; et al. Endoscopic screening for esophageal squamous-cell carcinoma in high-risk patients: A prospective study conducted in 62 French endoscopy centers. Endoscopy 2006, 38, 690–695. [Google Scholar] [CrossRef] [PubMed]
- Chang-Claude, J.; Becher, H.; Blettner, M.; Qiu, S.; Yang, G.; Wahrendorf, J. Familial aggregation of oesophageal cancer in a high incidence area in China. Int. J. Epidemiol. 1997, 26, 1159–1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, N.; Dawsey, S.M.; Wu, M.; Bonney, G.E.; He, L.J.; Han, X.Y.; Fu, M.; Taylor, P.R. Familial aggregation of oesophageal cancer in Yangcheng County, Shanxi Province, China. Int. J. Epidemiol. 1992, 21, 877–882. [Google Scholar] [CrossRef]
- Ko, J.M.; Ning, L.; Zhao, X.K.; Chai, A.W.Y.; Lei, L.C.; Choi, S.S.A.; Tao, L.; Law, S.; Kwong, A.; Lee, N.P.; et al. BRCA2 loss-of-function germline mutations are associated with esophageal squamous cell carcinoma risk in Chinese. Int. J. Cancer 2020, 146, 1042–1051. [Google Scholar] [CrossRef]
- Syed, A.; Tainer, J.A. The MRE11-RAD50-NBS1 Complex Conducts the Orchestration of Damage Signaling and Outcomes to Stress in DNA Replication and Repair. Annu. Rev. Biochem. 2018, 87, 263–294. [Google Scholar] [CrossRef]
- Van Den Bosch, M.; Bree, R.T.; Lowndes, N.F. The MRN complex: Coordinating and mediating the response to broken chromosomes. EMBO Rep. 2003, 4, 844–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robison, J.G.; Elliott, J.; Dixon, K.; Oakley, G.G. Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J. Biol. Chem. 2004, 279, 34802–34810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quennet, V.; Beucher, A.; Barton, O.; Takeda, S.; Lobrich, M. CtIP and MRN promote non-homologous end-joining of etoposide-induced DNA double-strand breaks in G1. Nucleic Acids Res. 2011, 39, 2144–2152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paull, T.T.; Lee, J.H. The Mre11/Rad50/Nbs1 complex and its role as a DNA double-strand break sensor for ATM. Cell Cycle 2005, 4, 737–740. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Paull, T.T. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 2005, 308, 551–554. [Google Scholar] [CrossRef]
- Ragamin, A.; Yigit, G.; Bousset, K.; Beleggia, F.; Verheijen, F.W.; De Wit, M.Y.; Strom, T.M.; Dork, T.; Wollnik, B.; Mancini, G.M.S. Human RAD50 deficiency: Confirmation of a distinctive phenotype. Am. J. Med. Genet. A 2020, 182, 1378–1386. [Google Scholar] [CrossRef] [Green Version]
- Barbi, G.; Scheres, J.M.; Schindler, D.; Taalman, R.D.; Rodens, K.; Mehnert, K.; Muller, M.; Seyschab, H. Chromosome instability and X-ray hypersensitivity in a microcephalic and growth-retarded child. Am. J. Med. Genet. 1991, 40, 44–50. [Google Scholar] [CrossRef]
- Waltes, R.; Kalb, R.; Gatei, M.; Kijas, A.W.; Stumm, M.; Sobeck, A.; Wieland, B.; Varon, R.; Lerenthal, Y.; Lavin, M.F.; et al. Human RAD50 deficiency in a Nijmegen breakage syndrome-like disorder. Am. J. Hum. Genet. 2009, 84, 605–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chansel-Da Cruz, M.; Hohl, M.; Ceppi, I.; Kermasson, L.; Maggiorella, L.; Modesti, M.; De Villartay, J.P.; Ileri, T.; Cejka, P.; Petrini, J.H.J.; et al. A Disease-Causing Single Amino Acid Deletion in the Coiled-Coil Domain of RAD50 Impairs MRE11 Complex Functions in Yeast and Humans. Cell Rep. 2020, 33, 108559. [Google Scholar] [CrossRef]
