Simple Summary
Despite being effective initially, almost all patients eventually develop resistance to EGFR-tyrosine kinase inhibitors (TKI). Changes in the histology of the tumor have been increasingly recognized as a critical mechanism of resistance to EGFR-directed therapies. This article summarizes histologic changes known to impart resistance to EGFR TKIs and discusses novel pathways to develop effective novel therapies.
Abstract
With the advent of potent EGFR tyrosine kinase inhibitors (TKIs), the treatment landscape of EGFR-mutant lung adenocarcinomas has changed drastically in recent years. However, the development of resistance to EGFR TKIs remains a critical barrier to improving survival in these patients. Histologic transformations to small cell lung carcinoma, large cell neuroendocrine carcinoma, squamous cell carcinoma, and the sarcomatoid phenotype have been increasingly recognized as important mechanisms of resistance. In this article, we summarize the known biological bases for such phenotypic switches in regard to EGFR TKIs and describe novel pathways that might be harnessed to develop effective novel therapies for patients with EGFR-mutant non-small cell lung cancers.
1. Introduction
With the application of advanced diagnostic techniques, our understanding of the molecular underpinnings of non-small cell lung cancer (NSCLC) has been transformed in the past two decades. Identification of oncogenic driver mutations has ushered in an era of genomic biomarker-directed therapies. Lung adenocarcinomas with activating mutations in the epidermal growth factor receptor (EGFR) gene represent the most common subset of targetable driver-positive non-small cell lung cancers (NSCLCs) and comprise ~10–15% of Caucasian patients and ~40–50% of East-Asian advanced NSCLC patients. There are higher incidences of such cancers in females and those who have either never smoked or are former light smokers [1]. The EGFR gene encodes the EGFR tyrosine kinase, which is a member of the human epidermal growth factor (HER) family of transmembrane receptors (HER1/EGFR, HER2, HER3, and HER4). EGFR tyrosine kinase exists as an inactive monomer that dimerizes with a receptor of the same type or another member of the HER family in response to ligand binding. Activating mutations of the EGFR gene, such as exon 19 deletions or exon 21 L858R point mutations, lead to the constitutional activation of the EGFR gene without ligand-induced stimulation and downstream signaling [2].
In the past few decades, various first (gefitinib and erlotinib) and second-generation (afatinib and dacomitinib) EGFR-tyrosine kinase inhibitors (TKIs) have been developed, which have been shown to be superior to chemotherapy in multiple phase 3 clinical trials, with response rates of ~70–75% and median progression-free survival (PFS) of 10–14 months [3]. These small-molecule TKIs bind the adenosine triphosphate pocket (ATP) of EGFR, thereby inhibiting its autophosphorylation and the resulting downstream signal transduction. Osimertinib, a third-generation EGFR TKI, has been shown to be superior to earlier-generation TKIs in both PFS (18.9 versus 10.2 months) and overall survival (38.6 versus 31.8 months) in a landmark phase 3 randomized clinical trial (FLAURA) [4]. More recently, osimertinib has been shown to be associated with disease-free survival (median was not reached in patients on the osimertinib arm compared with 27.5 months on the placebo arm (hazard ratio, 0.20; 95% confidence interval, 0.15 to 0.27; p < 0.0001)) in patients with stage IB to IIIA NSCLCs with completely resected tumors following standard platinum-based adjuvant chemotherapy, setting the scene for further changes in the treatment landscape of EGFR-mutant lung adenocarcinomas [5]. As a result of these advances, EGFR-mutant lung cancer patients are living longer than ever before [6].
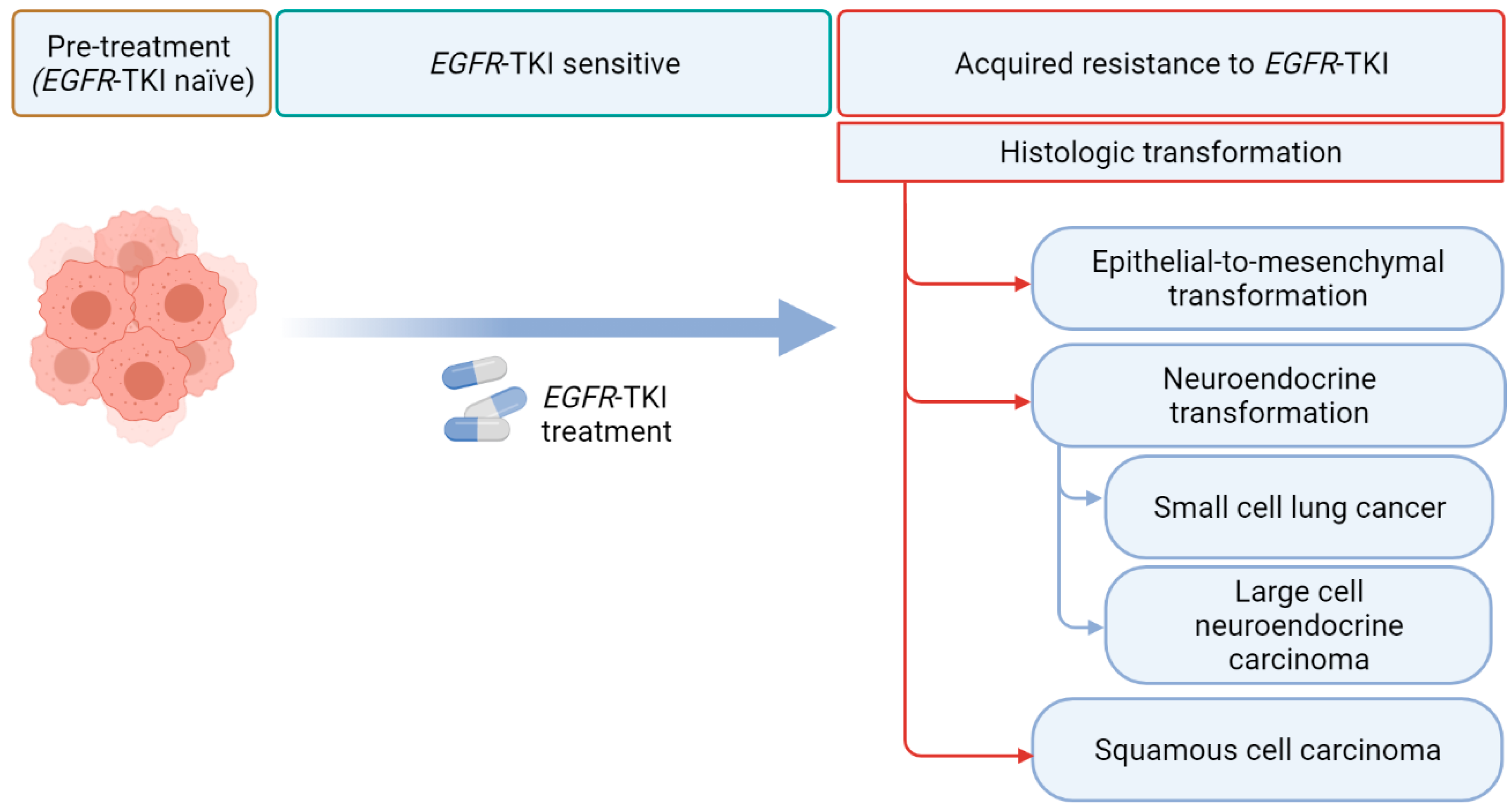
However, despite being effective, EGFR TKIs eventually lose efficacy in almost all EGFR TKI-treated patients [7]. Drug resistance is thought to arise from the clonal selection pressures exerted by EGFR TKIs with a selection of pre-existing subclones or a selection of randomly acquired alterations that develop over time [8]. Resistance to EGFR TKIs can be broadly classified as innate (defined as disease progression within the first 3 months after TKI initiation) or acquired [9]. Although most acquired resistance develops due to mutations in the EGFR receptor, activation of bypass intracellular signal pathways, or nongenetic adaptive changes, histologic transformation has been increasingly recognized as a crucial resistance mechanism [7,10]. In addition to the phenotypic transformation to small cell lung carcinoma (SCLC) [11,12], multiple recent reports have described transformations to rare histologic phenotypes, such as large cell neuroendocrine carcinomas (LCNECs) and squamous cell carcinomas (SCCs) (Figure 1) [13,14].
