Syndecan-4 in Tumor Cell Motility
Abstract
:Simple Summary
Abstract
1. Introduction
2. Cytoskeletal System during Cell Migration
2.1. Rearrangement of the Actin Cytoskeleton during Migration
2.2. The Role of Intermediate Filaments in Cell Motility
2.3. The Complex Function of Microtubules in Cell Migration
2.4. The Role of Septins in Cell Migration
3. Multiple Functions of Rho GTPases in Cell Motility
4. Front-Rear Polarity of Migrating Cells
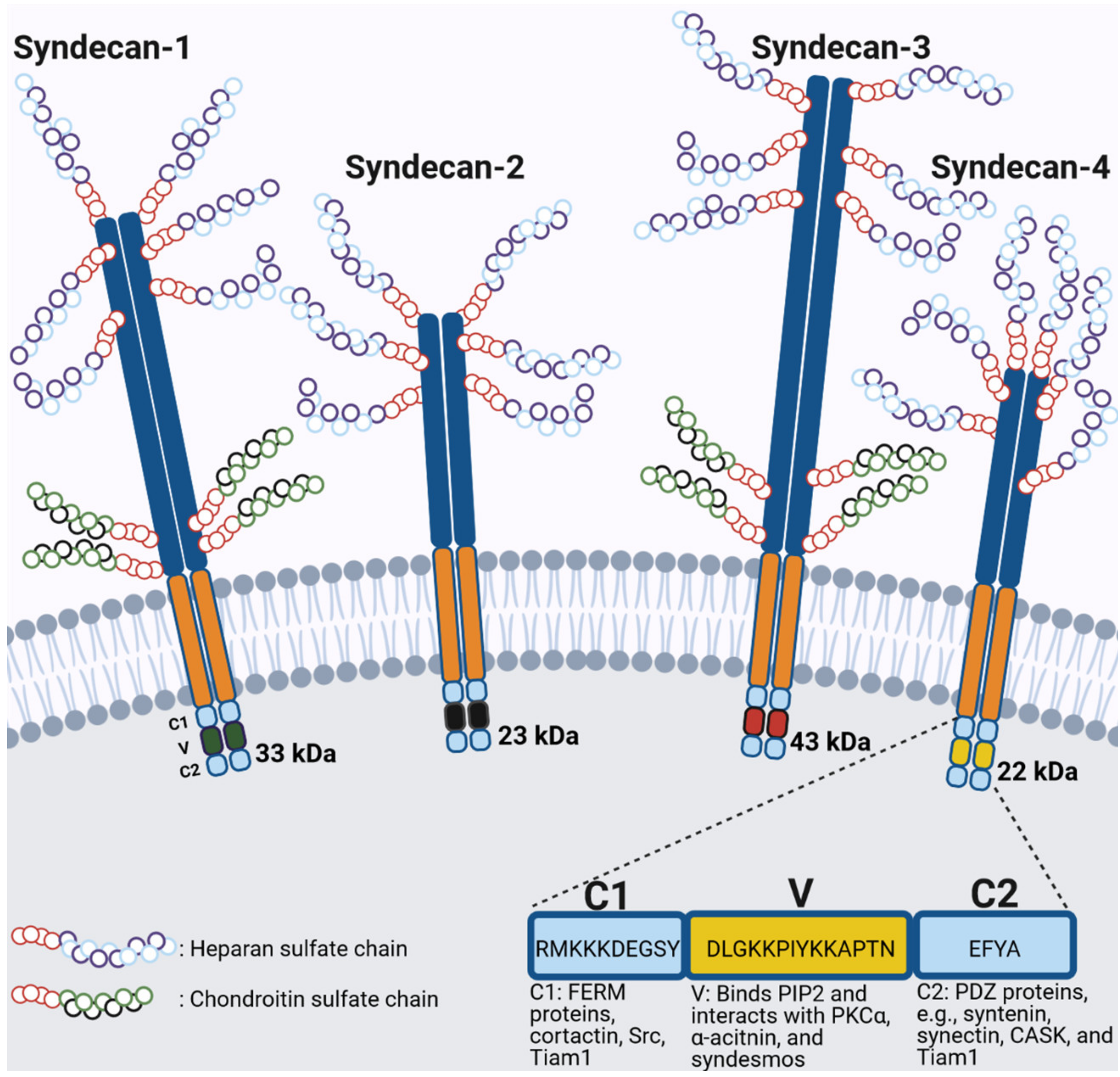
5. Syndecan Family of Transmembrane Proteoglycans
General Structure of Syndecans
6. Structure, Interacting Partners and Signaling of Syndecan-4
6.1. Syndecan-4 and the Regulation of Rac1/RhoA Activity
6.2. Syndecan-4 and Focal Adhesion Formation
7. SDC4 and Tumor Cell Migration
7.1. Melanoma
7.2. Breast Cancer
7.3. Lung Cancer
7.4. Other Tumor Types
| Cell Type | Migration Assay | Signaling Pathway | Biological Effect | Citation |
|---|---|---|---|---|
| 4T1 and MDA-MB-231 breast cancer cells | - | - | SDC4 has an anti-migratory, anti-invasive tumor suppressor role. | [161] |
| Colon carcinoma cells | - | SDC4 expression | SDC4 is downregulated in colon carcinoma cells. | [162] |
| Infiltrating breast carcinoma tissues | - | SDC4 expression | SDC4 is upregulated in normal breast tissue compared to malignant breast tissue | [202] |
| Human ovarian carcinoma cell line NIH:OVCAR5 | Modified Boyden chamber chemotaxis, Matrigel invasion assay | Carbohydrate modifications | The migration, invasion and tumor growth of ovarian carcinoma is mediated by the carbohydrate modifications of proteoglycans.SDC4 is upregulated in ovarian carcinoma. | [164] |
| Mesothelioma, fibrosarcoma | - | SDC4 expression | SDC4 is upregulated in mesothelioma and fibrosarcoma. | [165] |
| Breast carcinoma samples from patients | - | SDC4 expression | SDC4 is associated with high histological grade and a negative estrogen receptor status in breast carcinoma. | [166] |
| 4T1 mouse breast cancer cells | - | bone metastasis formation | SDC4-silenced breast carcinoma cells have decreased ability to form bone metastasis in mice. | [102] |
| JKT-1 human seminoma cell line, NTERA-2 human embryonal carcinoma cell line, NCCIT teratocarcinoma cell line | - | SDC4 expression—metastatic potential | Reduced SDC4 expression is associated with reduced metastatic potential in testicular germ cell tumors. | [167] |
| Patients with primary high grade intramedullary osteosarcoma, with low grade central osteosarcoma, with osteoid osteoma and normal bone tissues | - | SDC4 expression—metastasis formation, tumor size | Increased SDC4 expression is associated with the formation of distant metastasis and increased tumor size in osteosarcoma. | [168] |
| Renca (mouse), 786-O and Caki-2 (human) renal carcinoma cells | Wound scratch assay, Transwell assay | High SDC4 expression in renal cell carcinoma | High SDC4 expression determines increased patient survival in renal cell carcinoma. | [169] |
| M5 human metastatic melanoma cells | Chemotaxis assay, wound scratch assay | FGF-2/SDC4 | FGF-2 regulates melanoma cell migration in a SDC4-dependent manner. | [177] |
| MV3 human melanoma cell line | Wound scratch assay | Cyr61/SDC4 | Cyr61 is exocytosed by binding to SDC4. Cyr61 binds to and activates integrins, thus induce migration, metastasis formation and tumorigenicity. | [178] |
| Rat embryonic fibroblasts (REFs), A375 melanoma cells, B16F10 melanoma cells, C57BL/6 mice | Transwell migration assay, lung metastasis model | Syntenin-1/SDC4 SDC4—inhibition of cancer-associated melanoma migration | SDC4 overexpression decreases melanoma cell migration in vitro and reduces the metastatic potential of melanoma in vivo. Syntenin-1 negatively regulates SDC4-mediated inhibition of cell migration and SDC4-mediated tumor suppression in melanoma. | [179] |
| B16.F10 murine melanoma cells | Wound scratch assay | LysoPC/PKCδ/SDC4/PKCα/FAK | LysoPC C18:0 decreases the metastatic spread of melanoma cells. LysoPC activates PKCδ to phosphorylate SDC4 thereby deactivating PKCα and reducing FAK activity. | [180] |
| MDA-MB-231 and MCF7 human breast cancer cells | 2D: wound scratch assay 3D: Matrigel and Collagen Type I | ADAMTS-15/SDC4 | Inhibition of mammary cancer cell migration by ADAMTS-15 requires SDC4. | [184] |
| Human HaCat keratinocytes, A431 (human squamous skin epithelial) carcinoma cells, MCF10A (human mammary gland epithelial) cells | Wound scratch assay | HER1(EGFR)/α6β4 integrin/SDC4 | HER1-dependent activation of α6β4 integrin and α6β4 integrin-mediated cell invasion require SDC4. | [185] |
| MDA-MB-231 breast adenocarcinoma cells | Cell invasion into 3D collagen gel | Integrin α2β1/MT1-MMP/SDCs–K-Ras mutant cell invasion | K-Ras mutant cells show increased expression of SDC1 and SDC4. MT1-MMP and α2β1 integrin promote invasive phenotype, SDCs reduce invasion into collagen matrices. | [186] |
| MCF7, MDA-MB-435s and MDA-MB-231 breast cancer cells | Migration chamber (insert with polyethylene filter with 8 µM pores) | LL-37/SDC4LL-37/TRPV2/ic. Ca2+ LL-37/PI3K/AKT/motility | SDC4 is a receptor for LL-37 increasing Ca2+ levels via TRPV2 channels and increasing the motility of breast cancer cells via PI3K/AKT signaling. | [187] |
| Non-metastatic rat mammary R37 cells, highly metastatic KP1 cells (R37 cells transfected with S100A4) | Wound scratch assay | SDC4/α5β1 integrin/PKCα—TG2 and S100A4-mediated cell migration | S100A4 mediates migration of tumor cells via SDC4 and α5β1 integrin-mediated PKCα activation. | [188] |
| PANC-1 human pancreas adenocarcinoma cells, HT-29 human colon adenocarcinoma cells, MCF-7 and MDA-MB-231 human breast adenocarcinoma cells | - | NT4—SDC4 | The branched peptide NT4 inhibits cancer cell migration and FGF-induced invasion. NT4 binds to SDC4, the expression of SDC4 is upregulated breast cancer cells. | [189] |
| MCF-7 (low metastatic ERa+), MDA-MB-231 (highly invasive ERa-) breast cancer cells | Wound scratch assay | IGFR/SDC4 expression | IGFR regulates the expression of SDC4 both in the presence and in the absence of E2 in breast cancer cells.IGFR inhibitors reduced the migration of MCF-7 cells but did not have a significant effect on MDA-MB-231 cells. | [190] |
| C57Bl/6 mouse primary lymphatic endothelial cells, Lewis lung carcinoma cells, bone marrow–derived DCs (BMDCs) | Transwell migration assay, in vivo migration assay (BMDCs migration into lymph node), tumor growth studies | SDC4—dendritic cell maturation | SDC4-deficient mice exhibit impaired tumor growth and increased infiltration by mature dendritic cells. SDC4 is the dominant proteoglycan on dendritic cells. | [193] |
| Primary lung fibroblasts | Boyden chamber, chemotaxis assay | CXCL10—SDC4 | In response to lung injury, the expression of SDC4 is increased. SDC4 directly interacts with CXCL10 and they inhibit the migration of fibroblasts. SDC4 is required for the inhibitory effect of CXCL10 during fibrosis. | [194] |
| Human blood–derived monocytes, primary pulmonary endothelial cells, Lewis lung carcinoma cells (LLC1) | Boyden chamber, Transwell assay, spontaneous metastasis in mice | - | Increased expression of SDC4 is observed in endothelial cells after tumor cell seeding to the lungs. | [195] |
| Mouse lung endothelial cells | Random migration assay; ex vivo C57BL/6 mice aortic ring assay | ADAMTS-1—MMP9—SDC4 | ADAMTS-1 modulates the cell surface expression of SDC4 via MMP9. ADAMTS-1 and SDC4 inhibit cell migration, whilst their inhibition increase angiogenesis. | [126] |