- Heikkinen, K.; Rapakko, K.; Karppinen, S.M.; Erkko, H.; Knuutila, S.; Lundan, T.; Mannermaa, A.; Borresen-Dale, A.L.; Borg, A.; Barkardottir, R.B.; et al. RAD50 and NBS1 are breast cancer susceptibility genes associated with genomic instability. Carcinogenesis 2006, 27, 1593–1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, T.; King, M.C. Ten genes for inherited breast cancer. Cancer Cell 2007, 11, 103–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damiola, F.; Pertesi, M.; Oliver, J.; Le Calvez-Kelm, F.; Voegele, C.; Young, E.L.; Robinot, N.; Forey, N.; Durand, G.; Vallee, M.P.; et al. Rare key functional domain missense substitutions in MRE11A, RAD50, and NBN contribute to breast cancer susceptibility: Results from a Breast Cancer Family Registry case-control mutation-screening study. Breast Cancer Res. 2014, 16, R58. [Google Scholar] [CrossRef] [Green Version]
- Fan, C.; Zhang, J.; Ouyang, T.; Li, J.; Wang, T.; Fan, Z.; Fan, T.; Lin, B.; Xie, Y. RAD50 germline mutations are associated with poor survival in BRCA1/2-negative breast cancer patients. Int. J. Cancer 2018, 143, 1935–1942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bender, C.F.; Sikes, M.L.; Sullivan, R.; Huye, L.E.; Le Beau, M.M.; Roth, D.B.; Mirzoeva, O.K.; Oltz, E.M.; Petrini, J.H. Cancer predisposition and hematopoietic failure in Rad50(S/S) mice. Genes Dev. 2002, 16, 2237–2251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abuzeid, W.M.; Jiang, X.; Shi, G.; Wang, H.; Paulson, D.; Araki, K.; Jungreis, D.; Carney, J.; O’Malley, B.W., Jr.; Li, D. Molecular disruption of RAD50 sensitizes human tumor cells to cisplatin-based chemotherapy. J. Clin. Investig. 2009, 119, 1974–1985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Zheng, H.; Cheung, A.K.; Tang, C.S.; Ko, J.M.; Wong, B.W.; Leong, M.M.; Sham, P.C.; Cheung, F.; Kwong, D.L.; et al. Whole-exome sequencing identifies MST1R as a genetic susceptibility gene in nasopharyngeal carcinoma. Proc. Natl. Acad. Sci. USA 2016, 113, 3317–3322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, S.H.; Yee Ko, J.M.; Chan, K.W.; Chan, Y.P.; Tao, Q.; Hyytiainen, M.; Keski-Oja, J.; Law, S.; Srivastava, G.; Tang, J.; et al. The ECM protein LTBP-2 is a suppressor of esophageal squamous cell carcinoma tumor formation but higher tumor expression associates with poor patient outcome. Int. J. Cancer 2011, 129, 565–573. [Google Scholar] [CrossRef]
- Yu, V.Z.; Wong, V.C.; Dai, W.; Ko, J.M.; Lam, A.K.; Chan, K.W.; Samant, R.S.; Lung, H.L.; Shuen, W.H.; Law, S.; et al. Nuclear Localization of DNAJB6 Is Associated With Survival of Patients With Esophageal Cancer and Reduces AKT Signaling and Proliferation of Cancer Cells. Gastroenterology 2015, 149, 1825–1836. [Google Scholar] [CrossRef] [Green Version]
- Yu, V.Z.; Ko, J.M.Y.; Ning, L.; Dai, W.; Law, S.; Lung, M.L. Endoplasmic reticulum-localized ECM1b suppresses tumor growth and regulates MYC and MTORC1 through modulating MTORC2 activation in esophageal squamous cell carcinoma. Cancer Lett. 2019, 461, 56–64. [Google Scholar] [CrossRef]
- Ng, H.Y.; Ko, J.M.; Yu, V.Z.; Ip, J.C.; Dai, W.; Cal, S.; Lung, M.L. DESC1, a novel tumor suppressor, sensitizes cells to apoptosis by downregulating the EGFR/AKT pathway in esophageal squamous cell carcinoma. Int. J. Cancer 2016, 138, 2940–2951. [Google Scholar] [CrossRef] [Green Version]
- National Center for Biotechnology Information. ClinVar; [VCV000484641.5]. Available online: https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000484641.5 (accessed on 9 September 2021).
