LRG1 Promotes Metastatic Dissemination of Melanoma through Regulating EGFR/STAT3 Signalling
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Tissue Microarray and Immunohistochemistry
2.3. Cell Lines and Cell Culture Conditions
2.4. Chemicals
2.5. Molecular Biology Methods
2.6. Cell Viability Assay
2.7. Cell Proliferation Assay
2.8. Cell Migration and Invasion Assays
2.9. Transendothelial Migration Assay
2.10. Xenograft Tumour Model
2.11. Lung Metastasis Model
2.12. Extravasation Assay
2.13. Western Blot
2.14. Quantitative Real-Time PCR (qRT-PCR)
2.15. Histology and Immunofluorescence Staining
2.16. Statistical Analyses
3. Results
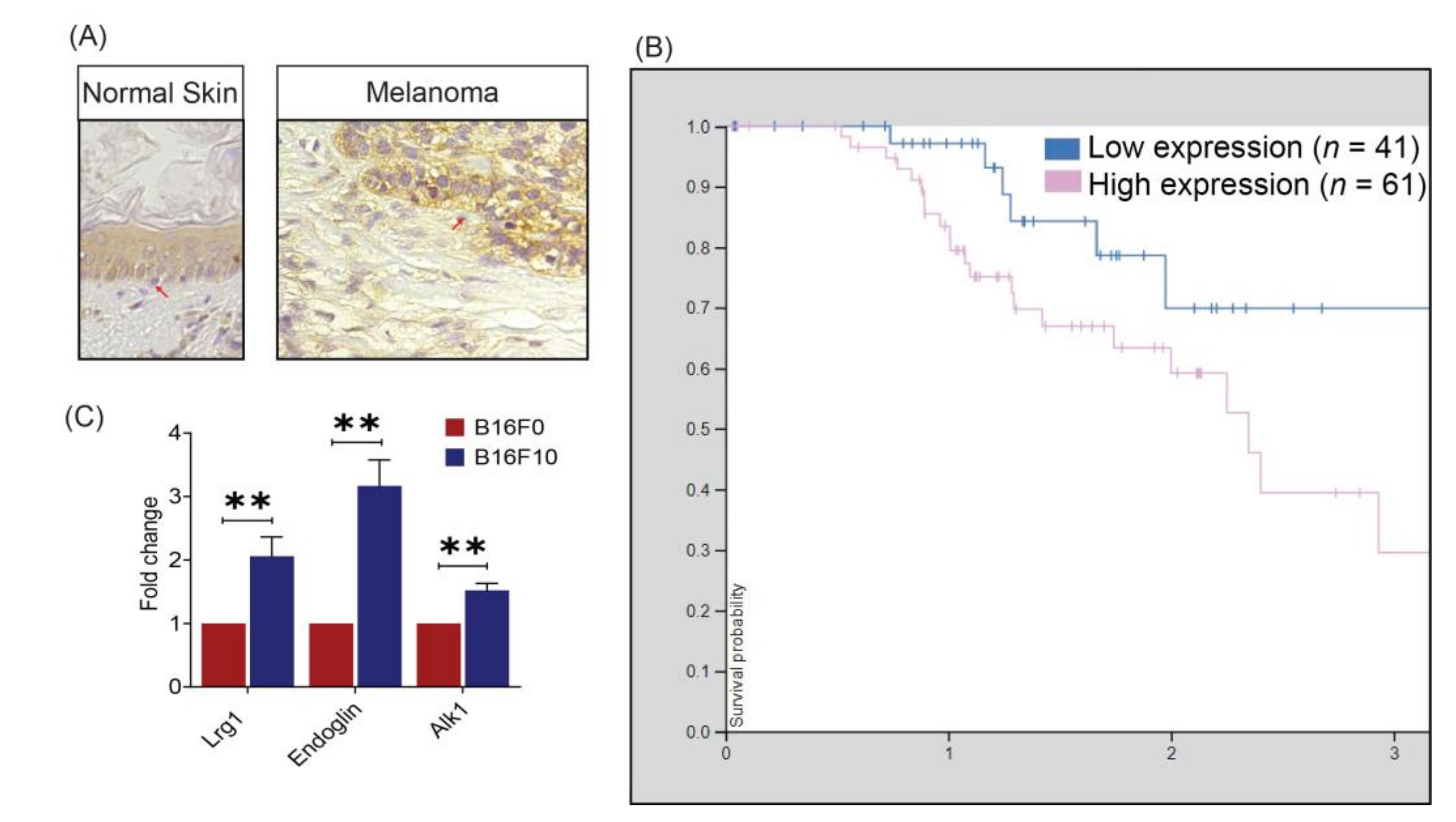
3.1. LRG1 Levels Are Highly Enhanced in Melanoma Cells
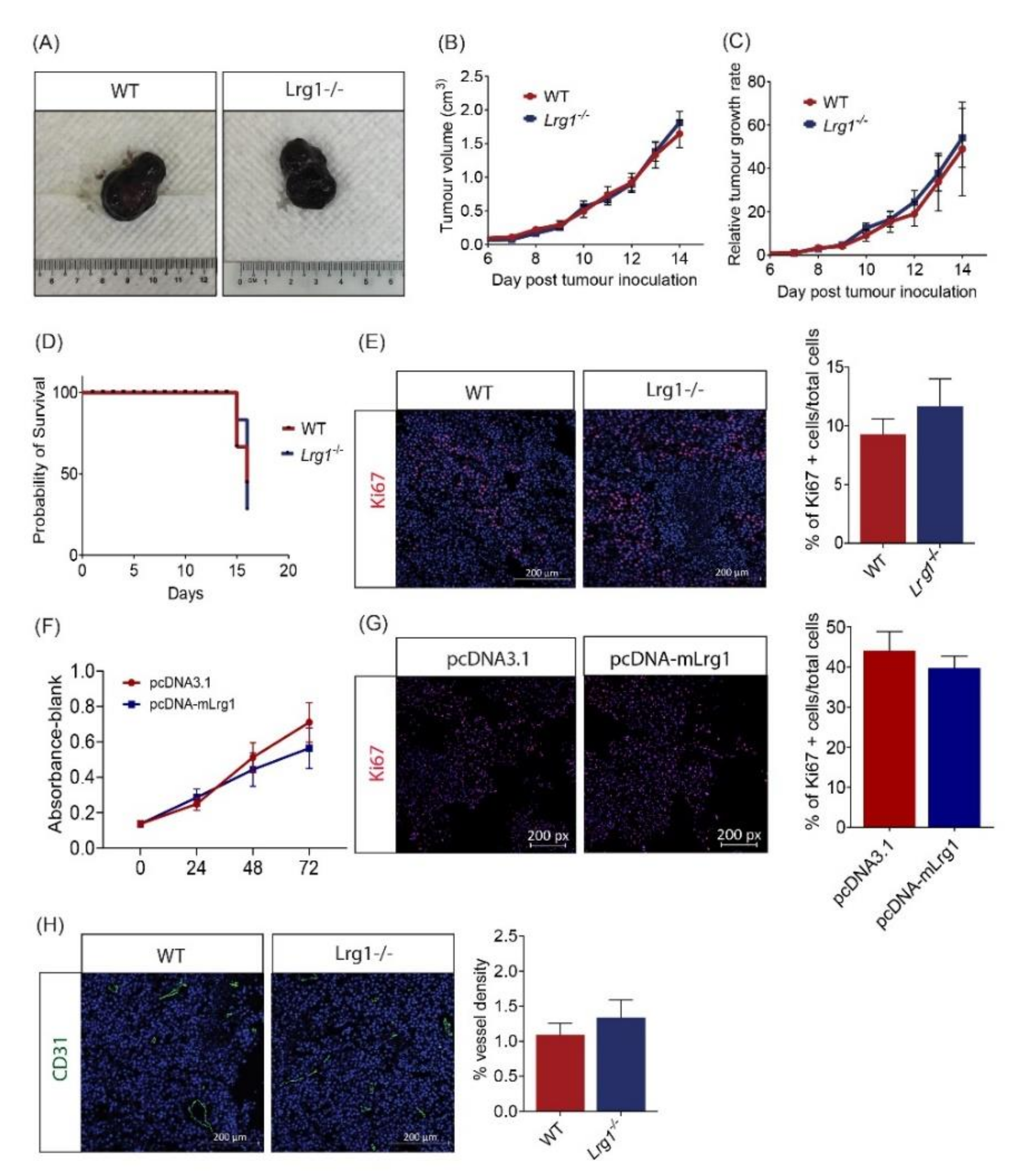
3.2. Host Lrg1 Deficiency Has No Impact on Melanoma Growth
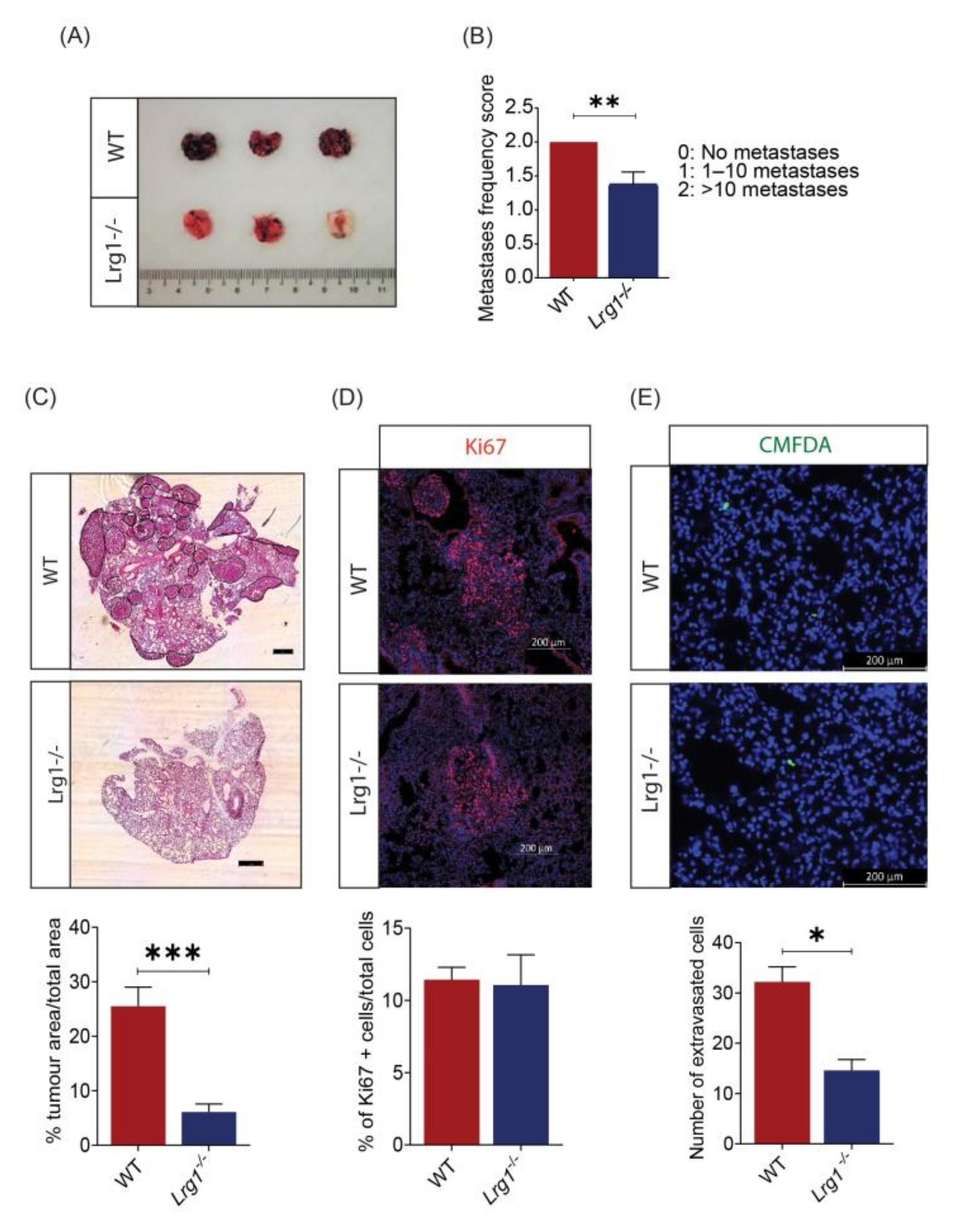