| A549 human lung adenocarcinoma cells | Wound scratch assay, transwell chemotaxis assay | SDC4/Snail/TGFβ1-induced EMT | SDC4 promotes migration and invasion of lung adenocarcinoma cells. SDC4 positively regulates TGFβ1-induced EMT (via Snail), consequently promoting a more motile phenotype. | [174] |
| A549 lung tumor epithelial cells | Wound scratch assay, matrigel invasion assay, in vivo lung tumor metastasis | ADAM17–SDC4 cleavage;SDC1—in vivo lung tumor metastasis | SDC1 tCFT was sufficient to induce lung metastasis formation in SCID mice, whilst SDC4 tCFT achieved as efficient wound closure as SDC1 tCFT. (tCTF = transmembrane C-terminal fragment) | [196] |
| JAR choriocarcinoma cells | Modified Boyden-chamber chemotactic assay | CXCL12/SDC4 | SDC4 binds to CXCL12 and regulates CXCL12-mediated cell migration and invasion. SDC4 plays a role in the invasiveness of extravillous cytotrophoblast in moles. | [197] |
| Huh7 human hepatoma cells | Bio-coat cell migration chambers, Matrigel invasion assay | SDF-1 (CXCL12)/ CXCR4/SDC4 | SDC4 is essential for SDF-1 (CXCL12) induced migration and invasion of hepatoma cells. | [198] |
| Human cervix epitheloid carcinoma (HeLa) cells | Bio-coat cell migration chambers, Matrigel invasion assay | SDC4–SDF-1/CXCL12– PKCδ, JNK/SAPK | SDC4 plays a role in SDF-1/CXCL12-mediated cell invasion and chemotaxis. PKCδ and c-jun NH2-terminal kinase/stress-activated protein kinase (JNK/SAPK) are involved in the SDF-1/CXCL12-induced cell invasion. | [199] |
| Human cervix epitheloid carcinoma (HeLa) cells | Wound scratch assay, Transwell assays | Calumenin–FN, SDC4, α5β1 integrin–ERK1/2 | Calumenin inhibits cell migration and tumor metastasis through FN, SDC4 and α5β1-integrin by the suppression of ERK1/2 signaling. | [200] |
| Papillary thyroid cancer cells K1, BCPAP, TPC-1 and IHH-4, normal thyroid Nthy-ori3-1 cells | Transwell assay, wound scratch assay | SDC4—Wnt/β-catenin signaling pathway | SDC4-silencing decreased papillary thyroid cancer cell migration and invasion and represses EMT. Furthermore, SDC4-silencing suppresses Wnt/βcatenin signaling, thus promoting apoptosis. | [176] |
| Huh7, HepG2and Hep3B human hepatoma cells | Bio-coat migration chambers, Matrigel invasion assay | RANTES/CCL5—SDC4 | SDC4 is essential in RANTES/CCL5-mediated hepatoma cell invasion and migration and its binding to the cell plasma membrane. | [201] |
8. Syndecan-4 and Non-Cancer Cell Migration
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Stuelten, C.H.; Parent, C.A.; Montell, D.J. Cell Motility in Cancer Invasion and Metastasis: Insights from Simple Model Organisms. Nat. Rev. Cancer 2018, 18, 296–312. [Google Scholar] [CrossRef]
- Welch, D.R.; Hurst, D.R. Defining the Hallmarks of Metastasis. Cancer Res. 2019, 79, 3011–3027. [Google Scholar] [CrossRef]
- Mittal, V. Epithelial Mesenchymal Transition in Tumor Metastasis. Annu. Rev. Pathol. 2018, 13, 395–412. [Google Scholar] [CrossRef]
- Zhang, Y.; Weinberg, R.A. Epithelial-to-Mesenchymal Transition in Cancer: Complexity and Opportunities. Front. Med. 2018, 12, 361–373. [Google Scholar] [CrossRef] [Green Version]
- Lu, W.; Kang, Y. Epithelial-Mesenchymal Plasticity in Cancer Progression and Metastasis. Dev. Cell 2019, 49, 361–374. [Google Scholar] [CrossRef]
- Seetharaman, S.; Etienne-Manneville, S. Cytoskeletal Crosstalk in Cell Migration. Trends Cell Biol. 2020, 30, 720–735. [Google Scholar] [CrossRef] [PubMed]
- Ribatti, D.; Tamma, R.; Annese, T. Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Transl. Oncol. 2020, 13, 100773. [Google Scholar] [CrossRef]
- Lindsey, S.; Langhans, S.A. Crosstalk of Oncogenic Signaling Pathways During Epithelial-Mesenchymal Transition. Front. Oncol. 2014, 4, 358. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular Mechanisms of Epithelial-Mesenchymal Transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [Green Version]
- Chhabra, E.S.; Higgs, H.N. The Many Faces of Actin: Matching Assembly Factors with Cellular Structures. Nat. Cell Biol. 2007, 9, 1110–1121. [Google Scholar] [CrossRef]
- Svitkina, T. The Actin Cytoskeleton and Actin-Based Motility. Cold Spring Harb. Perspect. Biol. 2018, 10, a018267. [Google Scholar] [CrossRef] [Green Version]
- Sackmann, E.; Tanaka, M. Critical Role of Lipid Membranes in Polarization and Migration of Cells: A Biophysical View. Biophys. Rev. 2021, 13, 123–138. [Google Scholar] [CrossRef]
- Zaidel-Bar, R.; Itzkovitz, S.; Ma’ayan, A.; Iyengar, R.; Geiger, B. Functional Atlas of the Integrin Adhesome. Nat. Cell Biol. 2007, 9, 858–867. [Google Scholar] [CrossRef] [PubMed]
- Ridley, A.J.; Schwartz, M.A.; Burridge, K.; Firtel, R.A.; Ginsberg, M.H.; Borisy, G.; Parsons, J.T.; Horwitz, A.R. Cell Migration: Integrating Signals from Front to Back. Science 2003, 302, 1704–1709. [Google Scholar] [CrossRef] [Green Version]
- Petrie, R.J.; Gavara, N.; Chadwick, R.S.; Yamada, K.M. Nonpolarized Signaling Reveals Two Distinct Modes of 3d Cell Migration. J. Cell Biol. 2012, 197, 439–455. [Google Scholar] [CrossRef] [Green Version]
- Franchi, M.; Masola, V.; Bellin, G.; Onisto, M.; Karamanos, K.A.; Piperigkou, Z. Collagen Fiber Array of Peritumoral Stroma Influences Epithelial-to-Mesenchymal Transition and Invasive Potential of Mammary Cancer Cells. J. Clin. Med. 2019, 8, 213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, K.M.; Collins, J.W.; Walma, D.A.C.; Doyle, A.D.; Morales, S.G.; Lu, J.; Matsumoto, K.; Nazari, S.S.; Sekiguchi, R.; Shinsato, Y.; et al. Extracellular Matrix Dynamics in Cell Migration, Invasion and Tissue Morphogenesis. Int. J. Exp. Pathol. 2019, 100, 144–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, K.M.; Sixt, M. Mechanisms of 3d Cell Migration. Nat. Rev. Mol. Cell Biol. 2019, 20, 738–752. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Condeelis, J. Regulation of the Actin Cytoskeleton in Cancer Cell Migration and Invasion. Biochim. Biophys. Acta 2007, 1773, 642–652. [Google Scholar] [CrossRef] [Green Version]
- Bachir, A.I.; Horwitz, A.R.; Nelson, W.J.; Bianchini, J.M. Actin-Based Adhesion Modules Mediate Cell Interactions with the Extracellular Matrix and Neighboring Cells. Cold Spring Harb. Perspect. Biol. 2017, 9, a023234. [Google Scholar] [CrossRef] [Green Version]
- Burridge, K.; Guilluy, C. Focal Adhesions, Stress Fibers and Mechanical Tension. Exp. Cell Res. 2016, 343, 14–20. [Google Scholar] [CrossRef] [Green Version]
- Kemp, J.P., Jr.; Brieher, W.M. The Actin Filament Bundling Protein Alpha-Actinin-4 Actually Suppresses Actin Stress Fibers by Permitting Actin Turnover. J. Biol. Chem. 2018, 293, 14520–14533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byers, H.R.; White, G.E.; Fujiwara, K. Organization and Function of Stress Fibers in Cells In Vitro and In Situ. A Review. Cell Muscle Motil. 1984, 5, 83–137. [Google Scholar] [PubMed]
- Hu, Y.L.; Li, S.; Miao, H.; Tsou, T.C.; del Pozo, M.A.; Chien, S. Roles of Microtubule Dynamics and Small Gtpase Rac in Endothelial Cell Migration and Lamellipodium Formation under Flow. J. Vasc. Res. 2002, 39, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Burnette, D.T.; Manley, S.; Sengupta, P.; Sougrat, R.; Davidson, M.W.; Kachar, B.; Lippincott-Schwartz, J. A Role for Actin Arcs in the Leading-Edge Advance of Migrating Cells. Nat. Cell Biol. 2011, 13, 371–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Small, J.V.; Herzog, M.; Anderson, K. Actin Filament Organization in the Fish Keratocyte Lamellipodium. J. Cell Biol. 1995, 129, 1275–1286. [Google Scholar] [CrossRef]