- National Center for Biotechnology Information. ClinVar; [VCV000408395.6]. Available online: https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000408395.6 (accessed on 9 September 2021).
- Gatei, M.; Kijas, A.W.; Biard, D.; Dork, T.; Lavin, M.F. RAD50 phosphorylation promotes ATR downstream signaling and DNA restart following replication stress. Hum. Mol. Genet. 2014, 23, 4232–4248. [Google Scholar] [CrossRef] [Green Version]
- National Center for Biotechnology Information. ClinVar; [VCV000141646.7]. Available online: https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000141646.7 (accessed on 9 September 2021).
- National Center for Biotechnology Information. ClinVar; [VCV000240219.5]. Available online: https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000240219.5 (accessed on 9 September 2021).
- National Center for Biotechnology Information. ClinVar; [VCV000486238.6]. Available online: https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000486238.6 (accessed on 9 September 2021).
- Lajud, S.A.; Nagda, D.A.; Yamashita, T.; Zheng, J.; Tanaka, N.; Abuzeid, W.M.; Civantos, A.; Bezpalko, O.; O’Malley, B.W., Jr.; Li, D. Dual disruption of DNA repair and telomere maintenance for the treatment of head and neck cancer. Clin. Cancer Res. 2014, 20, 6465–6478. [Google Scholar] [CrossRef] [Green Version]
- Al-Ahmadie, H.; Iyer, G.; Hohl, M.; Asthana, S.; Inagaki, A.; Schultz, N.; Hanrahan, A.J.; Scott, S.N.; Brannon, A.R.; Mcdermott, G.C.; et al. Synthetic lethality in ATM-deficient RAD50-mutant tumors underlies outlier response to cancer therapy. Cancer Discov. 2014, 4, 1014–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Populations | Phase | Definition | Number of Individuals | Number of Alleles (Number of Individuals × 2) |
|---|---|---|---|---|
| Family history-positive (FH+) ESCC | Discovery | ESCC patients with two generations and ≥2 family members diagnosed with ESCC including proband from Henan | 186 | 372 |
| FH+ ESCC | Validation | ESCC patients with one generation and ≥2 family members diagnosed with ESCC including proband from Henan | 858 | 1716 |