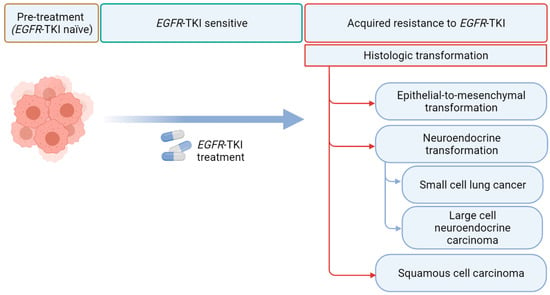
Figure 1.
Histological transformation via EGFR tyrosine kinase inhibitors (TKI). EGFR refers to epidermal growth factor receptor (created with Biorender.com).
In this article, we try to summarize the mechanisms of histologic transformations in EGFR-mutant NSCLCs and discuss potential therapeutic approaches to conquer this important resistance mechanism.
2. Targeting EGFR in Lung Cancer
Since the discovery of EGFR signaling in 1962, multiple attempts have been made to target this pathway, including the use of monoclonal antibodies such as cetuximab and panitumumab [15]. Further work in this space led to the identification of EGFR mutations in lung cancer, paving the way to precision medicine in lung cancer [16]. Somatic activating mutations in the EGFR gene result in ligand-independent receptor activation and downstream signaling promoting cell proliferation and survival [2]. The majority of EGFR mutations affect the intracellular tyrosine kinase domain of the receptor and include a heterogeneous group of mutations in exons 18 to 21. Exon 19 deletion and exon 21 L858R point mutations represent the majority of EGFR mutations that are sensitive to EGFR TKIs [17]. Uncommon EGFR mutations such as exon 18 (E709X and G719X), exon 19 (exon 19 insertions), exon 20 (exon 20 insertions and S768I), and exon 21 (L861Q) mutations constitute the remaining 10–15% of EGFR-mutated lung adenocarcinomas, which display variable sensitivities to EGFR TKIs [17].
3. Mechanisms of Resistance to EGFR TKIs
Selective pressures from TKI therapy can lead to the selection of cells lacking the original driver mutation or the acquisition of on and off-target resistance mechanisms. These can include the selection of preexisting resistant clones or induced adaptive resistance mechanisms [9]. The on-target (EGFR-dependent) and off-target (EGFR-independent) resistance mechanisms differ significantly according to the specific EGFR TKI used. For instance, patients receiving first or second-generation EGFR TKIs predominantly develop EGFR-dependent resistance mechanisms. Patients receiving the third-generation TKI osimertinib, as either first or second-line therapy, mostly develop off-target resistance, highlighting the differences in resistance mechanisms according to the type of TKI and line of therapy [7,18,19].
3.1. EGFR Target-Dependent Mechanisms of Resistance
Changes in EGFR to the critical amino acid residues can preclude the mechanisms of action of EGFR TKIs. The gatekeeper mutation in exon 20 of EGFR with the amino acid substitution of p.Thr790Met results in the steric hindering to the binding of first and second-generation EGFR TKIs to their ATP-binding site on EGFR [20]. Although seen in 50–60% of patients receiving gefitinib, erlotinib, or afatinib, no evidence of T790M mutation was seen to coincide with resistance from plasma genotyping when patients received first-line osimertinib [19]. Since osimertinib is selectively active against both EGFR-sensitizing mutations and the T790M mutations, the lack of emergence of a T790M mutation would not be surprising. With the approval of osimertinib in the first-line setting, the incidence of T790M as a resistance mechanism is expected to become less relevant over time. Although C797X (p.Cys797Ser or p.Cys797Gly) mutations have emerged as the most common EGFR-dependent mechanism of resistance to osimertinib in later lines of therapy, their incidence in the front-line setting is lower (only 7% patients out of 91 patients with paired plasma samples analyzed by next-generation sequencing) in the FLAURA trial [19]. It remains to be seen whether other EGFR-dependent mutations that were described on earlier-generation EGFR TKIs are relevant in the era of first-line osimertinib.
3.2. EGFR Target-Independent Mechanisms of Resistance
Continued EGFR TKI exposure can lead to rewiring of the non-EGFR cell signaling pathways that can lead to EGFR TKI resistance. Mesenchymal epithelial transition factor (MET) gene amplification has been reported in 5–22% of patients experiencing disease progressing in spite of first-generation EGFR TKIs [7,21]. Despite the challenges in harmonizing the definitions of MET amplification and the lack of consensus on the definition of MET amplification using liquid biopsies, MET receptor tyrosine kinase signaling is thought to be the most commonly altered pathway involved in EGFR TKI resistance, irrespective of the EGFR TKI used or the line of therapy [22]. HER2 amplification, PIK3CA, and rat sarcoma (RAS) mutations have also been described as possible mechanisms of resistance in liquid biopsy-based analyses from the FLAURA trial [19]. In addition to these mutations, numerous fusions/chromosomal rearrangements (such as the rearranged during transfection (RET), neurotrophic tropomyosin receptor kinase (NTRK), and ROS1), mutations in cell cycle-related genes (such as cyclin-dependent kinases 4 and 6, and cyclin-dependent kinase inhibitor 2A (CDKN2A)) have also been described as possible EGFR target-independent mechanisms of resistance to EGFR TKIs [22]. With the introduction of first-line osimertinib, further work is needed to understand the relative incidences of various types of EGFR-independent mechanisms of resistance in the Osimertinib era.
3.3. Histologic and Phenotypic Transformations
Histologic transformations in EGFR TKI treated patients have emerged as important mechanisms of resistance to EGFR TKIs. Although SCLC transformation has been considered to be the most common histologic transformation in EGFR-mutant NSCLCs in patients with disease progression during treatment with an earlier-generation EGFR TKI, [7] SCC transformation emerged as a potentially more common (15%) histologic transformation in front-line osimertinib-treated patients in a small cohort of patients with limited follow-up [18]. Transformed tumors generally retain the initial EGFR mutations [12,14]. An essential consideration in the diagnosis of histologic transformation is the possibility of preexisting initial mixed histology, which often cannot be precisely assessed on small biopsies/cytologic samples, highlighting the challenges of studying histologic transformations. Unfortunately, the current treatments for patients with histologic transformations are primarily driven by histology; further understanding of the biology and the mechanisms of histologic transformation are urgently needed to develop precision medicine for these patients.