- Svitkina, T.M.; Verkhovsky, A.B.; McQuade, K.M.; Borisy, G.G. Analysis of the Actin-Myosin Ii System in Fish Epidermal Keratocytes: Mechanism of Cell Body Translocation. J. Cell Biol. 1997, 139, 397–415. [Google Scholar] [CrossRef] [Green Version]
- Pollard, T.D.; Borisy, G.G. Cellular Motility Driven by Assembly and Disassembly of Actin Filaments. Cell 2003, 112, 453–465. [Google Scholar] [CrossRef] [Green Version]
- Huxley, A.F.; Niedergerke, R. Measurement of Muscle Striations in Stretch and Contraction. J. Physiol. 1954, 124, 46–47. [Google Scholar]
- Huxley, H.; Hanson, J. Changes in the Cross-Striations of Muscle During Contraction and Stretch and Their Structural Interpretation. Nature 1954, 173, 973–976. [Google Scholar] [CrossRef]
- Mullins, R.D.; Heuser, J.A.; Pollard, T.D. The Interaction of Arp2/3 Complex with Actin: Nucleation, High Affinity Pointed End Capping, and Formation of Branching Networks of Filaments. Proc. Natl. Acad. Sci. USA 1998, 95, 6181–6186. [Google Scholar] [CrossRef] [Green Version]
- Mullins, R.D.; Kelleher, J.F.; Xu, J.; Pollard, T.D. Arp2/3 Complex from Acanthamoeba Binds Profilin and Cross-Links Actin Filaments. Mol. Biol. Cell 1998, 9, 841–852. [Google Scholar] [CrossRef] [Green Version]
- Drees, F.; Pokutta, S.; Yamada, S.; Nelson, W.J.; Weis, W.I. Alpha-Catenin Is a Molecular Switch That Binds E-Cadherin-Beta-Catenin and Regulates Actin-Filament Assembly. Cell 2005, 123, 903–915. [Google Scholar] [CrossRef] [Green Version]
- Krause, M.; Dent, E.W.; Bear, J.E.; Loureiro, J.J.; Gertler, F.B. Ena/Vasp Proteins: Regulators of the Actin Cytoskeleton and Cell Migration. Annu. Rev. Cell Dev. Biol. 2003, 19, 541–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiering, D.; Hodgson, L. Dynamics of the Rho-Family Small Gtpases in Actin Regulation and Motility. Cell Adh. Migr. 2011, 5, 170–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksson, J.E.; Dechat, T.; Grin, B.; Helfand, B.; Mendez, M.; Pallari, H.M.; Goldman, R.D. Introducing Intermediate Filaments: From Discovery to Disease. J. Clin. Invest. 2009, 119, 1763–1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, B.M.; Rotty, J.D.; Coulombe, P.A. Networking Galore: Intermediate Filaments and Cell Migration. Curr. Opin. Cell Biol. 2013, 25, 600–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leduc, C.; Etienne-Manneville, S. Intermediate Filaments in Cell Migration and Invasion: The Unusual Suspects. Curr. Opin. Cell Biol. 2015, 32, 102–112. [Google Scholar] [CrossRef]
- Chernoivanenko, I.S.; Minin, A.A.; Minin, A.A. Role of Vimentin in Cell Migration. Ontogenez 2013, 44, 186–202. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Eriksson, J.E. Intermediate Filaments and the Regulation of Cell Motility During Regeneration and Wound Healing. Cold Spring Harb. Perspect. Biol. 2017, 9, a022046. [Google Scholar] [CrossRef] [PubMed]
- Helfand, B.T.; Mendez, M.G.; Murthy, S.N.; Shumaker, D.K.; Grin, B.; Mahammad, S.; Aebi, U.; Wedig, T.; Wu, Y.I.; Hahn, K.M.; et al. Vimentin Organization Modulates the Formation of Lamellipodia. Mol. Biol. Cell 2011, 22, 1274–1289. [Google Scholar] [CrossRef] [PubMed]
- Sonavane, P.R.; Wang, C.; Dzamba, B.; Weber, G.F.; Periasamy, A.; DeSimone, D.W. Mechanical and Signaling Roles for Keratin Intermediate Filaments in the Assembly and Morphogenesis of Xenopus Mesendoderm Tissue at Gastrulation. Development 2017, 144, 4363–4376. [Google Scholar]
- De Pascalis, C.; Pérez-González, C.; Seetharaman, S.; Boëda, B.; Vianay, B.; Burute, M.; Leduc, C.; Borghi, N.; Trepat, X.; Etienne-Manneville, S. Intermediate Filaments Control Collective Migration by Restricting Traction Forces and Sustaining Cell-Cell Contacts. J. Cell Biol. 2018, 217, 3031–3044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinogradova, T.; Miller, P.M.; Kaverina, I. Microtubule Network Asymmetry in Motile Cells: Role of Golgi-Derived Array. Cell Cycle 2009, 8, 2168–2174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Etienne-Manneville, S. Microtubules in Cell Migration. Annu. Rev. Cell Dev. Biol. 2013, 29, 471–499. [Google Scholar] [CrossRef]
- Garcin, C.; Straube, A. Microtubules in Cell Migration. Essays Biochem. 2019, 63, 509–520. [Google Scholar]
- Liao, G.; Mingle, L.; van de Water, L.; Liu, G. Control of Cell Migration through Mrna Localization and Local Translation. Wiley Interdiscip. Rev. RNA 2015, 6, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Laan, L.; Husson, J.; Munteanu, E.L.; Kerssemakers, J.W.; Dogterom, M. Force-Generation and Dynamic Instability of Microtubule Bundles. Proc. Natl. Acad. Sci. USA 2008, 105, 8920–8925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rooney, C.; White, G.; Nazgiewicz, A.; Woodcock, S.A.; Anderson, K.I.; Ballestrem, C.; Malliri, A. The Rac Activator Stef (Tiam2) Regulates Cell Migration by Microtubule-Mediated Focal Adhesion Disassembly. EMBO Rep. 2010, 11, 292–298. [Google Scholar] [CrossRef] [Green Version]
- Krylyshkina, O.; Anderson, K.I.; Kaverina, I.; Upmann, I.; Manstein, D.J.; Small, J.V.; Toomre, D.K. Nanometer Targeting of Microtubules to Focal Adhesions. J. Cell Biol. 2003, 161, 853–859. [Google Scholar] [CrossRef]
- Letort, G.; Nedelec, F.; Blanchoin, L.; Théry, M. Centrosome Centering and Decentering by Microtubule Network Rearrangement. Mol. Biol. Cell 2016, 27, 2833–2843. [Google Scholar] [CrossRef]
- Mostowy, S.; Cossart, P. Septins: The Fourth Component of the Cytoskeleton. Nat. Rev. Mol. Cell Biol. 2012, 13, 183–194. [Google Scholar] [CrossRef]
- Woods, B.L.; Gladfelter, A.S. The State of the Septin Cytoskeleton from Assembly to Function. Curr. Opin. Cell Biol. 2021, 68, 105–112. [Google Scholar] [CrossRef]
- Dolat, L.; Hunyara, J.L.; Bowen, J.R.; Karasmanis, E.P.; Elgawly, M.; Galkin, V.E.; Spiliotis, E.T. Septins Promote Stress Fiber-Mediated Maturation of Focal Adhesions and Renal Epithelial Motility. J. Cell Biol. 2014, 207, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Hall, A. Rho Gtpases and the Control of Cell Behaviour. Biochem. Soc. Trans. 2005, 33, 891–895. [Google Scholar] [CrossRef]
- Hall, A. Rho Family Gtpases. Biochem. Soc. Trans. 2012, 40, 1378–1382. [Google Scholar] [CrossRef] [Green Version]
- Hodge, R.G.; Ridley, A.J. Regulating Rho Gtpases and Their Regulators. Nat. Rev. Mol. Cell Biol. 2016, 17, 496–510. [Google Scholar] [CrossRef] [PubMed]
- Iden, S.; Collard, J.G. Crosstalk between Small Gtpases and Polarity Proteins in Cell Polarization. Nat. Rev. Mol. Cell Biol. 2008, 9, 846–859. [Google Scholar] [CrossRef]
- Bos, J.L.; Rehmann, H.; Wittinghofer, A. Gefs and Gaps: Critical Elements in the Control of Small G Proteins. Cell 2007, 129, 865–877. [Google Scholar] [CrossRef] [Green Version]
- Nobes, C.D.; Hall, A. Rho, Rac, and Cdc42 Gtpases Regulate the Assembly of Multimolecular Focal Complexes Associated with Actin Stress Fibers, Lamellipodia, and Filopodia. Cell 1995, 81, 53–62. [Google Scholar] [CrossRef] [Green Version]
- Itoh, R.E.; Kurokawa, K.; Ohba, Y.; Yoshizaki, H.; Mochizuki, N.; Matsuda, M. Activation of Rac and Cdc42 Video Imaged by Fluorescent Resonance Energy Transfer-Based Single-Molecule Probes in the Membrane of Living Cells. Mol. Cell Biol. 2002, 22, 6582–6591. [Google Scholar] [CrossRef] [Green Version]
- Ridley, A.J. Rho Gtpases and Actin Dynamics in Membrane Protrusions and Vesicle Trafficking. Trends Cell Biol. 2006, 16, 522–529. [Google Scholar] [CrossRef]
- Nobes, C.D.; Hawkins, P.; Stephens, L.; Hall, A. Activation of the Small Gtp-Binding Proteins Rho and Rac by Growth Factor Receptors. J. Cell Sci. 1995, 108, 225–233. [Google Scholar] [CrossRef]
