Validation of Gene Expression-Based Predictive Biomarkers for Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Microarray-Based Gene Expression Data Analysis
2.2. Selection of Microarray Datasets for Comprehensive Screening (the Discovery Cohorts)
2.3. Selection of Microarray Datasets for Validation (the Validation Cohorts)
2.4. Statistical Analysis
3. Results
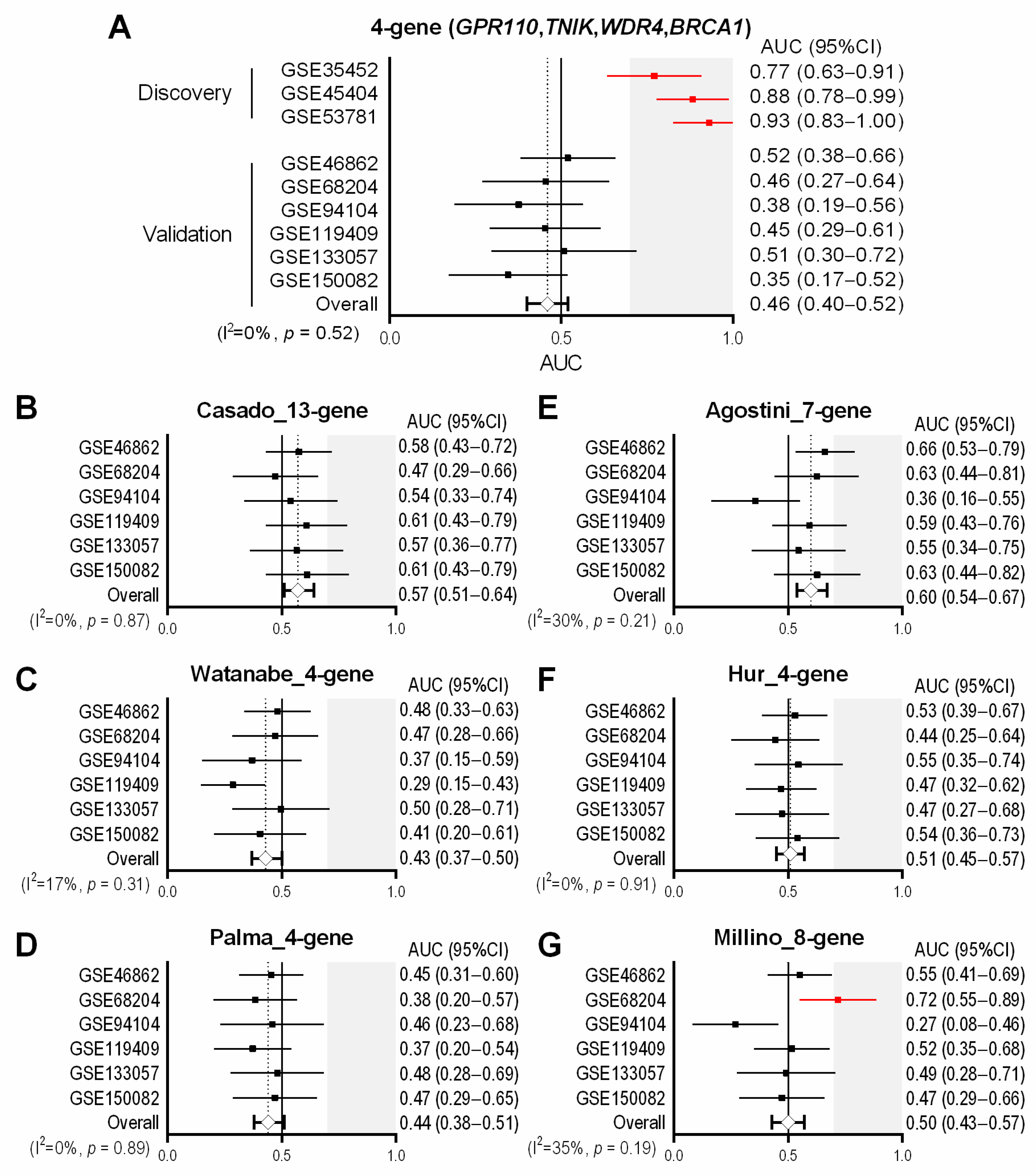
3.1. Identification of Four Common Genes Differentially Expressed between Non-Responders and Responders in the Discovery Datasets
3.2. Assessment of the Predictive Performance of the 4-Gene Signature in the Validation Cohorts
3.3. Previously-Published Multi-Gene Signatures in the Validation Cohorts
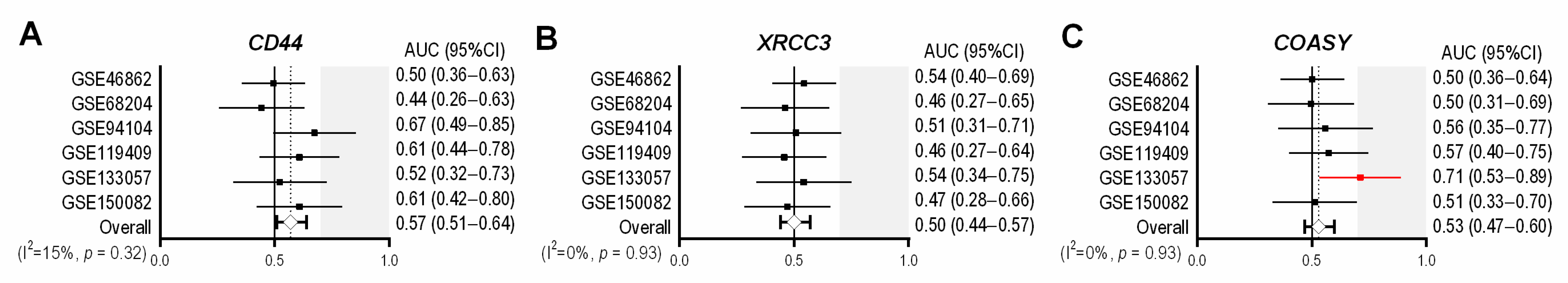
3.4. Previously-Published Single-Gene Signatures in the Validation Cohorts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Keller, D.S.; Berho, M.; Perez, R.O.; Wexner, S.D.; Chand, M. The multidisciplinary management of rectal cancer. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 414–429. [Google Scholar] [CrossRef]
- Smith, J.J.; Garcia-Aguilar, J. Advances and challenges in treatment of locally advanced rectal cancer. J. Clin. Oncol. 2015, 33, 1797–1808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benson, A.B.; Venook, A.P.; Al-Hawary, M.M.; Cederquist, L.; Chen, Y.J.; Ciombor, K.K.; Cohen, S.; Cooper, H.S.; Deming, D.; Engstrom, P.F.; et al. Rectal Cancer, Version 2.2018, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2018, 16, 874–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glynne-Jones, R.; Wyrwicz, L.; Tiret, E.; Brown, G.; Rodel, C.; Cervantes, A.; Arnold, D.; Committee, E.G. Rectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2017, 28, iv22–iv40. [Google Scholar] [CrossRef]
- Maas, M.; Nelemans, P.J.; Valentini, V.; Das, P.; Rodel, C.; Kuo, L.J.; Calvo, F.A.; Garcia-Aguilar, J.; Glynne-Jones, R.; Haustermans, K.; et al. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: A pooled analysis of individual patient data. Lancet Oncol. 2010, 11, 835–844. [Google Scholar] [CrossRef]
- Park, I.J.; You, Y.N.; Agarwal, A.; Skibber, J.M.; Rodriguez-Bigas, M.A.; Eng, C.; Feig, B.W.; Das, P.; Krishnan, S.; Crane, C.H.; et al. Neoadjuvant treatment response as an early response indicator for patients with rectal cancer. J. Clin. Oncol. 2012, 30, 1770–1776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fokas, E.; Liersch, T.; Fietkau, R.; Hohenberger, W.; Beissbarth, T.; Hess, C.; Becker, H.; Ghadimi, M.; Mrak, K.; Merkel, S.; et al. Tumor regression grading after preoperative chemoradiotherapy for locally advanced rectal carcinoma revisited: Updated results of the CAO/ARO/AIO-94 trial. J. Clin. Oncol. 2014, 32, 1554–1562. [Google Scholar] [CrossRef]