4. Histologic Transformations in EGFR TKIs Treated NSCLCs
4.1. Epithelial-to-Mesenchymal Transformation
Epithelial-to-mesenchymal transformation (EMT) refers to the process of epithelial cells transforming to mesenchymal cells by losing their polarity and intercellular adhesions [23]. “Sarcomatous/sarcomatoid dedifferentiation” is another term that is used by pathologists in clinical practice to imply EMT [24]. These changes are accompanied by anti-apoptotic signals and changes in molecular expression (downregulation of epithelial cytokeratin markers and upregulation of mesenchymal markers such as vimentin and fibronectin) [25]. The culmination of these changes imparts cancer cells with invasiveness, metastatic potential, and drug resistance. Thus, such changes mean poor prognoses for multiple types of cancer, including NSCLC [26,27].
Although some preclinical models have suggested EMT as a mechanism of acquired resistance to EGFR TKIs, [28] further work is needed to understand whether EMT is truly a driver of resistance, as opposed to a passenger event [29]. Early reports describing EMT imparting acquired resistance to EGFR TKIs, such as those by Uramoto et al. and Sequist et al., were based on the absence of any other mechanisms of resistance known at that time, and therefore may have overestimated the impact of EMT. Moreover, since early studies examining the associations of EMT signatures and sensitivity with EGFR TKIs were carried out in unselected patients irrespective of sensitizing EGFR mutations, [30] it is unclear whether those signatures remain valid for EGFR-mutant NSCLCs.
Although a relationship between sensitivity to EGFR TKIs and tumor E-cadherin expression has been proposed, depletion of cadherin 1 (CDH1) by itself in EGFR-mutant therapy-naïve NSCLC cells does not lead to a loss of EGFR TKI efficacy [31]. These observations suggest that both the downregulation of epithelial markers (that are repressed by several nuclear factors, such as zinc finger E-box binding homeobox 1 (ZEB1) (δEF1, ZFHX1A), ZEB2 (SIP1, ZFHX1B), snail family transcriptional repressor 1 (SNAI1) (Snail), SNAI2 (Slug), E12/E47, and TWIST) and upregulation of mesenchymal proteins are necessary to impart resistance to EGFR TKIs [32]. Preclinical studies have also revealed that the pro-EMT factor TWIST leads to inhibition of pro-apoptotic protein BCL2-interacting mediator of cell death (BIM) either by directly binding to its promoter or by inducing ZEB1 in EGFR-mutant NSCLC murine models [28,33]. EMT may, therefore, induce EGFR TKI resistance, at least in part, via transcriptional suppression of BIM-mediated apoptosis. Finally, silencing of microRNAs (such as miR-200) through promoter methylation has been shown to play a pivotal role in the development of EMT [34,35,36]. Ectopic expression of CRIPTO1 (an oncofetal, membrane-associated protein of the epidermal growth factor (EGF) cryptic family 1 gene (CFC) family) upregulated ZEB1 and activated the SRC pathway via microRNA-205 (miR-205) downregulation, thereby promoting EMT and erlotinib-resistance in EGFR-mutant NSCLC cell lines [37]. Anexelekto (AXL)-signaling has been associated with acquired EGFR TKI resistance through the induction of EMT [38]. Cancer-associated fibroblasts (CAFs) have been implicated in the induction of EMT via paracrine mechanisms, which might allow novel approaches to prevent the development of EMTs [39]. Recently, hypoxia has been shown to activate insulin-like growth factor 1 receptor (IGF1R), which can lead to the generation of primitive cancer stem cells and EMT [40].
Novel Therapies
EMT is believed to be associated with a reversible state of reduced drug sensitivity that precedes the onset of terminal resistance [41]. Drug tolerant persisters are associated with an altered chromatin state with high expression levels of lysine demethylase 5A (KDM5A), a histone demethylase H3 Lys 4 (H3K4), and other chromatin-modifying enzymes. Epigenetic therapies, such as histone deacetylase (HDAC) inhibitors, have been evaluated and shown to restore the expression of CDH1 and sensitivity to EGFR TKIs in preclinical models [41,42]. Trials combining epigenetic therapies such as HDAC inhibitors with EGFR TKIs in patients developing EMT are needed to test these hypotheses. In addition to epigenetic therapies, 1,25(OH)2D3-based combination therapies are currently being evaluated in preclinical models for EMT-associated EGFR TKI resistance [29]. Studies also suggest inhibition of BIM by the pro-EMT factor TWIST [28,33]. Hence, B-cell lymphoma 2 (BCL-2) homology 3 (BH3) mimetics, which target the B-cell lymphoma (BCL) family of proteins and leverage the apoptotic pathway, have been proposed as a means of overcoming EMT-associated EGFR TKI resistance [28,33].
Other approaches to reverse EMT-mediated EGFR TKI resistance include targeting specific bypass mutations (such as mitogen-activated protein kinase (MAPK) and fibroblast growth factor receptor 1 (FGFR1)), microRNAs [36], immune checkpoint inhibitors, [43] AXL receptor tyrosine kinase, [38] IGFR1, [40] etc. Targeting EGFR and SRC tyrosine kinase reduced the growth of CRIPTO1-positive, erlotinib-resistant, EGFR-mutant NSCLC cells, suggesting that this combination might be able to counteract intrinsic resistance to EGFR-TKIs in patients with CRIPTO1-positive, EGFR-mutant NSCLC undergoing EMT [37]. Pharmacological inhibition of neurogenic locus notch homolog protein 1 (NOTCH1) signaling pathway is currently being evaluated as a means of restoring sensitivity to EGFR TKIs (NCT01158404, NCT01653470). Finally, further understanding of the role of the MET pathway in EMT could pave the way towards the therapeutic application of MET inhibitors in EMT-driven resistant patients (Table 1) [44].

Table 1.
Novel pathways and potential therapies for histologically transformed EGFR-mutant lung cancers.
4.2. Neuroendocrine Transformation
Although SCLC transformation has been well described as a mechanism of acquired resistance in EGFR-mutant NSCLCs, several recent reports have also identified LCNEC as a means of resistance to EGFR TKIs.
4.2.1. SCLC
Studies have estimated that ~5–15% of EGFR-mutated lung adenocarcinomas treated with EGFR TKIs transform to SCLCs [7,10]. Patients with SCLC transformation tend to have poorer prognoses [12,45]. Preclinical studies in mice and cell lines have shown that RB1 inactivation is essential for SCLC development [46,47]. This observation has been further reinforced by the sequencing of human SCLCs showing mutation in or loss of retinoblastoma gene (RB1) in 100% of samples [48]. However, RB1 is necessary but not sufficient for SCLC development from adenocarcinomas. Additional pathways such as NOTCH-Achaete-scute homolog 1 (ASCL1) and TP53 perhaps play essential roles [49].