- Mullins, R.D. How Wasp-Family Proteins and the Arp2/3 Complex Convert Intracellular Signals into Cytoskeletal Structures. Curr. Opin. Cell Biol. 2000, 12, 91–96. [Google Scholar] [CrossRef]
- Machesky, L.M.; Gould, K.L. The Arp2/3 Complex: A Multifunctional Actin Organizer. Curr. Opin. Cell Biol. 1999, 11, 117–121. [Google Scholar] [CrossRef]
- Machesky, L.M.; Mullins, R.D.; Higgs, H.N.; Kaiser, D.A.; Blanchoin, L.; May, R.C.; Hall, M.E.; Pollard, T.D. Scar, a Wasp-Related Protein, Activates Nucleation of Actin Filaments by the Arp2/3 Complex. Proc. Natl. Acad. Sci. USA 1999, 96, 3739–3744. [Google Scholar] [CrossRef] [Green Version]
- Rohatgi, R.; Ma, L.; Miki, H.; Lopez, M.; Kirchhausen, T.; Takenawa, T.; Kirschner, M.W. The Interaction between N-Wasp and the Arp2/3 Complex Links Cdc42-Dependent Signals to Actin Assembly. Cell 1999, 97, 221–231. [Google Scholar] [CrossRef] [Green Version]
- Oleinik, N.V.; Helke, K.L.; Kistner-Griffin, E.; Krupenko, N.I.; Krupenko, S.A. Rho Gtpases Rhoa and Rac1 Mediate Effects of Dietary Folate on Metastatic Potential of A549 Cancer Cells through the Control of Cofilin Phosphorylation. J. Biol. Chem. 2014, 289, 26383–26394. [Google Scholar] [CrossRef] [Green Version]
- Assemat, E.; Bazellieres, E.; Pallesi-Pocachard, E.; le Bivic, A.; Massey-Harroche, D. Polarity Complex Proteins. Biochim. Biophys. Acta 2008, 1778, 614–630. [Google Scholar] [CrossRef] [Green Version]
- Insall, R.H.; Weiner, O.D. Pip3, Pip2, and Cell Movement—Similar Messages, Different Meanings? Dev. Cell 2001, 1, 743–747. [Google Scholar] [CrossRef] [Green Version]
- Mack, N.A.; Georgiou, M. The Interdependence of the Rho Gtpases and Apicobasal Cell Polarity. Small GTPases 2014, 5, 10. [Google Scholar] [CrossRef]
- Krause, M.; Gautreau, A. Steering Cell Migration: Lamellipodium Dynamics and the Regulation of Directional Persistence. Nat. Rev. Mol. Cell Biol. 2014, 15, 577–590. [Google Scholar] [CrossRef]
- Vicente-Manzanares, M.; Webb, D.J.; Horwitz, A.R. Cell Migration at a Glance. J. Cell Sci. 2005, 118, 4917–4919. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Wang, Y.L. Centrosome Defines the Rear of Cells During Mesenchymal Migration. Mol. Biol. Cell 2017, 28, 3240–3251. [Google Scholar] [CrossRef] [PubMed]
- Burridge, K. Crosstalk between Rac and Rho. Science 1999, 283, 2028–2029. [Google Scholar] [CrossRef]
- Xiang, B.; Liu, Y.; Zhao, W.; Zhao, H.; Yu, H. Extracellular Calcium Regulates the Adhesion and Migration of Osteoclasts Via Integrin Alphav Beta 3/Rho a/Cytoskeleton Signaling. Cell Biol. Int. 2019, 43, 1125–1136. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Ellenbroek, S.I.; Mertens, A.E.; van der Kammen, R.A.; de Rooij, J.; Collard, J.G. The Par-Tiam1 Complex Controls Persistent Migration by Stabilizing Microtubule-Dependent Front-Rear Polarity. Curr. Biol. 2007, 17, 1623–1634. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Watanabe, T.; Matsuzawa, K.; Katsumi, A.; Kakeno, M.; Matsui, T.; Ye, F.; Sato, K.; Murase, K.; Sugiyama, I.; et al. Tiam1 Interaction with the Par Complex Promotes Talin-Mediated Rac1 Activation During Polarized Cell Migration. J. Cell Biol. 2012, 199, 331–345. [Google Scholar] [CrossRef]
- Pasten, C.; Cerda, J.; Jausoro, I.; Court, F.A.; Caceres, A.; Marzolo, M.P. Apoer2 and Reelin Are Expressed in Regenerating Peripheral Nerve and Regulate Schwann Cell Migration by Activating the Rac1 Gef Protein, Tiam1. Mol. Cell Neurosci. 2015, 69, 1–11. [Google Scholar] [CrossRef]
- Oh, E.S.; Woods, A.; Couchman, J.R. Syndecan-4 Proteoglycan Regulates the Distribution and Activity of Protein Kinase C. J. Biol. Chem. 1997, 272, 8133–8136. [Google Scholar] [CrossRef] [Green Version]
- Putney, J.W. Capacitative Calcium Entry: From Concept to Molecules. Immunol. Rev. 2009, 231, 10–22. [Google Scholar] [CrossRef]
- Tsai, F.C.; Kuo, G.H.; Chang, S.W.; Tsai, P.J. Ca2+ Signaling in Cytoskeletal Reorganization, Cell Migration, and Cancer Metastasis. BioMed Res. Int. 2015, 2015, 409245. [Google Scholar] [CrossRef] [Green Version]
- Machaca, K. Ca2+ Signaling and Lipid Transfer ‘Pas a Deux’ at Er-Pm Contact Sites Orchestrate Cell Migration. Cell Calcium 2020, 89, 102226. [Google Scholar] [CrossRef]
- Schulman, H. Activity-Dependent Regulation of Calcium/Calmodulin-Dependent Protein Kinase Ii Localization. J. Neurosci. 2004, 24, 8399–8403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saneyoshi, T.; Hayashi, Y. The Ca2+ and Rho Gtpase Signaling Pathways Underlying Activity-Dependent Actin Remodeling at Dendritic Spines. Cytoskeleton 2012, 69, 545–554. [Google Scholar] [CrossRef]
- Bernfield, M.; Kokenyesi, R.; Kato, M.; Hinkes, M.T.; Spring, J.; Gallo, R.L.; Lose, E.J. Biology of the Syndecans: A Family of Transmembrane Heparan Sulfate Proteoglycans. Annu. Rev. Cell Biol. 1992, 8, 365–393. [Google Scholar] [CrossRef]
- Kim, C.W.; Goldberger, O.A.; Gallo, R.L.; Bernfield, M. Members of the Syndecan Family of Heparan Sulfate Proteoglycans Are Expressed in Distinct Cell-, Tissue-, and Development-Specific Patterns. Mol. Biol. Cell 1994, 5, 797–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carey, D.J. Syndecans: Multifunctional Cell-Surface Co-Receptors. Biochem. J. 1997, 327, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, P.; David, G. The Syndecans, Tuners of Transmembrane Signaling. FASEB J. 1999, 13, S91–S100. [Google Scholar] [CrossRef] [Green Version]
- Couchman, J.R. Syndecans: Proteoglycan Regulators of Cell-Surface Microdomains? Nat. Rev. Mol. Cell Biol. 2003, 4, 926–937. [Google Scholar] [CrossRef]
- Couchman, J.R.; Woods, A. Syndecans, Signaling, and Cell Adhesion. J. Cell Biochem. 1996, 61, 578–584. [Google Scholar] [CrossRef]
- Couchman, J.R.; Vogt, S.; Lim, S.T.; Lim, Y.; Oh, E.S.; Prestwich, G.D.; Theibert, A.; Lee, W.; Woods, A. Regulation of Inositol Phospholipid Binding and Signaling through Syndecan-4. J. Biol. Chem. 2002, 277, 49296–49303. [Google Scholar] [CrossRef] [Green Version]
- Manon-Jensen, T.; Itoh, Y.; Couchman, J.R. Proteoglycans in Health and Disease: The Multiple Roles of Syndecan Shedding. FEBS J. 2010, 277, 3876–3889. [Google Scholar] [CrossRef]
- Deepa, S.S.; Yamada, S.; Zako, M.; Goldberger, O.; Sugahara, K. Chondroitin Sulfate Chains on Syndecan-1 and Syndecan-4 from Normal Murine Mammary Gland Epithelial Cells Are Structurally and Functionally Distinct and Cooperate with Heparan Sulfate Chains to Bind Growth Factors. A Novel Function to Control Binding of Midkine, Pleiotrophin, and Basic Fibroblast Growth Factor. J. Biol. Chem. 2004, 279, 37368–37376. [Google Scholar]
- Bernfield, M.; Götte, M.; Park, P.W.; Reizes, O.; Fitzgerald, M.L.; Lincecum, J.; Zako, M. Functions of Cell Surface Heparan Sulfate Proteoglycans. Annu. Rev. Biochem. 1999, 68, 729–777. [Google Scholar] [CrossRef] [PubMed]
- Elfenbein, A.; Simons, M. Syndecan-4 Signaling at a Glance. J. Cell Sci. 2013, 126, 3799–3804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller-Pinter, A.; Szabo, K.; Kocsis, T.; Deak, F.; Ocsovszki, I.; Zvara, A.; Puskas, L.; Szilak, L.; Dux, L. Syndecan-4 Influences Mammalian Myoblast Proliferation by Modulating Myostatin Signalling and G1/S Transition. FEBS Lett. 2018, 592, 3139–3151. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, X.; Wang, G.; Cao, B.; Yang, H.; Jin, L.; Cui, M.; Mao, Y. Syndecan-1 Suppresses Cell Growth and Migration Via Blocking Jak1/Stat3 and Ras/Raf/Mek/Erk Pathways in Human Colorectal Carcinoma Cells. BMC Cancer 2019, 19, 1160. [Google Scholar] [CrossRef] [Green Version]