- Fokas, E.; Strobel, P.; Fietkau, R.; Ghadimi, M.; Liersch, T.; Grabenbauer, G.G.; Hartmann, A.; Kaufmann, M.; Sauer, R.; Graeven, U.; et al. Tumor Regression Grading After Preoperative Chemoradiotherapy as a Prognostic Factor and Individual-Level Surrogate for Disease-Free Survival in Rectal Cancer. J. Natl. Cancer Inst. 2017, 109, djx095. [Google Scholar] [CrossRef] [Green Version]
- Ghadimi, B.M.; Grade, M.; Difilippantonio, M.J.; Varma, S.; Simon, R.; Montagna, C.; Fuzesi, L.; Langer, C.; Becker, H.; Liersch, T.; et al. Effectiveness of gene expression profiling for response prediction of rectal adenocarcinomas to preoperative chemoradiotherapy. J. Clin. Oncol. 2005, 23, 1826–1838. [Google Scholar] [CrossRef] [Green Version]
- Kim, I.J.; Lim, S.B.; Kang, H.C.; Chang, H.J.; Ahn, S.A.; Park, H.W.; Jang, S.G.; Park, J.H.; Kim, D.Y.; Jung, K.H.; et al. Microarray gene expression profiling for predicting complete response to preoperative chemoradiotherapy in patients with advanced rectal cancer. Dis. Colon. Rectum. 2007, 50, 1342–1353. [Google Scholar] [CrossRef]
- Rimkus, C.; Friederichs, J.; Boulesteix, A.L.; Theisen, J.; Mages, J.; Becker, K.; Nekarda, H.; Rosenberg, R.; Janssen, K.P.; Siewert, J.R. Microarray-based prediction of tumor response to neoadjuvant radiochemotherapy of patients with locally advanced rectal cancer. Clin. Gastroenterol. Hepatol. 2008, 6, 53–61. [Google Scholar] [CrossRef]
- Brettingham-Moore, K.H.; Duong, C.P.; Greenawalt, D.M.; Heriot, A.G.; Ellul, J.; Dow, C.A.; Murray, W.K.; Hicks, R.J.; Tjandra, J.; Chao, M.; et al. Pretreatment transcriptional profiling for predicting response to neoadjuvant chemoradiotherapy in rectal adenocarcinoma. Clin. Cancer Res. 2011, 17, 3039–3047. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, T.; Kobunai, T.; Akiyoshi, T.; Matsuda, K.; Ishihara, S.; Nozawa, K. Prediction of response to preoperative chemoradiotherapy in rectal cancer by using reverse transcriptase polymerase chain reaction analysis of four genes. Dis. Colon. Rectum. 2014, 57, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Palma, P.; Cano, C.; Conde-Muino, R.; Comino, A.; Bueno, P.; Ferron, J.A.; Cuadros, M. Expression profiling of rectal tumors defines response to neoadjuvant treatment related genes. PLoS ONE 2014, 9, e112189. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.; Janssen, K.P.; Kim, I.J.; D’Angelo, E.; Pizzini, S.; Zangrando, A.; Zanon, C.; Pastrello, C.; Maretto, I.; Digito, M.; et al. An integrative approach for the identification of prognostic and predictive biomarkers in rectal cancer. Oncotarget 2015, 6, 32561–32574. [Google Scholar] [CrossRef]