In a study aimed at characterizing the molecular changes in transformed SCLCs, Niederst et al. observed that transformed SCLCs share many features of classical SCLCs, such as universal loss tumor suppression via RB1 and TP53 inactivation, reduced or absent EGFR protein expression, epigenetic changes, and increased sensitivity to BCL-2 inhibition. All patients with transformed SCLCs harbored the original activating EGFR mutation [50]. Kim et al. showed that the divergence of SCLC clones from adenocarcinomas occurred early on, even before the initiation of EGFR TKIs. The authors also found that complete inactivation of both RB1 and TP53 increased the risk of SCLC transformation by >42 fold [51]. Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC)-induced hypermutation was frequent in the branches toward SCLC transformation [51]. The loss of sensitivity to EGFR TKIs following SCLC transformation seems to be mediated partly by the downregulation of the expression of EGFR protein, although alterations in EGFR-dependent and EGFR-independent pathways might also contribute [52]. This has been reflected in clinical experience from two large case series. Ferrer et al. reported a multicenter retrospective study evaluating clinical outcomes of SCLC transformation in 48 EGFR mutant NSCLCs between 2005 and 2017 [11]. The median time to SCLC transformation was 16 months. The median overall survival after transformation was only nine months, which mirrors survival seen in de novo extensive-stage SCLC patients. Marcoux et al. reported another multicenter retrospective study evaluating clinical outcomes of 58 patients with transformed SCLC from adenocarcinomas [12]. The median time to transformation was similar to the Ferrer et al. series. A few responses were seen in patients who had concurrently active NSCLC clones. Median survival after transformation was 10.9 months [12].
Data implicating RB1 loss as a driver of SCLC transformation is further supported by Offin et al., who found that “triple-positive” patients with EGFR/RB1/TP53 mutations had an at least 6-fold higher risk of SCLC transformation compared to EGFR-mutant lung adenocarcinomas without RB1/TP53 mutations. The authors also noted that triple-mutant tumors had enrichment of whole-genome doubling and an APOBEC signature, which may represent early genomic determinants of lineage plasticity [53]. However, not all EGFR/ RB1/TP53 triple-mutant NSCLCs undergo SCLC transformation, highlighting the importance of additional pathways such as epigenetic changes [29]. These data suggest that multiple genetic hits are probably needed to transform adenocarcinomas to SCLCs, which has made the development of pre-clinical models of transformed SCLCs challenging.
Although biopsy is essential to establishing the diagnosis of SCLC transformation, serial elevations of blood neuron-specific enolase (NSE) or pro-gastrin-releasing peptide (GRP) might help detect the emergence of SCLC clones earlier before they manifest clinically [54]. Serial rises in pro-GRP and NSE levels have been noted to mirror SCLC transformation in numerous case reports and deserve further studies to determine their clinical utility in patients EGFR TKI-treated NSCLCs [55,56,57].
Novel Therapies
Various strategies to overcome acquired resistance due to SCLC transformation are currently being investigated. The combination of EGFR TKIs with upfront platinum-etoposide chemotherapy is being evaluated in patients with triple-mutant cancers at high risk of transformation (NCT03567642). In addition to cytotoxic chemotherapy, approaches utilizing BCL-2 family inhibitors may be an attractive target in transformed SCLC patients [50]. Targeting cell cycle vulnerabilities created due to RB1 loss, such as sensitivity to checkpoint kinase 1 (CHK1) and polo-like kinase 1 (PLK1) inhibitors (which target DNA damage checkpoints), might yield synthetic lethality in transformed SCLCs [58]. Similarly, dependence on aurora kinases (AURK) arising from RB1 loss [59] (coupled with AURKA mediated acquired resistance to EGFR TKIs in RB1 proficient clones [60]) may open doors to combination therapies with aurora kinase inhibitors, e.g., a phase 1/1b clinical trial of AURKA inhibitor alisertib with osimertinib in metastatic EGFR-mutant lung cancer (NCT04085315). Modulation of epigenetic pathways (such as enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2)) has been proposed as a means of not only treating patients with transformed SCLC but also preventing SCLC transformation. EZH2 inhibitors have been hypothesized to prevent SCLC transformation in high-risk triple-mutant cancers, based on SCLC data and small cell transformation in other cancers [29,47,61]. Further studies which examine signaling pathways in transformed SCLCs might pave the way to novel targets in future.
4.2.2. LCNEC
Although LCNECs share many morphologic, phenotypic, and genetic features of SCLC, they also differ in many important ways [62,63]. Only a handful of LCNEC transformations have been reported in EGFR-mutant NSCLCs, thereby limiting our understanding of the mechanisms of this phenotypic change [13,64,65]. Case reports have suggested that in some cases, subclonal cell populations harboring molecular markers of neuroendocrine differentiation such as TP53 and RB1 loss could be present at baseline, which may be selected under the EGFR TKI treatment pressure leading to pathologically evident transformation in due course [64]. Further research is needed to understand the biology and mechanisms of LCNEC transformation in EGFR-mutant NSCLCs.
4.3. SCC Transformation
Adenocarcinoma to SCC lineage transformation is the least common type of acquired resistance to EGFR TKIs, although the incidence of SCC transformation during first-line osimertinib therapy might be higher than previously seen with later-line osimertinib or earlier-generation EGFR TKIs [18,66]. Although multiple case reports have suggested resistance to EGFR TKIs after developing SCC transformation, few case reports have suggested durable responses to TKIs, highlighting that SCC transformation may not always contribute to EGFR TKI resistance [14,67]. Patients with de-novo EGFR-mutated SCC tend to have a shorter PFS than their adenocarcinoma counterparts [68]. However, the biology of SCC transformation remains poorly defined, and it is not uncommon for NSCLCs to have coexistent adenocarcinoma and squamous cell carcinoma components [69]. Therefore, squamous “transformation” after treatment could reflect a shift in the predominant histology of a mixed tumor rather than a true lineage shift (as could be the case with other histologic transformations)—especially in cases of small biopsy/cytologic samples where histology may not have been accurately assessed.
Higher frequencies of mutations in neurofibromatosis (NF1), ataxia telangiectasia and Rad3 related (ATR), and breast cancer (BRCA1) genes have been reported in EGFR-mutant SCCs compared to EGFR-mutant adenocarcinomas [68]. However, despite comprehensive genomic analyses, no consistent molecular signature has been identified in SCC transformation; considerable genomic complexity is seen in these patients with mutations in phosphoinositide 3-kinase (PI3K)/AKT/mechanistic target of rapamycin (mTOR) pathways, chromosome 31 amplification, and FGF amplification [66]. In a study examining the acquired resistance mechanisms to osimertinib using paired pre and posttreatment tissue samples, patients with SCC transformation did not have any overarching genomic correlate; patients had the PIK3CA E726K mutation, chromosome 3q gain (and FGF amplification), and PIK3CA copy number gains [18]. Genetically engineered mouse models have shown that the loss of liver kinase B1 (LKB1, also known as serine/threonine kinase (STK11)) can lead to lineage plasticity and SCC transformation in Kirsten rat sarcoma 2 viral oncogene homolog (KRAS) mutant lung cancers [70]. However, concurrent LKB1 loss is rare in human EGFR-mutant NSCLCs [71,72]. Further studies are therefore needed to understand the gene expression of the transformed cases and to assess the nongenomic processes, such as transcription factor and epigenome changes, to determine the biological basis of SCC transformation in EGFR-mutant NSCLCs.
Novel Therapies
Therapies targeting the PI3K/AKT/mTOR pathway need to be studied systematically in the setting of clinical trials. Further work is urgently needed to understand novel pathways in transformed SCC, including the epigenetic pathways which may help discover new therapies for these patients [66].