- Maeda, T.; Alexander, C.M.; Friedl, A. Induction of Syndecan-1 Expression in Stromal Fibroblasts Promotes Proliferation of Human Breast Cancer Cells. Cancer Res. 2004, 64, 612–621. [Google Scholar] [CrossRef] [Green Version]
- Park, H.; Kim, Y.; Lim, Y.; Han, I.; Oh, E.S. Syndecan-2 Mediates Adhesion and Proliferation of Colon Carcinoma Cells. J. Biol. Chem. 2002, 277, 29730–29736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leblanc, R.; Sahay, D.; Houssin, A.; Machuca-Gayet, I.; Peyruchaud, O. Autotaxin-Beta Interaction with the Cell Surface Via Syndecan-4 Impacts on Cancer Cell Proliferation and Metastasis. Oncotarget 2018, 9, 33170–33185. [Google Scholar] [CrossRef]
- Huang, W.; Chiquet-Ehrismann, R.; Moyano, J.V.; Garcia-Pardo, A.; Orend, G. Interference of Tenascin-C with Syndecan-4 Binding to Fibronectin Blocks Cell Adhesion and Stimulates Tumor Cell Proliferation. Cancer Res. 2001, 61, 8586–8594. [Google Scholar]
- Fitzgerald, M.L.; Wang, Z.; Park, P.W.; Murphy, G.; Bernfield, M. Shedding of Syndecan-1 and -4 Ectodomains Is Regulated by Multiple Signaling Pathways and Mediated by a Timp-3-Sensitive Metalloproteinase. J. Cell Biol. 2000, 148, 811–824. [Google Scholar] [CrossRef]
- Park, P.W.; Foster, T.J.; Nishi, E.; Duncan, S.J.; Klagsbrun, M.; Chen, Y. Activation of Syndecan-1 Ectodomain Shedding by Staphylococcus Aureus Alpha-Toxin and Beta-Toxin. J. Biol. Chem. 2004, 279, 251–258. [Google Scholar] [CrossRef] [Green Version]
- Yuan, K.; Hong, T.M.; Chen, J.J.; Tsai, W.H.; Lin, M.T. Syndecan-1 up-Regulated by Ephrinb2/Ephb4 Plays Dual Roles in Inflammatory Angiogenesis. Blood 2004, 104, 1025–1033. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Adenekan, B.; Chen, L.; Vaughan, E.D.; Gerald, W.; Feng, Z.; Knudsen, B.S. Syndecan-1 Expression in Locally Invasive and Metastatic Prostate Cancer. Urology 2004, 63, 402–407. [Google Scholar] [CrossRef]
- Letoha, T.; Keller-Pinter, A.; Kusz, E.; Kolozsi, C.; Bozso, Z.; Toth, G.; Vizler, C.; Olah, Z.; Szilak, L. Cell-Penetrating Peptide Exploited Syndecans. Biochim. Biophys. Acta 2010, 1798, 2258–2265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehman, Z.U.; Sjollema, K.A.; Kuipers, J.; Hoekstra, D.; Zuhorn, I.S. Nonviral Gene Delivery Vectors Use Syndecan-Dependent Transport Mechanisms in Filopodia to Reach the Cell Surface. ACS Nano 2012, 6, 7521–7532. [Google Scholar] [CrossRef]
- Letoha, T.; Kolozsi, C.; Ekes, C.; Keller-Pinter, A.; Kusz, E.; Szakonyi, G.; Duda, E.; Szilak, L. Contribution of Syndecans to Lipoplex-Mediated Gene Delivery. Eur. J. Pharm. Sci. 2013, 49, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Adepu, S.; Rosman, C.W.; Dam, W.; van Dijk, M.C.; Navis, G.; van Goor, H.; Bakker, S.J.; van den Born, J. Incipient Renal Transplant Dysfunction Associates with Tubular Syndecan-1 Expression and Shedding. Am. J. Physiol. Renal Physiol. 2015, 309, F137–F145. [Google Scholar] [CrossRef] [PubMed]
- Vuong, T.T.; Reine, T.M.; Sudworth, A.; Jenssen, T.G.; Kolset, S.O. Syndecan-4 Is a Major Syndecan in Primary Human Endothelial Cells in Vitro, Modulated by Inflammatory Stimuli and Involved in Wound Healing. J. Histochem. Cytochem. 2015, 63, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Lee, E.; Kwon, S.; Park, H.; Yi, J.Y.; Kim, S.; Han, I.O.; Yun, Y.; Oh, E.S. Transmembrane Domain-Induced Oligomerization Is Crucial for the Functions of Syndecan-2 and Syndecan-4. J. Biol. Chem. 2005, 280, 42573–42579. [Google Scholar] [CrossRef] [Green Version]
- Dews, I.C.; Mackenzie, K.R. Transmembrane Domains of the Syndecan Family of Growth Factor Coreceptors Display a Hierarchy of Homotypic and Heterotypic Interactions. Proc. Natl. Acad. Sci. USA 2007, 104, 20782–20787. [Google Scholar] [CrossRef] [Green Version]
- Couchman, J.R.; Chen, L.; Woods, A. Syndecans and Cell Adhesion. Int. Rev. Cytol. 2001, 207, 113–150. [Google Scholar]
- Granes, F.; Berndt, C.; Roy, C.; Mangeat, P.; Reina, M.; Vilaro, S. Identification of a Novel Ezrin-Binding Site in Syndecan-2 Cytoplasmic Domain. FEBS Lett. 2003, 547, 212–216. [Google Scholar] [CrossRef]
- Multhaupt, H.A.; Yoneda, A.; Whiteford, J.R.; Oh, E.S.; Lee, W.; Couchman, J.R. Syndecan Signaling: When, Where and Why? J. Physiol. Pharmacol. 2009, 60, 31–38. [Google Scholar]
- Keller-Pinter, A.; Ughy, B.; Domoki, M.; Pettko-Szandtner, A.; Letoha, T.; Tovari, J.; Timar, J.; Szilak, L. The Phosphomimetic Mutation of Syndecan-4 Binds and Inhibits Tiam1 Modulating Rac1 Activity in Pdz Interaction-Dependent Manner. PLoS ONE 2017, 12, e0187094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becsky, D.; Gyulai-Nagy, S.; Balind, A.; Horvath, P.; Dux, L.; Keller-Pinter, A. Myoblast Migration and Directional Persistence Affected by Syndecan-4-Mediated Tiam-1 Expression and Distribution. Int. J. Mol. Sci. 2020, 21, 823. [Google Scholar] [CrossRef] [Green Version]
- Gopal, S.; Multhaupt, H.A.B.; Pocock, R.; Couchman, J.R. Cell-Extracellular Matrix and Cell-Cell Adhesion Are Linked by Syndecan-4. Matrix Biol. 2017, 60–61, 57–69. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Takeuchi, T.; Kuwata, K.; Chiba, J.; Hatanaka, Y.; Nakase, I.; Futaki, S. Syndecan-4 Is a Receptor for Clathrin-Mediated Endocytosis of Arginine-Rich Cell-Penetrating Peptides. Bioconjug. Chem. 2016, 27, 1119–1130. [Google Scholar] [CrossRef]
- Bellin, R.M.; Kubicek, J.D.; Frigault, M.J.; Kamien, A.J.; Steward, R.L., Jr.; Barnes, H.M.; Digiacomo, M.B.; Duncan, L.J.; Edgerly, C.K.; Morse, E.M.; et al. Defining the Role of Syndecan-4 in Mechanotransduction Using Surface-Modification Approaches. Proc. Natl. Acad. Sci. USA 2009, 106, 22102–22107. [Google Scholar] [CrossRef] [Green Version]
- Keller-Pinter, A.; Bottka, S.; Timar, J.; Kulka, J.; Katona, R.; Dux, L.; Deak, F.; Szilak, L. Syndecan-4 Promotes Cytokinesis in a Phosphorylation-Dependent Manner. Cell. Mol. Life Sci. 2010, 67, 1881–1894. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, J.; Partovian, C.; Sellke, F.W.; Simons, M. Syndecan-4 Modulates Basic Fibroblast Growth Factor 2 Signaling in Vivo. Am. J. Physiol. Heart Circ. Physiol. 2003, 284, H2078–H2082. [Google Scholar] [CrossRef] [Green Version]
- Cornelison, D.D.; Wilcox-Adelman, S.A.; Goetinck, P.F.; Rauvala, H.; Rapraeger, A.C.; Olwin, B.B. Essential and Separable Roles for Syndecan-3 and Syndecan-4 in Skeletal Muscle Development and Regeneration. Genes Dev. 2004, 18, 2231–2236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambert, J.; Makin, K.; Akbareian, S.; Johnson, R.; Alghamdi, A.A.A.; Robinson, S.D.; Edwards, D.R. Adamts-1 and Syndecan-4 Intersect in the Regulation of Cell Migration and Angiogenesis. J. Cell Sci. 2020, 133, jcs235762. [Google Scholar] [CrossRef] [Green Version]
- Slimani, H.; Charnaux, N.; Mbemba, E.; Saffar, L.; Vassy, R.; Vita, C.; Gattegno, L. Interaction of Rantes with Syndecan-1 and Syndecan-4 Expressed by Human Primary Macrophages. Biochim. Biophys. Acta 2003, 1617, 80–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charnaux, N.; Brule, S.; Hamon, M.; Chaigneau, T.; Saffar, L.; Prost, C.; Lievre, N.; Gattegno, L. Syndecan-4 Is a Signaling Molecule for Stromal Cell-Derived Factor-1 (Sdf-1)/Cxcl12. FEBS J. 2005, 272, 1937–1951. [Google Scholar] [CrossRef]
- Tumova, S.; Woods, A.; Couchman, J.R. Heparan Sulfate Chains from Glypican and Syndecans Bind the Hep Ii Domain of Fibronectin Similarly Despite Minor Structural Differences. J. Biol. Chem. 2000, 275, 9410–9417. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Kim, S.; Lee, J.; Ko, S.G.; Lee, W.; Han, I.O.; Woods, A.; Oh, E.S. The Oligomeric Status of Syndecan-4 Regulates Syndecan-4 Interaction with Alpha-Actinin. Eur. J. Cell Biol. 2008, 87, 807–815. [Google Scholar] [CrossRef]