- Casado, E.; Garcia, V.M.; Sanchez, J.J.; Blanco, M.; Maurel, J.; Feliu, J.; Fernandez-Martos, C.; de Castro, J.; Castelo, B.; Belda-Iniesta, C.; et al. A combined strategy of SAGE and quantitative PCR Provides a 13-gene signature that predicts preoperative chemoradiotherapy response and outcome in rectal cancer. Clin. Cancer Res. 2011, 17, 4145–4154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agostini, M.; Zangrando, A.; Pastrello, C.; D’Angelo, E.; Romano, G.; Giovannoni, R.; Giordan, M.; Maretto, I.; Bedin, C.; Zanon, C.; et al. A functional biological network centered on XRCC3: A new possible marker of chemoradiotherapy resistance in rectal cancer patients. Cancer Biol. Ther. 2015, 16, 1160–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millino, C.; Maretto, I.; Pacchioni, B.; Digito, M.; De Paoli, A.; Canzonieri, V.; D’Angelo, E.; Agostini, M.; Rizzolio, F.; Giordano, A.; et al. Gene and MicroRNA Expression Are Predictive of Tumor Response in Rectal Adenocarcinoma Patients Treated With Preoperative Chemoradiotherapy. J. Cell. Physiol. 2017, 232, 426–435. [Google Scholar] [CrossRef]
- Hur, H.; Tulina, I.; Cho, M.S.; Min, B.S.; Koom, W.S.; Lim, J.S.; Ahn, J.B.; Kim, N.K. Biomarker-Based Scoring System for Prediction of Tumor Response After Preoperative Chemoradiotherapy in Rectal Cancer by Reverse Transcriptase Polymerase Chain Reaction Analysis. Dis. Colon. Rectum. 2016, 59, 1174–1182. [Google Scholar] [CrossRef]
- Huh, J.W.; Lee, J.H.; Kim, H.R. Pretreatment expression of 13 molecular markers as a predictor of tumor responses after neoadjuvant chemoradiation in rectal cancer. Ann. Surg. 2014, 259, 508–515. [Google Scholar] [CrossRef]
- Ferrandon, S.; DeVecchio, J.; Duraes, L.; Chouhan, H.; Karagkounis, G.; Davenport, J.; Orloff, M.; Liska, D.; Kalady, M.F. CoA Synthase (COASY) Mediates Radiation Resistance via PI3K Signaling in Rectal Cancer. Cancer Res. 2020, 80, 334–346. [Google Scholar] [CrossRef] [Green Version]
- Gim, J.; Cho, Y.B.; Hong, H.K.; Kim, H.C.; Yun, S.H.; Wu, H.G.; Jeong, S.Y.; Joung, J.G.; Park, T.; Park, W.Y.; et al. Predicting multi-class responses to preoperative chemoradiotherapy in rectal cancer patients. Radiat. Oncol. 2016, 11, 50. [Google Scholar] [CrossRef] [Green Version]