| FH+ ESCC | Combined discovery and validation | ESCC patients with one or two generations and ≥2 family members diagnosed with ESCC including proband from Henan | 1044 | 2088 |
| Sporadic ESCC | Validation | ESCC patients without known history of ESCC in the family members from Henan | 1074 | 2148 |
| Control | Validation | Non-ESCC individuals from Henan | 1171 | 2342 |
| Non-FH+ control population | Validation | Combined sporadic ESCC and control populations | 2245 | 4490 |
| gnomAD East Asian | - | East Asian population from gnomAD, https://gnomad.broadinstitute.org/ accessed on 5 June 2020 | 9977 | 19,954 |
| gnomAD All | - | All populations from gnomAD including African, Latino, Jewish, European, East and South Asian https://gnomad.broadinstitute.org/ accessed on 5 June 2020 | 141,335 | 282,670 |
| Chr5 (hg19) | Mutation (DNA) a | Exon | Protein Change | Familial ESCC Cases (1044) Discovery FH+ (186) (n = 372) Validation FH+ (858) (n = 1716) | Validation Sporadic ESCC Cases (1074) (n = 2148) | Validation Controls (1177) (n = 2342) | gnomAD b East Asian (9977) (n = 19954) | p | OR |
|---|---|---|---|---|---|---|---|---|---|
| Whole-exome sequencing of discovery cohort of 186 FH+ ESCC involved two generations | |||||||||
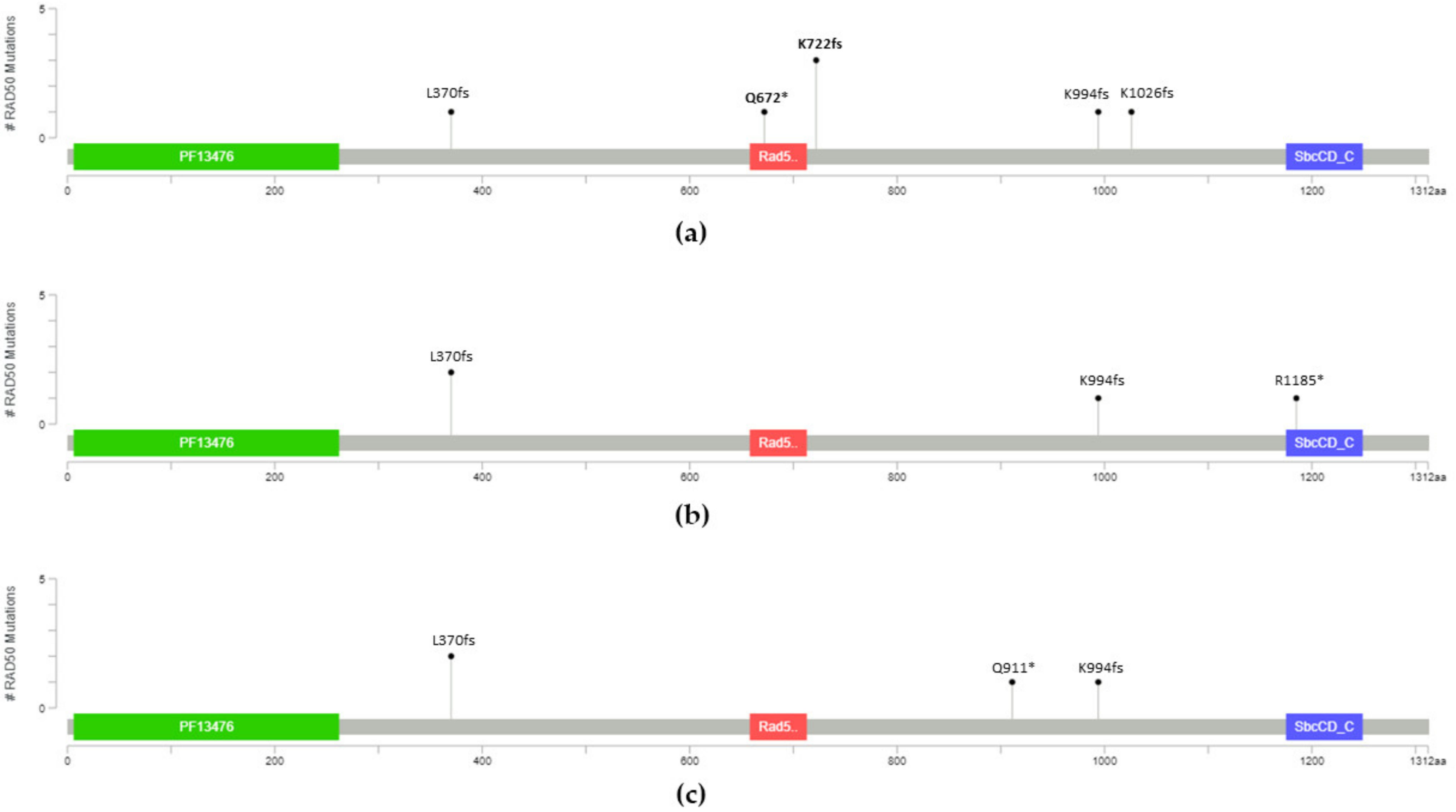
| 131,931,309 | c.C2014T | 13 | p.Q672X | 1 | NA | NA | 0 | ||
| 131,931,460 | c.2165_2166insT | 13 | p.K722fs | 1 | NA | NA | 4 | ||
| 131,945,032 | c.2980_2983del | 19 | p.K994fs | 1 | NA | NA | 8 | ||
| 131,951,735 | c.3077_3080del | 20 | p.K1026fs | 1 | NA | NA | Not reported | ||
| Total Discovery | 4/372 | 30/19954 | 3.3 × 10−3 c | 7.22 c | |||||
| Target whole-gene sequencing of RAD50 in validation cohort of 3103 individuals containing 858 FH+ and 1074 sporadic ESCC and 1171 controls | |||||||||
| 131,924,437 | c.1110delA | 8 | p.L370fs | 1 | 2 | 2 | Not reported g | 0.37 d | 1.97 d |
| 13,193,1460 [28] | c.2165_2166insT | 13 | p.K722fs | 2 | 0 | 0 | 4 | ||
| 131,944,319 | c.C2731T | 17 | p.Q911X | 0 | 0 | 1 | Not reported | 0.38 e | 1.80 e |
| 131,945,032 | c.2980_2983del | 19 | p.K994fs | 0 | 1 | 1 | 8 | ||