- Greene, D.K.; Tumova, S.; Couchman, J.R.; Woods, A. Syndecan-4 Associates with Alpha-Actinin. J. Biol. Chem. 2003, 278, 7617–7623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tkachenko, E.; Simons, M. Clustering Induces Redistribution of Syndecan-4 Core Protein into Raft Membrane Domains. J. Biol. Chem. 2002, 277, 19946–19951. [Google Scholar] [CrossRef] [Green Version]
- Baciu, P.C.; Goetinck, P.F. Protein Kinase C Regulates the Recruitment of Syndecan-4 into Focal Contacts. Mol. Biol. Cell 1995, 6, 1503–1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horowitz, A.; Simons, M. Regulation of Syndecan-4 Phosphorylation In Vivo. J. Biol. Chem. 1998, 273, 10914–10918. [Google Scholar] [CrossRef] [Green Version]
- Koo, B.K.; Jung, Y.S.; Shin, J.; Han, I.; Mortier, E.; Zimmermann, P.; Whiteford, J.R.; Couchman, J.R.; Oh, E.S.; Lee, W. Structural Basis of Syndecan-4 Phosphorylation as a Molecular Switch to Regulate Signaling. J. Mol. Biol. 2006, 355, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Gopal, S.; Sogaard, P.; Multhaupt, H.A.; Pataki, C.; Okina, E.; Xian, X.; Pedersen, M.E.; Stevens, T.; Griesbeck, O.; Park, P.W.; et al. Transmembrane Proteoglycans Control Stretch-Activated Channels to Set Cytosolic Calcium Levels. J. Cell Biol. 2015, 210, 1199–1211. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Echtermeyer, F.; Thilo, F.; Theilmeier, G.; Schmidt, A.; Schulein, R.; Jensen, B.L.; Loddenkemper, C.; Jankowski, V.; Marcussen, N.; et al. The Proteoglycan Syndecan 4 Regulates Transient Receptor Potential Canonical 6 Channels Via Rhoa/Rho-Associated Protein Kinase Signaling. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 378–385. [Google Scholar] [CrossRef] [Green Version]
- Becsky, D.; Szabo, K.; Gyulai-Nagy, S.; Gajdos, T.; Bartos, Z.; Balind, A.; Dux, L.; Horvath, P.; Erdelyi, M.; Homolya, L.; et al. Syndecan-4 Modulates Cell Polarity and Migration by Influencing Centrosome Positioning and Intracellular Calcium Distribution. Front. Cell Dev. Biol. 2020, 8, 575227. [Google Scholar] [CrossRef]
- Tkachenko, E.; Elfenbein, A.; Tirziu, D.; Simons, M. Syndecan-4 Clustering Induces Cell Migration in a Pdz-Dependent Manner. Circ. Res. 2006, 98, 1398–1404. [Google Scholar] [CrossRef] [Green Version]
- Bass, M.D.; Morgan, M.R.; Humphries, M.J. Integrins and Syndecan-4 Make Distinct, but Critical, Contributions to Adhesion Contact Formation. Soft Matter 2007, 3, 372–376. [Google Scholar] [CrossRef] [Green Version]
- Mertens, A.E.; Pegtel, D.M.; Collard, J.G. Tiam1 Takes Part in Cell Polarity. Trends Cell Biol. 2006, 16, 308–316. [Google Scholar] [CrossRef]
- Ten Klooster, J.P.; Evers, E.E.; Janssen, L.; Machesky, L.M.; Michiels, F.; Hordijk, P.; Collard, J.G. Interaction between Tiam1 and the Arp2/3 Complex Links Activation of Rac to Actin Polymerization. Biochem. J. 2006, 397, 39–45. [Google Scholar] [CrossRef]
- Saoncella, S.; Echtermeyer, F.; Denhez, F.; Nowlen, J.K.; Mosher, D.F.; Robinson, S.D.; Hynes, R.O.; Goetinck, P.F. Syndecan-4 Signals Cooperatively with Integrins in a Rho-Dependent Manner in the Assembly of Focal Adhesions and Actin Stress Fibers. Proc. Natl. Acad. Sci. USA 1999, 96, 2805–2810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dovas, A.; Yoneda, A.; Couchman, J.R. Pkcbeta-Dependent Activation of Rhoa by Syndecan-4 During Focal Adhesion Formation. J. Cell Sci. 2006, 119, 2837–2846. [Google Scholar] [CrossRef] [Green Version]
- Elfenbein, A.; Rhodes, J.M.; Meller, J.; Schwartz, M.A.; Matsuda, M.; Simons, M. Suppression of Rhog Activity Is Mediated by a Syndecan 4-Synectin-Rhogdi1 Complex and Is Reversed by Pkcalpha in a Rac1 Activation Pathway. J. Cell Biol. 2009, 186, 75–83. [Google Scholar] [CrossRef] [Green Version]
- Bass, M.D.; Humphries, M.J. Cytoplasmic Interactions of Syndecan-4 Orchestrate Adhesion Receptor and Growth Factor Receptor Signalling. Biochem. J. 2002, 368, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Mostafavi-Pour, Z.; Askari, J.A.; Parkinson, S.J.; Parker, P.J.; Ng, T.T.; Humphries, M.J. Integrin-Specific Signaling Pathways Controlling Focal Adhesion Formation and Cell Migration. J. Cell Biol. 2003, 161, 155–167. [Google Scholar] [CrossRef] [Green Version]
- Woods, A.; Couchman, J.R. Syndecan-4 and Focal Adhesion Function. Curr. Opin. Cell Biol. 2001, 13, 578–583. [Google Scholar] [CrossRef]
- Brunton, V.G.; Avizienyte, E.; Fincham, V.J.; Serrels, B.; Metcalf, C.A., 3rd; Sawyer, T.K.; Frame, M.C. Identification of Src-Specific Phosphorylation Site on Focal Adhesion Kinase: Dissection of the Role of Src Sh2 and Catalytic Functions and Their Consequences for Tumor Cell Behavior. Cancer Res. 2005, 65, 1335–1342. [Google Scholar] [CrossRef] [Green Version]
- Wilcox-Adelman, S.A.; Denhez, F.; Goetinck, P.F. Syndecan-4 Modulates Focal Adhesion Kinase Phosphorylation. J. Biol. Chem. 2002, 277, 32970–32977. [Google Scholar] [CrossRef] [Green Version]
- Parsons, M.; Keppler, M.D.; Kline, A.; Messent, A.; Humphries, M.J.; Gilchrist, R.; Hart, I.R.; Quittau-Prevostel, C.; Hughes, W.E.; Parker, P.J.; et al. Site-Directed Perturbation of Protein Kinase C- Integrin Interaction Blocks Carcinoma Cell Chemotaxis. Mol. Cell Biol. 2002, 22, 5897–5911. [Google Scholar] [CrossRef] [Green Version]
- Denhez, F.; Wilcox-Adelman, S.A.; Baciu, P.C.; Saoncella, S.; Lee, S.; French, B.; Neveu, W.; Goetinck, P.F. Syndesmos, a Syndecan-4 Cytoplasmic Domain Interactor, Binds to the Focal Adhesion Adaptor Proteins Paxillin and Hic-5. J. Biol. Chem. 2002, 277, 12270–12274. [Google Scholar] [CrossRef] [Green Version]
- Brown, M.C.; Turner, C.E. Paxillin: Adapting to Change. Physiol. Rev. 2004, 84, 1315–1339. [Google Scholar] [CrossRef]
- Pataki, C.A.; Couchman, J.R.; Brábek, J. Wnt Signaling Cascades and the Roles of Syndecan Proteoglycans. J. Histochem. Cytochem. 2015, 63, 465–480. [Google Scholar] [CrossRef] [Green Version]
- Cavalheiro, R.P.; Lima, M.A.; Jarrouge-Boucas, T.R.; Viana, G.M.; Lopes, C.C.; Coulson-Thomas, V.J.; Dreyfuss, J.L.; Yates, E.A.; Tersariol, I.L.S.; Nader, H.B. Coupling of Vinculin to F-Actin Demands Syndecan-4 Proteoglycan. Matrix Biol. 2017, 63, 23–37. [Google Scholar] [CrossRef]
- Yoo, J.; Jeong, M.J.; Cho, H.J.; Oh, E.S.; Han, M.Y. Dynamin Ii Interacts with Syndecan-4, a Regulator of Focal Adhesion and Stress-Fiber Formation. Biochem. Biophys. Res. Commun. 2005, 328, 424–431. [Google Scholar] [CrossRef]
- Yip, G.W.; Smollich, M.; Gotte, M. Therapeutic Value of Glycosaminoglycans in Cancer. Mol. Cancer Ther. 2006, 5, 2139–2148. [Google Scholar] [CrossRef] [Green Version]
- Espinoza-Sanchez, N.A.; Gotte, M. Role of Cell Surface Proteoglycans in Cancer Immunotherapy. Semin. Cancer Biol. 2020, 62, 48–67. [Google Scholar] [CrossRef]
- Hassan, N.; Greve, B.; Espinoza-Sanchez, N.A.; Gotte, M. Cell-Surface Heparan Sulfate Proteoglycans as Multifunctional Integrators of Signaling in Cancer. Cell. Signal. 2021, 77, 109822. [Google Scholar] [CrossRef]
- Onyeisi, J.O.S.; Lopes, C.C.; Gotte, M. Syndecan-4 as a Pathogenesis Factor and Therapeutic Target in Cancer. Biomolecules 2021, 11, 503. [Google Scholar] [CrossRef]
- Liao, W.C.; Yen, H.R.; Chen, C.H.; Chu, Y.H.; Song, Y.C.; Tseng, T.J.; Liu, C.H. Chpf Promotes Malignancy of Breast Cancer Cells by Modifying Syndecan-4 and the Tumor Microenvironment. Am. J. Cancer Res. 2021, 11, 812–826. [Google Scholar]