- Alderdice, M.; Richman, S.D.; Gollins, S.; Stewart, J.P.; Hurt, C.; Adams, R.; McCorry, A.M.; Roddy, A.C.; Vimalachandran, D.; Isella, C.; et al. Prospective patient stratification into robust cancer-cell intrinsic subtypes from colorectal cancer biopsies. J. Pathol. 2018, 245, 19–28. [Google Scholar] [CrossRef] [Green Version]
- Ji, D.; Song, C.; Li, Y.; Xia, J.; Wu, Y.; Jia, J.; Cui, X.; Yu, S.; Gu, J. Combination of radiotherapy and suppression of Tregs enhances abscopal antitumor effect and inhibits metastasis in rectal cancer. J. Immunother. Cancer 2020, 8, e000826. [Google Scholar] [CrossRef]
- Sendoya, J.M.; Iseas, S.; Coraglio, M.; Golubicki, M.; Robbio, J.; Salanova, R.; Kujaruk, M.; Mikolaitis, V.; Rizzolo, M.; Ruiz, G.; et al. Pre-Existing Tumoral B Cell Infiltration and Impaired Genome Maintenance Correlate with Response to Chemoradiotherapy in Locally Advanced Rectal Cancer. Cancers 2020, 12, 2227. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, R.D.; Quinn, J.E.; Mullan, P.B.; Johnston, P.G.; Harkin, D.P. The role of BRCA1 in the cellular response to chemotherapy. J. Natl. Cancer Inst. 2004, 96, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Chalasani, P.; Livingston, R. Differential chemotherapeutic sensitivity for breast tumors with “BRCAness”: A review. Oncologist 2013, 18, 909–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Zhu, J.; Zhang, X.; Wu, Q.; Jiang, S.; Liu, Y.; Hu, Z.; Liu, B.; Chen, X. BRCA1 mRNA expression as a predictive and prognostic marker in advanced esophageal squamous cell carcinoma treated with cisplatin- or docetaxel-based chemotherapy/chemoradiotherapy. PLoS ONE 2013, 8, e52589. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Yu, Y.; Rong, J.; Hu, D.; Zhang, L.; Fu, S.; Yang, H.; Fan, J.; Yang, L.; Wu, J. Expression of BRCA1 and ERCC1 as predictive clinical outcome after radiochemotherapy in patients with locoregionally moderate-advanced nasopharyngeal carcinoma. Oncotarget 2017, 8, 31355–31367. [Google Scholar] [CrossRef] [Green Version]
- He, Q.; Zhang, M.; Zhang, J.; Zhong, S.; Liu, Y.; Shen, J.; He, J.; Jiang, L.; Yang, C.; Zeng, Y.; et al. Predictive value of BRCA1 expression on the efficacy of chemotherapy based on anti-microtubule agents: A pooled analysis across different malignancies and agents. Ann. Transl. Med. 2016, 4, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdulkareem, N.M.; Bhat, R.; Qin, L.; Vasaikar, S.; Gopinathan, A.; Mitchell, T.; Shea, M.J.; Nanda, S.; Thangavel, H.; Zhang, B.; et al. A novel role of ADGRF1 (GPR110) in promoting cellular quiescence and chemoresistance in human epidermal growth factor receptor 2-positive breast cancer. FASEB J. 2021, 35, e21719. [Google Scholar] [CrossRef] [PubMed]