3.3. Host Lrg1 Deficiency Leads to Reduced Pulmonary Metastasis of Melanoma In Vivo
3.4. Lrg1 Promotes B16F10 Cell Invasiveness In Vitro
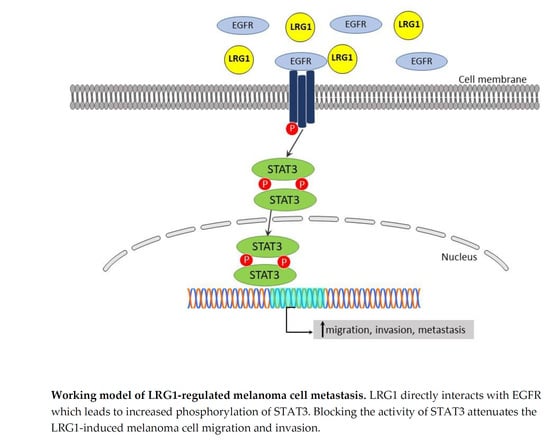
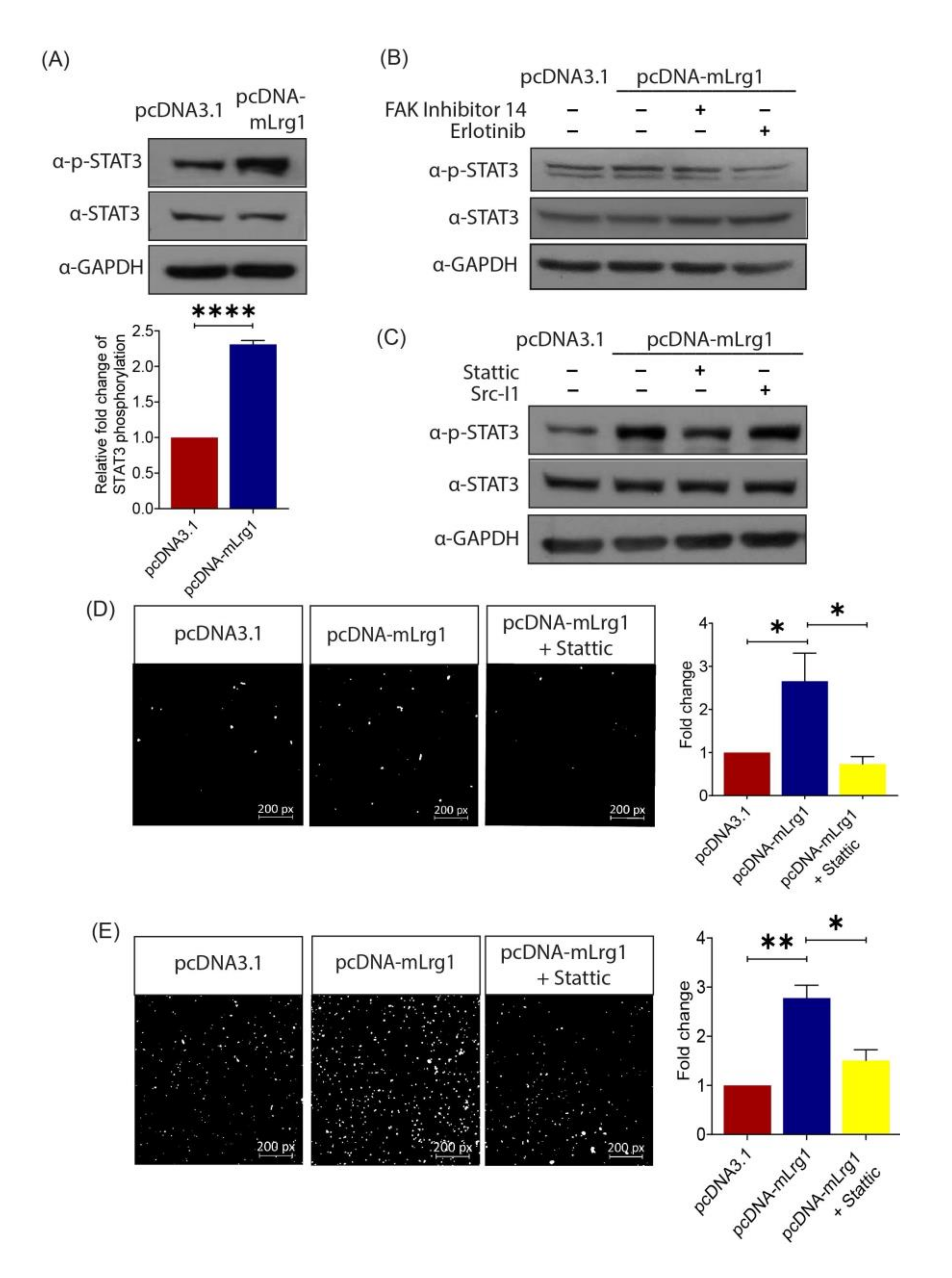
3.5. Lrg1-Induced Activation of the EGFR/STAT3 Pathway Is Required for Melanoma Cell Invasiveness
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Crowson, A.N.; Magro, C.; Miller, A.; Mihm, M.C., Jr. The molecular basis of melanomagenesis and the metastatic phenotype. Semin. Oncol. 2007, 34, 476–490. [Google Scholar] [CrossRef]
- Miller, A.J.; Mihm, M.C., Jr. Melanoma. New Engl. J. Med. 2006, 355, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Global Cancer Observatory—Projected 2025 Data from ‘Cancer Tomorrow’. Available online: https://gco.iarc.fr/tomorrow/en/dataviz/isotype?cancers=16&single_unit=10000&years=2025&types=1 (accessed on 2 February 2021).