- Jayson, G.C.; Vives, C.; Paraskeva, C.; Schofield, K.; Coutts, J.; Fleetwood, A.; Gallagher, J.T. Coordinated Modulation of the Fibroblast Growth Factor Dual Receptor Mechanism During Transformation from Human Colon Adenoma to Carcinoma. Int. J. Cancer 1999, 82, 298–304. [Google Scholar] [CrossRef]
- Warner, T.F.; Wrone, D.A.; Williams, E.C.; Cripps, D.J.; Mundhenke, C.; Friedl, A. Heparan Sulphate Proteoglycan in Scleromyxedema Promotes Fgf-2 Activity. Pathol. Res. Pract. 2002, 198, 701–707. [Google Scholar] [CrossRef]
- Casey, R.C.; Oegema, T.R., Jr.; Skubitz, K.M.; Pambuccian, S.E.; Grindle, S.M.; Skubitz, A.P. Cell Membrane Glycosylation Mediates the Adhesion, Migration, and Invasion of Ovarian Carcinoma Cells. Clin. Exp. Metastasis 2003, 20, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Szatmari, T.; Dobra, K. The Role of Syndecan-1 in Cellular Signaling and Its Effects on Heparan Sulfate Biosynthesis in Mesenchymal Tumors. Front. Oncol. 2013, 3, 310. [Google Scholar] [CrossRef] [Green Version]
- Baba, F.; Swartz, K.; van Buren, R.; Eickhoff, J.; Zhang, Y.; Wolberg, W.; Friedl, A. Syndecan-1 and Syndecan-4 Are Overexpressed in an Estrogen Receptor-Negative, Highly Proliferative Breast Carcinoma Subtype. Breast Cancer Res. Treat. 2006, 98, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Labropoulou, V.T.; Skandalis, S.S.; Ravazoula, P.; Perimenis, P.; Karamanos, N.K.; Kalofonos, H.P.; Theocharis, A.D. Expression of Syndecan-4 and Correlation with Metastatic Potential in Testicular Germ Cell Tumours. BioMed Res. Int. 2013, 2013, 214864. [Google Scholar] [CrossRef] [PubMed]
- Na, K.Y.; Bacchini, P.; Bertoni, F.; Kim, Y.W.; Park, Y.K. Syndecan-4 and Fibronectin in Osteosarcoma. Pathology 2012, 44, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Majo, S.; Courtois, S.; Souleyreau, W.; Bikfalvi, A.; Auguste, P. Impact of Extracellular Matrix Components to Renal Cell Carcinoma Behavior. Front. Oncol. 2020, 10, 625. [Google Scholar] [CrossRef]
- Wang, X.; He, J.; Zhao, X.; Qi, T.; Zhang, T.; Kong, C. Syndecan-1 Suppresses Epithelial-Mesenchymal Transition and Migration in Human Oral Cancer Cells. Oncol. Rep. 2018, 39, 1835–1842. [Google Scholar] [CrossRef] [Green Version]
- Fujii, T.; Shimada, K.; Tatsumi, Y.; Tanaka, N.; Fujimoto, K.; Konishi, N. Syndecan-1 up-Regulates Microrna-331-3p and Mediates Epithelial-to-Mesenchymal Transition in Prostate Cancer. Mol. Carcinog. 2016, 55, 1378–1386. [Google Scholar] [CrossRef]
- Contreras, H.R.; Ledezma, R.A.; Vergara, J.; Cifuentes, F.; Barra, C.; Cabello, P.; Gallegos, I.; Morales, B.; Huidobro, C.; Castellón, E.A. The Expression of Syndecan-1 and -2 Is Associated with Gleason Score and Epithelial-Mesenchymal Transition Markers, E-Cadherin and Beta-Catenin, in Prostate Cancer. Urol. Oncol. 2010, 28, 534–540. [Google Scholar] [CrossRef]
- Hua, R.; Yu, J.; Yan, X.; Ni, Q.; Zhi, X.; Li, X.; Jiang, B.; Zhu, J. Syndecan-2 in Colorectal Cancer Plays Oncogenic Role Via Epithelial-Mesenchymal Transition and Mapk Pathway. Biomed. Pharmacother. 2020, 121, 109630. [Google Scholar] [CrossRef] [PubMed]
- Toba-Ichihashi, Y.; Yamaoka, T.; Ohmori, T.; Ohba, M. Up-Regulation of Syndecan-4 Contributes to Tgf-Beta1-Induced Epithelial to Mesenchymal Transition in Lung Adenocarcinoma A549 Cells. Biochem. Biophys. Rep. 2016, 5, 1–7. [Google Scholar] [PubMed] [Green Version]
- Cevikbas, F.; Schaefer, L.; Uhlig, P.; Robenek, H.; Theilmeier, G.; Echtermeyer, F.; Bruckner, P. Unilateral Nephrectomy Leads to up-Regulation of Syndecan-2- and Tgf-Beta-Mediated Glomerulosclerosis in Syndecan-4 Deficient Male Mice. Matrix Biol. 2008, 27, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L.; Gao, G.X.; Shen, F.X.; Chen, X.; Gong, X.H.; Wu, W.J. Sdc4 Gene Silencing Favors Human Papillary Thyroid Carcinoma Cell Apoptosis and Inhibits Epithelial Mesenchymal Transition Via Wnt/Beta-Catenin Pathway. Mol. Cells 2018, 41, 853–867. [Google Scholar]
- Chalkiadaki, G.; Nikitovic, D.; Berdiaki, A.; Sifaki, M.; Krasagakis, K.; Katonis, P.; Karamanos, N.K.; Tzanakakis, G.N. Fibroblast Growth Factor-2 Modulates Melanoma Adhesion and Migration through a Syndecan-4-Dependent Mechanism. Int. J. Biochem. Cell Biol. 2009, 41, 1323–1331. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, P.; Gerber, U.; Schutze, N.; Jungel, E.; Blaheta, R.; Naggi, A.; Torri, G.; Bendas, G. Cyr61 Is a Target for Heparin in Reducing Mv3 Melanoma Cell Adhesion and Migration Via the Integrin Vla-4. Thromb. Haemost. 2013, 110, 1046–1054. [Google Scholar]
- Choi, Y.; Yun, J.H.; Yoo, J.; Lee, I.; Kim, H.; Son, H.N.; Kim, I.S.; Yoon, H.S.; Zimmermann, P.; Couchman, J.R.; et al. New Structural Insight of C-Terminal Region of Syntenin-1, Enhancing the Molecular Dimerization and Inhibitory Function Related on Syndecan-4 Signaling. Sci. Rep. 2016, 6, 36818. [Google Scholar] [CrossRef]
- Ross, T.; Jakubzig, B.; Grundmann, M.; Massing, U.; Kostenis, E.; Schlesinger, M.; Bendas, G. The Molecular Mechanism by Which Saturated Lysophosphatidylcholine Attenuates the Metastatic Capacity of Melanoma Cells. FEBS Open Bio 2016, 6, 1297–1309. [Google Scholar] [CrossRef]
- Karamanou, K.; Franchi, M.; Proult, I.; Rivet, R.; Vynios, D.; Brézillon, S. Lumican Inhibits in Vivo Melanoma Metastasis by Altering Matrix-Effectors and Invadopodia Markers. Cells 2021, 10, 841. [Google Scholar] [CrossRef] [PubMed]
- Afratis, N.A.; Karamanou, K.; Piperigkou, Z.; Vynios, D.H.; Theocharis, A.D. The Role of Heparins and Nano-Heparins as Therapeutic Tool in Breast Cancer. Glycoconj. J. 2017, 34, 299–307. [Google Scholar] [CrossRef]
- Filou, S.; Korpetinou, A.; Kyriakopoulou, D.; Bounias, D.; Stavropoulos, M.; Ravazoula, P.; Papachristou, D.J.; Theocharis, A.D.; Vynios, D.H. Adamts Expression in Colorectal Cancer. PLoS ONE 2015, 10, e0121209. [Google Scholar] [CrossRef] [Green Version]
- Kelwick, R.; Wagstaff, L.; Decock, J.; Roghi, C.; Cooley, L.S.; Robinson, S.D.; Arnold, H.; Gavrilovic, J.; Jaworski, D.M.; Yamamoto, K.; et al. Metalloproteinase-Dependent and -Independent Processes Contribute to Inhibition of Breast Cancer Cell Migration, Angiogenesis and Liver Metastasis by a Disintegrin and Metalloproteinase with Thrombospondin Motifs-15. Int. J. Cancer 2015, 136, E14–E26. [Google Scholar] [CrossRef]
- Wang, H.; Jin, H.; Beauvais, D.M.; Rapraeger, A.C. Cytoplasmic Domain Interactions of Syndecan-1 and Syndecan-4 with Alpha6beta4 Integrin Mediate Human Epidermal Growth Factor Receptor (Her1 and Her2)-Dependent Motility and Survival. J. Biol. Chem. 2014, 289, 30318–30332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuoriluoto, K.; Hognas, G.; Meller, P.; Lehti, K.; Ivaska, J. Syndecan-1 and -4 Differentially Regulate Oncogenic K-Ras Dependent Cell Invasion into Collagen through Alpha2beta1 Integrin and Mt1-Mmp. Matrix Biol. 2011, 30, 207–217. [Google Scholar] [CrossRef]
- Habes, C.; Weber, G.; Goupille, C. Sulfated Glycoaminoglycans and Proteoglycan Syndecan-4 Are Involved in Membrane Fixation of Ll-37 and Its Pro-Migratory Effect in Breast Cancer Cells. Biomolecules 2019, 9, 481. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Griffin, M. The Role of Tg2 in Regulating S100a4-Mediated Mammary Tumour Cell Migration. PLoS ONE 2013, 8, e57017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunetti, J.; Riolo, G.; Depau, L.; Mandarini, E.; Bernini, A.; Karousou, E.; Passi, A.; Pini, A.; Bracci, L.; Falciani, C. Unraveling Heparan Sulfate Proteoglycan Binding Motif for Cancer Cell Selectivity. Front. Oncol. 2019, 9, 843. [Google Scholar] [CrossRef] [PubMed]