- Masuda, M.; Uno, Y.; Ohbayashi, N.; Ohata, H.; Mimata, A.; Kukimoto-Niino, M.; Moriyama, H.; Kashimoto, S.; Inoue, T.; Goto, N.; et al. TNIK inhibition abrogates colorectal cancer stemness. Nat. Commun. 2016, 7, 12586. [Google Scholar] [CrossRef] [Green Version]
- Hirozane, T.; Masuda, M.; Sugano, T.; Sekita, T.; Goto, N.; Aoyama, T.; Sakagami, T.; Uno, Y.; Moriyama, H.; Sawa, M.; et al. Direct conversion of osteosarcoma to adipocytes by targeting TNIK. JCI Insight. 2021, 6. [Google Scholar] [CrossRef] [PubMed]
- Petrelli, F.; Sgroi, G.; Sarti, E.; Barni, S. Increasing the Interval Between Neoadjuvant Chemoradiotherapy and Surgery in Rectal Cancer: A Meta-analysis of Published Studies. Ann. Surg. 2016, 263, 458–464. [Google Scholar] [CrossRef]
- Lefevre, J.H.; Mineur, L.; Kotti, S.; Rullier, E.; Rouanet, P.; de Chaisemartin, C.; Meunier, B.; Mehrdad, J.; Cotte, E.; Desrame, J.; et al. Effect of Interval (7 or 11 weeks) Between Neoadjuvant Radiochemotherapy and Surgery on Complete Pathologic Response in Rectal Cancer: A Multicenter, Randomized, Controlled Trial (GRECCAR-6). J. Clin. Oncol. 2016, 34, 3773–3780. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Chang, H.J.; Kim, D.Y.; Park, J.W.; Baek, J.Y.; Kim, S.Y.; Park, S.C.; Oh, J.H.; Yu, A.; Nam, B.H. What Is the Ideal Tumor Regression Grading System in Rectal Cancer Patients after Preoperative Chemoradiotherapy? Cancer Res. Treat. 2016, 48, 998–1009. [Google Scholar] [CrossRef] [Green Version]
- De Palma, F.D.E.; Luglio, G.; Tropeano, F.P.; Pagano, G.; D’Armiento, M.; Kroemer, G.; Maiuri, M.C.; De Palma, G.D. The Role of Micro-RNAs and Circulating Tumor Markers as Predictors of Response to Neoadjuvant Therapy in Locally Advanced Rectal Cancer. Int. J. Mol. Sci. 2020, 21, 7040. [Google Scholar] [CrossRef]
- Cho, E.; Park, I.J.; Yeom, S.S.; Hong, S.M.; Lee, J.B.; Kim, Y.W.; Kim, M.J.; Lim, H.M.; Lim, S.B.; Yu, C.S.; et al. A Multigene Model for Predicting Tumor Responsiveness After Preoperative Chemoradiotherapy for Rectal Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2019, 105, 834–842. [Google Scholar] [CrossRef]
- Park, I.J.; Yu, Y.S.; Mustafa, B.; Park, J.Y.; Seo, Y.B.; Kim, G.D.; Kim, J.; Kim, C.M.; Noh, H.D.; Hong, S.M.; et al. A Nine-Gene Signature for Predicting the Response to Preoperative Chemoradiotherapy in Patients with Locally Advanced Rectal Cancer. Cancers 2020, 12, 800. [Google Scholar] [CrossRef] [Green Version]