5. Conclusions and Future Directions
Improved understanding of the biology and molecular underpinnings of histologic transformation in EGFR-mutated NSCLCs is urgently needed to help develop reliable biomarkers and effective novel therapies. Although EGFR/RB1/TP53 triple mutation seems to identify a group of patients at the highest risk for SCLC transformation, biomarkers to predict transformation to SCC, EMT, and LCNEC are currently lacking. Studies incorporating mid-treatment serial biopsies might help identify clones that drive eventual resistance: the information gained from evaluating these clones might identify potential biomarkers. Bioinformatics and computational biology techniques might enable the development of novel multiomics biomarkers that can identify patients at risk of transformation during early, tailored combination therapies can then be introduced early to eradicate the seeds of resistance before they manifest clinically.
Identification of epigenetic alterations and novel pathways seen with the transformed phenotypes (such as FGFR for EMT or AURKA for RB1-deficient, transformed SCLC) can help develop novel therapies. Some of these therapies might be applied early to block lineage plasticity and prevent the development of abnormal phenotypes or allow restoration of EGFR dependency and EGFR TKI sensitivity once transformation emerges. In addition, multimodality approaches such as the early introduction of stereotactic ablative radiotherapy of metastatic sites should be evaluated to prevent the development of transformed clones in patients deemed high risk for histologic transformation. Multi-arm trials such as the ORCHARD trial (NCT03944772) might provide a unique opportunity to explore treatment options for patients with SCC and SCLC transformation through histology-driven cohorts.
Blood-based tumor analysis offers a non-invasive and easily accessible modality for cancer diagnostics [73]. Despite the rising popularity of circulating tumor DNA (ctDNA) based testing, clinicians should be mindful that histological transformation cannot be detected by blood-based testing alone. ctDNA at the time of progression, however, might help identify patients with RB1 and TP53 alterations, who are at higher risk of SCLC transformation and will benefit from further investigation of the tissue. Histologic and ctDNA analyses should be used as complementary modalities to further our understanding of histologic transformation in EGFR-mutated NSCLC patients. Dynamic monitoring of ctDNA offers an attractive tool to investigate the emergence of resistance mutations and subclonal events. This approach in clinical practice is being evaluated in the APPLE trial, which aims to compare the initiation of the treatment of EGFRT790M based on ctDNA against radiological evidence of disease progression [74]. Although data on osimertinib are still evolving, histologic transformation has been seen in up to 15% of osimertinib-treated patients in early analyses [18]. Further investigation is, therefore, urgently needed to understand the mechanisms of histologic transformation in response to newer-generation EGFR TKIs, such as osimertinib. Finally, further work is needed to understand the incidence of histologic transformation in patients receiving adjuvant osimertinib who only have microscopic disease.
In conclusion, histologic transformation is an emerging and important mechanism of resistance to EGFR TKIs with limited therapeutic options. Although the overall survival of EGFR-mutant NSCLC patients has improved in recent decades, much work needs to be done to understand various mechanisms of resistance so that effective therapies can be developed. Serial mid-treatment biopsies of tumor tissue and ctDNA at baseline and progression to detect histologic transformation should be incorporated in all future clinical trials with EGFR TKIs, whenever possible. Further studies of signaling pathways in tumors with histologic transformation will hopefully help elucidate novel mechanisms that can help develop new biomarkers and discover new therapies.
Author Contributions
Conceptualization, R.P. and V.M.V.; methodology, R.P. and V.M.V.; validation, R.P. and V.M.V.; formal analysis, R.P. and V.M.V.; data curation, R.P. and V.M.V.; writing—original draft preparation, R.P.; writing—review and editing, R.P. and V.M.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, Y.-L.; Yuan, J.-Q.; Wang, K.-F.; Fu, X.-H.; Han, X.-R.; Threapleton, D.; Yang, Z.-Y.; Mao, C.; Tang, J.-L. The prevalence of EGFR mutation in patients with non-small cell lung cancer: A systematic review and meta-analysis. Oncotarget 2016, 7, 78985–78993. [Google Scholar] [CrossRef] [PubMed]
- Mass, R.D. The HER receptor family: A rich target for therapeutic development. Int. J. Radiat. Oncol. Biol. Phys. 2004, 58, 932–940. [Google Scholar] [CrossRef]
- Lee, C.K.; Davies, L.; Wu, Y.-L.; Mitsudomi, T.; Inoue, A.; Rosell, R.; Zhou, C.; Nakagawa, K.; Thongprasert, S.; Fukuoka, M.; et al. Gefitinib or erlotinib vs chemotherapy for EGFR Mutation-positive lung cancer: Individual patient data meta-analysis of overall survival. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, S.S.; Vansteenkiste, J.; Planchard, D.; Cho, B.C.; Gray, J.E.; Ohe, Y.; Zhou, C.; Reungwetwattana, T.; Cheng, Y.; Chewaskulyong, B.; et al. Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N. Engl. J. Med. 2020, 382, 41–50. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Tsuboi, M.; He, J.; John, T.; Grohe, C.; Majem, M.; Goldman, J.W.; Laktionov, K.; Kim, S.-W.; Kato, T.; et al. Osimertinib in resected EGFR-mutated non–small-cell lung cancer. N. Engl. J. Med. 2020, 383, 1711–1723. [Google Scholar] [CrossRef]
- Howlader, N.; Forjaz, G.; Mooradian, M.J.; Meza, R.; Kong, C.Y.; Cronin, K.A.; Mariotto, A.B.; Lowy, D.R.; Feuer, E.J. The effect of advances in lung-cancer treatment on population mortality. N. Engl. J. Med. 2020, 387, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Sequist, L.V.; Waltman, B.A.; Dias-Santagata, D.; Digumarthy, S.; Turke, A.B.; Fidias, P.; Bergethon, K.; Shaw, A.T.; Gettinger, S.; Cosper, A.K.; et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 2011, 3, 75ra26. [Google Scholar] [CrossRef] [PubMed]