- Tsonis, A.I.; Afratis, N.; Gialeli, C.; Ellina, M.I.; Piperigkou, Z.; Skandalis, S.S.; Theocharis, A.D.; Tzanakakis, G.N.; Karamanos, N.K. Evaluation of the Coordinated Actions of Estrogen Receptors with Epidermal Growth Factor Receptor and Insulin-Like Growth Factor Receptor in the Expression of Cell Surface Heparan Sulfate Proteoglycans and Cell Motility in Breast Cancer Cells. FEBS J. 2013, 280, 2248–2259. [Google Scholar] [CrossRef]
- Karamanou, K.; Franchi, M.; Onisto, M.; Passi, A.; Vynios, D.H.; Brézillon, S. Evaluation of Lumican Effects on Morphology of Invading Breast Cancer Cells, Expression of Integrins and Downstream Signaling. FEBS J. 2020, 287, 4862–4880. [Google Scholar] [CrossRef]
- Karamanou, K.; Franchi, M.; Vynios, D.; Brézillon, S. Epithelial-to-Mesenchymal Transition and Invadopodia Markers in Breast Cancer: Lumican a Key Regulator. Semin. Cancer Biol. 2020, 62, 125–133. [Google Scholar] [CrossRef]
- El Ghazal, R.; Yin, X.; Johns, S.C.; Swanson, L.; Macal, M.; Ghosh, P.; Zuniga, E.I.; Fuster, M.M. Glycan Sulfation Modulates Dendritic Cell Biology and Tumor Growth. Neoplasia 2016, 18, 294–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, D.; Liang, J.; Campanella, G.S.; Guo, R.; Yu, S.; Xie, T.; Liu, N.; Jung, Y.; Homer, R.; Meltzer, E.B.; et al. Inhibition of Pulmonary Fibrosis in Mice by Cxcl10 Requires Glycosaminoglycan Binding and Syndecan-4. J. Clin. Invest. 2010, 120, 2049–2057. [Google Scholar] [CrossRef]
- Roblek, M.; Strutzmann, E.; Zankl, C.; Adage, T.; Heikenwalder, M.; Atlic, A.; Weis, R.; Kungl, A.; Borsig, L. Targeting of Ccl2-Ccr2-Glycosaminoglycan Axis Using a Ccl2 Decoy Protein Attenuates Metastasis through Inhibition of Tumor Cell Seeding. Neoplasia 2016, 18, 49–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasqualon, T.; Pruessmeyer, J.; Weidenfeld, S.; Babendreyer, A.; Groth, E.; Schumacher, J.; Schwarz, N.; Denecke, B.; Jahr, H.; Zimmermann, P.; et al. A Transmembrane C-Terminal Fragment of Syndecan-1 Is Generated by the Metalloproteinase Adam17 and Promotes Lung Epithelial Tumor Cell Migration and Lung Metastasis Formation. Cell Mol. Life Sci. 2015, 72, 3783–3801. [Google Scholar] [CrossRef] [PubMed]
- Schanz, A.; Baston-Bust, D.; Krussel, J.S.; Heiss, C.; Janni, W.; Hess, A.P. Cxcr7 and Syndecan-4 Are Potential Receptors for Cxcl12 in Human Cytotrophoblasts. J. Reprod. Immunol. 2011, 89, 18–25. [Google Scholar] [CrossRef]
- Sutton, A.; Friand, V.; Brule-Donneger, S.; Chaigneau, T.; Ziol, M.; Sainte-Catherine, O.; Poire, A.; Saffar, L.; Kraemer, M.; Vassy, J.; et al. Stromal Cell-Derived Factor-1/Chemokine (C-X-C Motif) Ligand 12 Stimulates Human Hepatoma Cell Growth, Migration, and Invasion. Mol. Cancer Res. 2007, 5, 21–33. [Google Scholar] [CrossRef] [Green Version]
- Brule, S.; Friand, V.; Sutton, A.; Baleux, F.; Gattegno, L.; Charnaux, N. Glycosaminoglycans and Syndecan-4 Are Involved in Sdf-1/Cxcl12-Mediated Invasion of Human Epitheloid Carcinoma Hela Cells. Biochim. Biophys. Acta 2009, 1790, 1643–1650. [Google Scholar] [CrossRef]
- Wang, Q.; Shen, B.; Chen, L.; Zheng, P.; Feng, H.; Hao, Q.; Liu, X.; Liu, L.; Xu, S.; Chen, J.; et al. Extracellular Calumenin Suppresses Erk1/2 Signaling and Cell Migration by Protecting Fibulin-1 from Mmp-13-Mediated Proteolysis. Oncogene 2015, 34, 1006–1018. [Google Scholar] [CrossRef]
- Charni, F.; Friand, V.; Haddad, O.; Hlawaty, H.; Martin, L.; Vassy, R.; Oudar, O.; Gattegno, L.; Charnaux, N.; Sutton, A. Syndecan-1 and Syndecan-4 Are Involved in Rantes/Ccl5-Induced Migration and Invasion of Human Hepatoma Cells. Biochim. Biophys. Acta 2009, 1790, 1314–1326. [Google Scholar] [CrossRef]
- Mundhenke, C.; Meyer, K.; Drew, S.; Friedl, A. Heparan Sulfate Proteoglycans as Regulators of Fibroblast Growth Factor-2 Receptor Binding in Breast Carcinomas. Am. J. Pathol. 2002, 160, 185–194. [Google Scholar] [CrossRef]
- Chaudhuri, P.; Colles, S.M.; Fox, P.L.; Graham, L.M. Protein Kinase Cdelta-Dependent Phosphorylation of Syndecan-4 Regulates Cell Migration. Circ. Res. 2005, 97, 674–681. [Google Scholar] [CrossRef] [Green Version]
- Yin, L.; Qi, Y.; Xu, Y.; Xu, L.; Han, X.; Tao, X.; Song, S.; Peng, J. Dioscin Inhibits Hsc-T6 Cell Migration Via Adjusting Sdc-4 Expression: Insights from Itraq-Based Quantitative Proteomics. Front. Pharmacol. 2017, 8, 665. [Google Scholar] [CrossRef] [Green Version]
- Endo, T.; Ito, K.; Morimoto, J.; Kanayama, M.; Ota, D.; Ikesue, M.; Kon, S.; Takahashi, D.; Onodera, T.; Iwasaki, N.; et al. Syndecan 4 Regulation of the Development of Autoimmune Arthritis in Mice by Modulating B Cell Migration and Germinal Center Formation. Arthritis Rheumatol. 2015, 67, 2512–2522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeyarajah, M.J.; Bhattad, G.J.; Kops, B.F.; Renaud, S.J. Syndecan-4 Regulates Extravillous Trophoblast Migration by Coordinating Protein Kinase C Activation. Sci. Rep. 2019, 9, 10175. [Google Scholar] [CrossRef] [Green Version]
- Averbeck, M.; Gebhardt, C.; Anderegg, U.; Termeer, C.; Sleeman, J.P.; Simon, J.C. Switch in Syndecan-1 and Syndecan-4 Expression Controls Maturation Associated Dendritic Cell Motility. Exp. Dermatol. 2007, 16, 580–589. [Google Scholar] [CrossRef]
- Frohling, M.; Tepasse, P.; Intemann, J.; Sambale, M.; Sherwood, J.; Paruzel, P.; Tiemeyer, N.M.; Nowacki, T.M.; Bruckner, M.; Mennigen, R.; et al. Syndecan-4 Modulates Epithelial Gut Barrier Function and Epithelial Regeneration in Experimental Colitis. Inflamm. Bowel Dis. 2018, 24, 2579–2589. [Google Scholar] [CrossRef] [PubMed]
- Matsui, Y.; Ikesue, M.; Danzaki, K.; Morimoto, J.; Sato, M.; Tanaka, S.; Kojima, T.; Tsutsui, H.; Uede, T. Syndecan-4 Prevents Cardiac Rupture and Dysfunction after Myocardial Infarction. Circ. Res. 2011, 108, 1328–1339. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Wu, H.; Xie, J.; Li, G.; Gu, R.; Kang, L.; Wang, L.; Xu, B. Syndecan-4 Regulates the Bfgf-Induced Chemotactic Migration of Endothelial Cells. J. Mol. Histol. 2016, 47, 503–509. [Google Scholar] [CrossRef]
- Jang, E.; Albadawi, H.; Watkins, M.T.; Edelman, E.R.; Baker, A.B. Syndecan-4 Proteoliposomes Enhance Fibroblast Growth Factor-2 (Fgf-2)-Induced Proliferation, Migration, and Neovascularization of Ischemic Muscle. Proc. Natl. Acad. Sci. USA 2012, 109, 1679–1684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, J.; McFarland, D.C.; Velleman, S.G. Migration of Turkey Muscle Satellite Cells Is Enhanced by the Syndecan-4 Cytoplasmic Domain through the Activation of Rhoa. Mol. Cell. Biochem. 2013, 375, 115–130. [Google Scholar] [CrossRef]
- Qin, Y.; Zhu, Y.; Luo, F.; Chen, C.; Chen, X.; Wu, M. Killing Two Birds with One Stone: Dual Blockade of Integrin and Fgf Signaling through Targeting Syndecan-4 in Postoperative Capsular Opacification. Cell Death Dis. 2017, 8, e2920. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keller-Pinter, A.; Gyulai-Nagy, S.; Becsky, D.; Dux, L.; Rovo, L. Syndecan-4 in Tumor Cell Motility. Cancers 2021, 13, 3322. https://doi.org/10.3390/cancers13133322
Keller-Pinter A, Gyulai-Nagy S, Becsky D, Dux L, Rovo L. Syndecan-4 in Tumor Cell Motility. Cancers. 2021; 13(13):3322. https://doi.org/10.3390/cancers13133322
Chicago/Turabian StyleKeller-Pinter, Aniko, Szuzina Gyulai-Nagy, Daniel Becsky, Laszlo Dux, and Laszlo Rovo. 2021. "Syndecan-4 in Tumor Cell Motility" Cancers 13, no. 13: 3322. https://doi.org/10.3390/cancers13133322
APA StyleKeller-Pinter, A., Gyulai-Nagy, S., Becsky, D., Dux, L., & Rovo, L. (2021). Syndecan-4 in Tumor Cell Motility. Cancers, 13(13), 3322. https://doi.org/10.3390/cancers13133322