| 131,973,850 | c.C3553T | 23 | p.R1185X | 0 | 1 | 0 | 1 | 1.00 f | 1.09 f |
| Combined Total | 7/2088 | 4/2148 | 4/2342 | ||||||
| Mutation | Familial ESCC (n = 2088) | Sporadic ESCC (n = 2148) | Controls (n = 2342) | pa | OR a | gnomAD d East Asian (n = 19,954) | pb | OR b | gnomAD d All (n = 282,670) | pc | OR c |
|---|---|---|---|---|---|---|---|---|---|---|---|
| p.Q672X | 0.045% (1) | 0% | 0% | 0.32 | inf | 0% | 0.095 | inf | 0.00035% (1) | 0.015 | 135.44 |
| p.K722fs | 0.14% (3) | 0% | 0% | 0.032 | inf | 0.02% (4) | 0.022 | 7.18 | 0.0014% (4) | 1.3 × 10−5 | 101.73 |
| p.Q672X/ p.K722fs | 0.19% (4) | 0% | 0% | 0.010 | inf | 0.02% (4) | 4.1 × 10−3 | 9.57 | 0.0018% (5) | 3.5 × 10−7 | 108.51 |
| All RAD50 LOF | 0.34% (7) | 0.19% (4) | 0.17% (4) | 0.33 | 1.88 | 0.15% (30) | 0.092 | 2.23 | 0.17% (427) | 0.062 | 2.22 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ko, J.M.Y.; Lam, S.Y.; Ning, L.; Chai, A.W.Y.; Lei, L.C.; Choi, S.S.A.; Wong, C.W.Y.; Lung, M.L. RAD50 Loss of Function Variants in the Zinc Hook Domain Associated with Higher Risk of Familial Esophageal Squamous Cell Carcinoma. Cancers 2021, 13, 4715. https://doi.org/10.3390/cancers13184715
Ko JMY, Lam SY, Ning L, Chai AWY, Lei LC, Choi SSA, Wong CWY, Lung ML. RAD50 Loss of Function Variants in the Zinc Hook Domain Associated with Higher Risk of Familial Esophageal Squamous Cell Carcinoma. Cancers. 2021; 13(18):4715. https://doi.org/10.3390/cancers13184715
Chicago/Turabian StyleKo, Josephine Mun Yee, Shiu Yeung Lam, Lvwen Ning, Annie Wai Yeeng Chai, Lisa Chan Lei, Sheyne Sta Ana Choi, Carissa Wing Yan Wong, and Maria Li Lung. 2021. "RAD50 Loss of Function Variants in the Zinc Hook Domain Associated with Higher Risk of Familial Esophageal Squamous Cell Carcinoma" Cancers 13, no. 18: 4715. https://doi.org/10.3390/cancers13184715
APA StyleKo, J. M. Y., Lam, S. Y., Ning, L., Chai, A. W. Y., Lei, L. C., Choi, S. S. A., Wong, C. W. Y., & Lung, M. L. (2021). RAD50 Loss of Function Variants in the Zinc Hook Domain Associated with Higher Risk of Familial Esophageal Squamous Cell Carcinoma. Cancers, 13(18), 4715. https://doi.org/10.3390/cancers13184715