- Vasan, N.; Baselga, J.; Hyman, D.M. A view on drug resistance in cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef]
- Santoni-Rugiu, E.; Melchior, L.C.; Urbanska, E.M.; Jakobsen, J.N.; de Stricker, K.; Grauslund, M.; Sørensen, J.B. Intrinsic resistance to EGFR-tyrosine kinase inhibitors in EGFR-mutant non-small cell lung cancer: Differences and similarities with acquired resistance. Cancers 2019, 11, 923. [Google Scholar] [CrossRef]
- Yu, H.A.; Arcila, M.E.; Rekhtman, N.; Sima, C.S.; Zakowski, M.F.; Pao, W.; Kris, M.G.; Miller, V.A.; Ladanyi, M.; Riely, G.J. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 2240–2247. [Google Scholar] [CrossRef]
- Ferrer, L.; Giaj Levra, M.; Brevet, M.; Antoine, M.; Mazieres, J.; Rossi, G.; Chiari, R.; Westeel, V.; Poudenx, M.; Letreut, J.; et al. A Brief report of transformation from NSCLC to SCLC: Molecular and therapeutic characteristics. J. Thorac. Oncol. 2019, 14, 130–134. [Google Scholar] [CrossRef]
- Marcoux, N.; Gettinger, S.N.; O’Kane, G.; Arbour, K.C.; Neal, J.W.; Husain, H.; Evans, T.L.; Brahmer, J.R.; Muzikansky, A.; Bonomi, P.D.; et al. EGFR-mutant adenocarcinomas that transform to small-cell lung cancer and other neuroendocrine carcinomas: Clinical outcomes. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2019, 37, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Moriya, R.; Hokari, S.; Shibata, S.; Koizumi, T.; Tetsuka, T.; Ito, K.; Hashidate, H.; Tsukada, H. Histological transformation to large cell neuroendocrine carcinoma from lung adenocarcinoma harboring an EGFR mutation: An autopsy case report. Intern. Med. 2017, 56, 2013–2017. [Google Scholar] [CrossRef][Green Version]
- Izumi, H.; Yamasaki, A.; Ueda, Y.; Sumikawa, T.; Maeta, H.; Nakamoto, S.; Shimizu, E. Squamous cell carcinoma transformation from EGFR-mutated lung adenocarcinoma: A case report and literature review. Clin. Lung Cancer 2018, 19, e63–e66. [Google Scholar] [CrossRef] [PubMed]
- Khalil, M.Y.; Grandis, J.R.; Shin, D.M. Targeting epidermal growth factor receptor: Novel therapeutics in the management of cancer. Expert Rev. Anticancer Ther. 2003, 3, 367–380. [Google Scholar] [CrossRef]
- Paez, J.G.; Jänne, P.A.; Lee, J.C.; Tracy, S.; Greulich, H.; Gabriel, S.; Herman, P.; Kaye, F.J.; Lindeman, N.; Boggon, T.J.; et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 2004, 304, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Khaddour, K.; Jonna, S.; Deneka, A.; Patel, J.D.; Abazeed, M.E.; Golemis, E.; Borghaei, H.; Boumber, Y. Targeting the epidermal growth factor receptor in EGFR-mutated lung cancer: Current and emerging therapies. Cancers 2021, 13, 3164. [Google Scholar] [CrossRef]
- Schoenfeld, A.J.; Chan, J.M.; Kubota, D.; Sato, H.; Rizvi, H.; Daneshbod, Y.; Chang, J.C.; Paik, P.K.; Offin, M.; Arcila, M.E.; et al. Tumor analyses reveal squamous transformation and off-target alterations as early resistance mechanisms to first-line osimertinib in EGFR-mutant lung cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 2654–2663. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, S.S.; Cheng, Y.; Zhou, C.; Ohe, Y.; Imamura, F.; Cho, B.C.; Lin, M.-C.; Majem, M.; Shah, R.; Rukazenkov, Y.; et al. Mechanisms of acquired resistance to first-line osimertinib: Preliminary data from the phase III FLAURA study. Ann. Oncol. 2018, 29, viii740. [Google Scholar] [CrossRef]
- Yun, C.-H.; Mengwasser, K.E.; Toms, A.V.; Woo, M.S.; Greulich, H.; Wong, K.-K.; Meyerson, M.; Eck, M.J. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc. Natl. Acad. Sci. USA 2008, 105, 2070–2075. [Google Scholar] [CrossRef]
- Engelman, J.A.; Zejnullahu, K.; Mitsudomi, T.; Song, Y.; Hyland, C.; Park, J.O.; Lindeman, N.; Gale, C.-M.; Zhao, X.; Christensen, J.; et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007, 316, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Passaro, A.; Jänne, P.A.; Mok, T.; Peters, S. Overcoming therapy resistance in EGFR -mutant lung cancer. Nat. Cancer 2021, 2, 377–391. [Google Scholar] [CrossRef]
- Shibue, T.; Weinberg, R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef]
- Marck, V.L.V.; Bracke, M.E. Epithelial-Mesenchymal Transitions in Human Cancer; Landes Bioscience: Austin, TX, USA, 2013. [Google Scholar]
- Jakobsen, K.R.; Demuth, C.; Sorensen, B.S.; Nielsen, A.L. The role of epithelial to mesenchymal transition in resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Transl. Lung Cancer Res. 2016, 5, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Prudkin, L.; Liu, D.D.; Ozburn, N.C.; Sun, M.; Behrens, C.; Tang, X.; Brown, K.C.; Bekele, B.N.; Moran, C.; Wistuba, I.I. Epithelial-to-mesenchymal transition in the development and progression of adenocarcinoma and squamous cell carcinoma of the lung. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc 2009, 22, 668–678. [Google Scholar] [CrossRef] [PubMed]
- Thomson, S.; Buck, E.; Petti, F.; Griffin, G.; Brown, E.; Ramnarine, N.; Iwata, K.K.; Gibson, N.; Haley, J.D. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005, 65, 9455–9462. [Google Scholar] [CrossRef]
- Song, K.-A.; Niederst, M.J.; Lochmann, T.L.; Hata, A.N.; Kitai, H.; Ham, J.; Floros, K.V.; Hicks, M.A.; Hu, H.; Mulvey, H.E.; et al. Epithelial-to-mesenchymal transition antagonizes response to targeted therapies in lung cancer by suppressing BIM. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 197–208. [Google Scholar] [CrossRef]
- Shaurova, T.; Zhang, L.; Goodrich, D.W.; Hershberger, P.A. Understanding lineage plasticity as a path to targeted therapy failure in EGFR-mutant non-small cell lung cancer. Front. Genet. 2020, 11, 281. [Google Scholar] [CrossRef]
- Richardson, F.; Young, G.D.; Sennello, R.; Wolf, J.; Argast, G.M.; Mercado, P.; Davies, A.; Epstein, D.M.; Wacker, B. The evaluation of E-Cadherin and vimentin as biomarkers of clinical outcomes among patients with non-small cell lung cancer treated with erlotinib as second- or third-line therapy. Anticancer Res. 2012, 32, 537–552. [Google Scholar]
- Lee, A.-F.; Chen, M.-C.; Chen, C.-J.; Yang, C.-J.; Huang, M.-S.; Liu, Y.-P. Reverse epithelial-mesenchymal transition contributes to the regain of drug sensitivity in tyrosine kinase inhibitor-resistant non-small cell lung cancer cells. PLoS ONE 2017, 12, e0180383. [Google Scholar] [CrossRef]