- Kamran, S.C.; Lennerz, J.K.; Margolis, C.A.; Liu, D.; Reardon, B.; Wankowicz, S.A.; Van Seventer, E.E.; Tracy, A.; Wo, J.Y.; Carter, S.L.; et al. Integrative Molecular Characterization of Resistance to Neoadjuvant Chemoradiation in Rectal Cancer. Clin. Cancer Res. 2019, 25, 5561–5571. [Google Scholar] [CrossRef]
- Garcia-Aguilar, J.; Chen, Z.; Smith, D.D.; Li, W.; Madoff, R.D.; Cataldo, P.; Marcet, J.; Pastor, C. Identification of a biomarker profile associated with resistance to neoadjuvant chemoradiation therapy in rectal cancer. Ann. Surg. 2011, 254, 486–492, discussion 492–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toomey, S.; Gunther, J.; Carr, A.; Weksberg, D.C.; Thomas, V.; Salvucci, M.; Bacon, O.; Sherif, E.M.; Fay, J.; Kay, E.W.; et al. Genomic and Transcriptomic Characterisation of Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Cancers 2020, 12, 1808. [Google Scholar] [CrossRef]
- do Canto, L.M.; Barros-Filho, M.C.; Rainho, C.A.; Marinho, D.; Kupper, B.E.C.; Begnami, M.; Scapulatempo-Neto, C.; Havelund, B.M.; Lindebjerg, J.; Marchi, F.A.; et al. Comprehensive Analysis of DNA Methylation and Prediction of Response to NeoadjuvantTherapy in Locally Advanced Rectal Cancer. Cancers 2020, 12, 3079. [Google Scholar] [CrossRef] [PubMed]
- Akiyoshi, T.; Tanaka, N.; Kiyotani, K.; Gotoh, O.; Yamamoto, N.; Oba, K.; Fukunaga, Y.; Ueno, M.; Mori, S. Immunogenomic profiles associated with response to neoadjuvant chemoradiotherapy in patients with rectal cancer. Br. J. Surg. 2019, 106, 1381–1392. [Google Scholar] [CrossRef]
- El Sissy, C.; Kirilovsky, A.; Van den Eynde, M.; Musina, A.M.; Anitei, M.G.; Romero, A.; Marliot, F.; Junca, A.; Doyen, J.; Mlecnik, B.; et al. A Diagnostic Biopsy-Adapted Immunoscore Predicts Response to Neoadjuvant Treatment and Selects Patients with Rectal Cancer Eligible for a Watch-and-Wait Strategy. Clin. Cancer Res. 2020, 26, 5198–5207. [Google Scholar] [CrossRef]
- Goncalves-Ribeiro, S.; Sanz-Pamplona, R.; Vidal, A.; Sanjuan, X.; Guillen Diaz-Maroto, N.; Soriano, A.; Guardiola, J.; Albert, N.; Martinez-Villacampa, M.; Lopez, I.; et al. Prediction of pathological response to neoadjuvant treatment in rectal cancer with a two-protein immunohistochemical score derived from stromal gene-profiling. Ann. Oncol. 2017, 28, 2160–2168. [Google Scholar] [CrossRef]

| Purpose | GEO Accession | Platform | Total | Non- Responder | Responder | References |
|---|---|---|---|---|---|---|
| Discovery | GSE35452 | Affymetrix Human Genome U133 Plus 2.0 Array | 46 | 22 | 24 | [14] |
| GSE45404 | Affymetrix Human Genome U133 Plus 2.0 Array | 42 | 23 | 19 | [18] | |
| GSE53781 | CodeLink Human Whole Genome Array | 26 | 16 | 10 | [15] | |