- Xie, M.; Zhang, L.; He, C.; Xu, F.; Liu, J.; Hu, Z.; Zhao, L.; Tian, Y. Activation of Notch-1 enhances epithelial-mesenchymal transition in gefitinib-acquired resistant lung cancer cells. J. Cell. Biochem. 2012, 113, 1501–1513. [Google Scholar] [CrossRef]
- Yochum, Z.A.; Cades, J.; Wang, H.; Chatterjee, S.; Simons, B.W.; O’Brien, J.P.; Khetarpal, S.K.; Lemtiri-Chlieh, G.; Myers, K.V.; Huang, E.H.-B.; et al. Targeting the EMT transcription factor TWIST1 overcomes resistance to EGFR inhibitors in EGFR-mutant non-small-cell lung cancer. Oncogene 2019, 38, 656–670. [Google Scholar] [CrossRef]
- Izumchenko, E.; Chang, X.; Michailidi, C.; Kagohara, L.; Ravi, R.; Paz, K.; Brait, M.; Hoque, M.O.; Ling, S.; Bedi, A.; et al. The TGFβ-miR200-MIG6 pathway orchestrates the EMT-associated kinase switch that induces resistance to EGFR inhibitors. Cancer Res. 2014, 74, 3995–4005. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Shien, K.; Tomida, S.; Okayasu, K.; Suzawa, K.; Hashida, S.; Torigoe, H.; Watanabe, M.; Yamamoto, H.; Soh, J.; et al. Targeting the miR-200c/LIN28B axis in acquired EGFR-TKI resistance non-small cell lung cancer cells harboring EMT features. Sci. Rep. 2017, 7, 40847. [Google Scholar] [CrossRef] [PubMed]
- Legras, A.; Pécuchet, N.; Imbeaud, S.; Pallier, K.; Didelot, A.; Roussel, H.; Gibault, L.; Fabre, E.; Le Pimpec-Barthes, F.; Laurent-Puig, P.; et al. Epithelial-to-mesenchymal transition and microRNAs in lung cancer. Cancers 2017, 9, 101. [Google Scholar] [CrossRef]
- Park, K.-S.; Raffeld, M.; Moon, Y.W.; Xi, L.; Bianco, C.; Pham, T.; Lee, L.C.; Mitsudomi, T.; Yatabe, Y.; Okamoto, I.; et al. CRIPTO1 expression in EGFR-mutant NSCLC elicits intrinsic EGFR-inhibitor resistance. J. Clin. Invest. 2014, 124, 3003–3015. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lee, J.C.; Lin, L.; Olivas, V.; Au, V.; LaFramboise, T.; Abdel-Rahman, M.; Wang, X.; Levine, A.D.; Rho, J.K.; et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat. Genet. 2012, 44, 852–860. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; Zeng, S.; Wang, Z.; Wu, M.; Ma, Y.; Ye, X.; Zhang, B.; Liu, H. Cancer-associated fibroblasts promote epithelial-mesenchymal transition and EGFR-TKI resistance of non-small cell lung cancers via HGF/IGF-1/ANXA2 signaling. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Nurwidya, F.; Takahashi, F.; Kobayashi, I.; Murakami, A.; Kato, M.; Minakata, K.; Nara, T.; Hashimoto, M.; Yagishita, S.; Baskoro, H.; et al. Treatment with insulin-like growth factor 1 receptor inhibitor reverses hypoxia-induced epithelial-mesenchymal transition in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2014, 455, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.V.; Lee, D.Y.; Li, B.; Quinlan, M.P.; Takahashi, F.; Maheswaran, S.; McDermott, U.; Azizian, N.; Zou, L.; Fischbach, M.A.; et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010, 141, 69–80. [Google Scholar] [CrossRef]
- Weng, C.-H.; Chen, L.-Y.; Lin, Y.-C.; Shih, J.-Y.; Lin, Y.-C.; Tseng, R.-Y.; Chiu, A.-C.; Yeh, Y.-H.; Liu, C.; Lin, Y.-T.; et al. Epithelial-mesenchymal transition (EMT) beyond EGFR mutations per se is a common mechanism for acquired resistance to EGFR TKI. Oncogene 2019, 38, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Diao, L.; Cuentas, E.R.P.; Denning, W.L.; Chen, L.; Fan, Y.; Byers, L.A.; Wang, J.; Papadimitrakopoulou, V.; Behrens, C.; et al. Epithelial-mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 3630–3642. [Google Scholar] [CrossRef]
- Hsieh, M.-S.; Lin, M.-W.; Lee, Y.-H. Lung adenocarcinoma with sarcomatoid transformation after tyrosine kinase inhibitor treatment and chemotherapy. Lung Cancer 2019, 137, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Roca, E.; Gurizzan, C.; Amoroso, V.; Vermi, W.; Ferrari, V.; Berruti, A. Outcome of patients with lung adenocarcinoma with transformation to small-cell lung cancer following tyrosine kinase inhibitors treatment: A systematic review and pooled analysis. Cancer Treat. Rev. 2017, 59, 117–122. [Google Scholar] [CrossRef]
- Meuwissen, R.; Linn, S.C.; Linnoila, R.I.; Zevenhoven, J.; Mooi, W.J.; Berns, A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 2003, 4, 181–189. [Google Scholar] [CrossRef]
- Byers, L.A.; Wang, J.; Nilsson, M.B.; Fujimoto, J.; Saintigny, P.; Yordy, J.; Giri, U.; Peyton, M.; Fan, Y.H.; Diao, L.; et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2012, 2, 798–811. [Google Scholar] [CrossRef]
- Peifer, M.; Fernández-Cuesta, L.; Sos, M.L.; George, J.; Seidel, D.; Kasper, L.H.; Plenker, D.; Leenders, F.; Sun, R.; Zander, T.; et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat. Genet. 2012, 44, 1104–1110. [Google Scholar] [CrossRef]
- Meder, L.; König, K.; Ozretić, L.; Schultheis, A.M.; Ueckeroth, F.; Ade, C.P.; Albus, K.; Boehm, D.; Rommerscheidt-Fuss, U.; Florin, A.; et al. NOTCH, ASCL1, p53 and RB alterations define an alternative pathway driving neuroendocrine and small cell lung carcinomas. Int. J. Cancer 2016, 138, 927–938. [Google Scholar] [CrossRef]
- Niederst, M.J.; Sequist, L.V.; Poirier, J.T.; Mermel, C.H.; Lockerman, E.L.; Garcia, A.R.; Katayama, R.; Costa, C.; Ross, K.N.; Moran, T.; et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat. Commun. 2015, 6, 6377. [Google Scholar] [CrossRef]
- Lee, J.-K.; Lee, J.; Kim, S.; Kim, S.; Youk, J.; Park, S.; An, Y.; Keam, B.; Kim, D.-W.; Heo, D.S.; et al. Clonal history and genetic predictors of transformation into small-cell carcinomas from lung adenocarcinomas. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2017, 35, 3065–3074. [Google Scholar] [CrossRef] [PubMed]
- Oser, M.G.; Niederst, M.J.; Sequist, L.V.; Engelman, J.A. Transformation from non-small-cell lung cancer to small-cell lung cancer: Molecular drivers and cells of origin. Lancet Oncol. 2015, 16, e165–e172. [Google Scholar] [CrossRef]
- Offin, M.; Chan, J.M.; Tenet, M.; Rizvi, H.A.; Shen, R.; Riely, G.J.; Rekhtman, N.; Daneshbod, Y.; Quintanal-Villalonga, A.; Penson, A.; et al. Concurrent RB1 and TP53 Alterations define a subset of EGFR-mutant lung cancers at risk for histologic transformation and inferior clinical outcomes. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2019, 14, 1784–1793. [Google Scholar] [CrossRef]
- Gong, Z.; Lu, R.; Xie, S.; Jiang, M.; Liu, K.; Xiao, R.; Shen, J.; Wang, Y.; Guo, L. Overexpression of pro-gastrin releasing peptide promotes the cell proliferation and progression in small cell lung cancer. Biochem. Biophys. Res. Commun. 2016, 479, 312–318. [Google Scholar] [CrossRef]
- Liu, Y. Small cell lung cancer transformation from EGFR-mutated lung adenocarcinoma: A case report and literatures review. Cancer Biol. Ther. 2018, 19, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, X.-Y.; Tang, Y.; Xu, Y.; Guo, W.-H.; Li, Y.-C.; Liu, X.-K.; Huang, C.-Y.; Wang, Y.-S.; Wei, Y.-Q. Rapid increase of serum neuron specific enolase level and tachyphylaxis of EGFR-tyrosine kinase inhibitor indicate small cell lung cancer transformation from EGFR positive lung adenocarcinoma? Lung Cancer Amst. Neth. 2013, 81, 302–305. [Google Scholar] [CrossRef]
- Kato, Y.; Tanaka, Y.; Hino, M.; Gemma, A. ProGRP as early predictive marker of non–small-cell lung cancer to small-cell lung cancer transformation after EGFR-TKI treatment. Respir. Med. Case Rep. 2019, 27, 100837. [Google Scholar] [CrossRef] [PubMed]
- Witkiewicz, A.K.; Chung, S.; Brough, R.; Vail, P.; Franco, J.; Lord, C.J.; Knudsen, E.S. Targeting the vulnerability of RB tumor suppressor loss in triple-negative breast cancer. Cell Rep. 2018, 22, 1185–1199. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Du, J.; Parsons, S.H.; Merzoug, F.F.; Webster, Y.; Iversen, P.W.; Chio, L.-C.; Van Horn, R.D.; Lin, X.; Blosser, W.; et al. Aurora a kinase inhibition is synthetic lethal with loss of the RB1 tumor suppressor gene. Cancer Discov. 2019, 9, 248–263. [Google Scholar] [CrossRef]
- Shah, K.N.; Bhatt, R.; Rotow, J.; Rohrberg, J.; Olivas, V.; Wang, V.E.; Hemmati, G.; Martins, M.M.; Maynard, A.; Kuhn, J.; et al. Aurora kinase A drives the evolution of resistance to third-generation EGFR inhibitors in lung cancer. Nat. Med. 2019, 25, 111–118. [Google Scholar] [CrossRef]
- Ku, S.Y.; Rosario, S.; Wang, Y.; Mu, P.; Seshadri, M.; Goodrich, Z.W.; Goodrich, M.M.; Labbé, D.P.; Gomez, E.C.; Wang, J.; et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017, 355, 78–83. [Google Scholar] [CrossRef]
- Lantuejoul, S.; Fernandez-Cuesta, L.; Damiola, F.; Girard, N.; McLeer, A. New molecular classification of large cell neuroendocrine carcinoma and small cell lung carcinoma with potential therapeutic impacts. Transl. Lung Cancer Res. 2020, 9, 2233–2244. [Google Scholar] [CrossRef]
- George, J.; Walter, V.; Peifer, M.; Alexandrov, L.B.; Seidel, D.; Leenders, F.; Maas, L.; Müller, C.; Dahmen, I.; Delhomme, T.M.; et al. Integrative genomic profiling of large-cell neuroendocrine carcinomas reveals distinct subtypes of high-grade neuroendocrine lung tumors. Nat. Commun. 2018, 9, 1048. [Google Scholar] [CrossRef]
- Belluomini, L.; Caliò, A.; Giovannetti, R.; Motton, M.; Mazzarotto, R.; Micheletto, C.; Infante, M.V.; Scarpa, A.; Milella, M.; Pilotto, S. Molecular predictors of EGFR-mutant NSCLC transformation into LCNEC after frontline osimertinib: Digging under the surface. ESMO Open 2021, 6, 100028. [Google Scholar] [CrossRef] [PubMed]
- Baglivo, S.; Ludovini, V.; Sidoni, A.; Metro, G.; Ricciuti, B.; Siggillino, A.; Rebonato, A.; Messina, S.; Crinò, L.; Chiari, R. Large cell neuroendocrine carcinoma transformation and EGFR-T790M mutation as coexisting mechanisms of acquired resistance to EGFR-TKIs in lung cancer. Mayo Clin. Proc. 2017, 92, 1304–1311. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Shim, J.H.; Lee, B.; Cho, I.; Park, W.-Y.; Kim, Y.; Lee, S.-H.; Choi, Y.L.; Han, J.; Ahn, J.S.; et al. Paired genomic analysis of squamous cell carcinoma transformed from EGFR-mutated lung adenocarcinoma. Lung Cancer Amst. Neth. 2019, 134, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Matsui, A.; Shimoyama, Y.; Omote, N.; Morise, M.; Hase, T.; Tanaka, I.; Suzuki, K.; Hasegawa, Y. An EGFR-mutated lung adenocarcinoma undergoing squamous cell carcinoma transformation exhibited a durable response to afatinib. Intern. Med. 2018, 57, 3429–3432. [Google Scholar] [CrossRef]
- Jin, R.; Peng, L.; Shou, J.; Wang, J.; Jin, Y.; Liang, F.; Zhao, J.; Wu, M.; Li, Q.; Zhang, B.; et al. EGFR-mutated squamous cell lung cancer and its association with outcomes. Front. Oncol. 2021, 11, 2262. [Google Scholar] [CrossRef]
- Ruffini, E.; Rena, O.; Oliaro, A.; Filosso, P.L.; Bongiovanni, M.; Arslanian, A.; Papalia, E.; Maggi, G. Lung tumors with mixed histologic pattern. Clinico-pathologic characteristics and prognostic significance. Eur. J. Cardio-Thorac. Surg. Off. J. Eur. Assoc. Cardio-Thorac. Surg. 2002, 22, 701–707. [Google Scholar] [CrossRef]
- Han, X.; Li, F.; Fang, Z.; Gao, Y.; Li, F.; Fang, R.; Yao, S.; Sun, Y.; Li, L.; Zhang, W.; et al. Transdifferentiation of lung adenocarcinoma in mice with Lkb1 deficiency to squamous cell carcinoma. Nat. Commun. 2014, 5, 3261. [Google Scholar] [CrossRef]
- Koivunen, J.P.; Kim, J.; Lee, J.; Rogers, A.M.; Park, J.O.; Zhao, X.; Naoki, K.; Okamoto, I.; Nakagawa, K.; Yeap, B.Y.; et al. Mutations in the LKB1 tumour suppressor are frequently detected in tumours from Caucasian but not Asian lung cancer patients. Br. J. Cancer 2008, 99, 245–252. [Google Scholar] [CrossRef]
- Gao, B.; Sun, Y.; Zhang, J.; Ren, Y.; Fang, R.; Han, X.; Shen, L.; Liu, X.-Y.; Pao, W.; Chen, H.; et al. Spectrum of LKB1, EGFR, and KRAS mutations in chinese lung adenocarcinomas. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2010, 5, 1130–1135. [Google Scholar] [CrossRef] [PubMed]
- Cescon, D.W.; Bratman, S.V.; Chan, S.M.; Siu, L.L. Circulating tumor DNA and liquid biopsy in oncology. Nat. Cancer 2020, 1, 276–290. [Google Scholar] [CrossRef]
- Remon, J.; Menis, J.; Hasan, B.; Peric, A.; De Maio, E.; Novello, S.; Reck, M.; Berghmans, T.; Wasag, B.; Besse, B.; et al. The APPLE trial: Feasibility and activity of AZD9291 (osimertinib) treatment on positive PLASMA T790M in EGFR-mutant NSCLC patients. EORTC 1613. Clin. Lung Cancer 2017, 18, 583–588. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).