| Validation | GSE46862 | Affymetrix Human Gene 1.0 ST Array | 69 | 41 | 28 | [23] |
| GSE68204 | Agilent-014850 Whole Human Genome Microarray 4x44K G4112F | 38 | 22 | 16 | [19] | |
| GSE94104 | Illumina HumanHT-12 WG-DASL V4.0 R2 expression beadchip | 40 | 29 | 11 | [24] | |
| GSE119409 | Affymetrix Human Genome U133 Plus 2.0 Array | 56 | 41 | 15 | [25] | |
| GSE133057 | Illumina human-6 v2.0 expression beadchip | 33 | 20 | 13 | [22] | |
| GSE150082 | Agilent-026652 Whole Human Genome Microarray 4x44K v2 | 39 | 23 | 16 | [26] |
| Signature | Genes Upregulated in Non-Responders vs. Responders | Genes Downregulated in Non-Responders vs. Responders | References |
|---|---|---|---|
| 4-gene | GPR110 (ADGRF1), TNIK | WDR4, BRCA1 | This study |
| Casado_13-gene | BAK1, MLH1, TYMS, CKB, GPX2, HIG2 (HILPDA), PH4 (P4HTM) | ALDH1A1, CDKN1A, FOS, RELB, STAT3, TFF3 | [17] |
| Watanabe_4-gene | FRMD3, SAMD5, TMC7 | LRRIQ3 (LRRC44) | [14] |
| Palma_4-gene | - | GNG4, MYC, POLA1, RRM1 | [15] |
| Agostini_7-gene | AKR1C3 | CXCL11, CXCL10, IDO1, CXCL9, MMP12, HLA-DRA | [16] |
| Hur_4-gene | TP53 | MKI67, CDKN1A, CD133 (PROM1) | [20] |
| Millino_8-gene | ITGA2, NRG1, KLF7 | TMEM188 (CNEP1R1), TRAM1, BCL2L13, MYO1B, GTSE1 | [19] |
| CD44 | CD44 | - | [21] |
| XRCC3 | - | XRCC3 | [18] |
| COASY | COASY | - | [22] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Momma, T.; Okayama, H.; Kanke, Y.; Fukai, S.; Onozawa, H.; Fujita, S.; Sakamoto, W.; Saito, M.; Ohki, S.; Kono, K. Validation of Gene Expression-Based Predictive Biomarkers for Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Cancers 2021, 13, 4642. https://doi.org/10.3390/cancers13184642
Momma T, Okayama H, Kanke Y, Fukai S, Onozawa H, Fujita S, Sakamoto W, Saito M, Ohki S, Kono K. Validation of Gene Expression-Based Predictive Biomarkers for Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Cancers. 2021; 13(18):4642. https://doi.org/10.3390/cancers13184642
Chicago/Turabian StyleMomma, Tomoyuki, Hirokazu Okayama, Yasuyuki Kanke, Satoshi Fukai, Hisashi Onozawa, Shotaro Fujita, Wataru Sakamoto, Motonobu Saito, Shinji Ohki, and Koji Kono. 2021. "Validation of Gene Expression-Based Predictive Biomarkers for Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer" Cancers 13, no. 18: 4642. https://doi.org/10.3390/cancers13184642
APA StyleMomma, T., Okayama, H., Kanke, Y., Fukai, S., Onozawa, H., Fujita, S., Sakamoto, W., Saito, M., Ohki, S., & Kono, K. (2021). Validation of Gene Expression-Based Predictive Biomarkers for Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Cancers, 13(18), 4642. https://doi.org/10.3390/cancers13184642