- Global Cancer Observatory—Projected 2040 Data from ‘Cancer Tomorrow’. Available online: https://gco.iarc.fr/tomorrow/en/dataviz/isotype?cancers=16&single_unit=10000&years=2040&types=1 (accessed on 2 February 2021).
- Domingues, B.; Lopes, J.M.; Soares, P.; Pópulo, H. Melanoma treatment in review. Immunotargets Ther. 2018, 7, 35–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Abraham, S.; McKenzie, J.; Jeffs, N.; Swire, M.; Tripathi, V.B.; Luhmann, U.; Lange, C.A.K.; Zhai, Z.; Arthur, H.M.; et al. LRG1 promotes angiogenesis by modulating endothelial TGF-β signalling. Nature 2013, 499, 306–311. [Google Scholar] [CrossRef] [Green Version]
- Zhang, A.; Fang, H.; Chen, J.; He, L.; Chen, Y. Role of VEGF-A and LRG1 in abnormal angiogenesis associated with diabetic nephropathy. Front. Physiol. 2020, 11, 1064. [Google Scholar] [CrossRef]
- Meng, H.; Song, Y.; Zhu, J.; Liu, Q.; Lu, P.; Ye, N.; Zhang, Z.; Pang, Y.; Qi, J.; Wu, H. LRG1 promotes angiogenesis through upregulating the TGF-β1 pathway in ischemic rat brain. Mol. Med. Rep. 2016, 14, 5535–5543. [Google Scholar] [CrossRef] [Green Version]
- Song, S.; Cheng, J.; Yu, B.J.; Zhou, L.; Xu, H.F.; Yang, L.L. LRG1 promotes corneal angiogenesis and lymphangiogenesis in a corneal alkali burn mouse model. Int. J. Ophthalmol. 2020, 13, 365–373. [Google Scholar] [CrossRef]
- Liu, C.; Teo, M.H.Y.; Pek, S.L.T.; Wu, X.; Leong, M.L.; Tay, H.M.; Hou, H.W.; Ruedl, C.; Moss, S.E.; Greenwood, J.; et al. A Multifunctional Role of Leucine-Rich α-2-Glycoprotein 1 in Cutaneous Wound Healing Under Normal and Diabetic Conditions. Diabetes 2020, 69, 2467–2480. [Google Scholar] [CrossRef] [PubMed]
- Ziyad, S.; Iruela-Arispe, M.L. Molecular mechanisms of tumor angiogenesis. Genes Cancer 2011, 12, 1085–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katt, M.E.; Wong, A.D.; Searson, P.C. Dissemination from a solid tumor: Examining the multiple parallel pathways. Trends Cancer 2018, 4, 20–37. [Google Scholar] [CrossRef]
- Uen, Y.H.; Lin, K.Y.; Sun, D.P.; Liao, C.C.; Hsieh, M.S.; Huang, Y.K.; Chen, Y.W.; Huang, P.H.; Chen, W.J.; Tai, C.C.; et al. Comparative proteomics, network analysis and post-translational modification identification reveal differential profiles of plasma con a-bound glycoprotein biomarkers in gastric cancer. J. Proteom. 2013, 83, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Xie, X.; Nie, S.; Buckanovich, R.J.; Lubman, D.M. Altered expression of sialylated glycoproteins in ovarian cancer sera using lectinbased elisa assay and quantitative glycoproteomics analysis. J. Proteome Res. 2013, 12, 3342–3352. [Google Scholar] [CrossRef] [PubMed]
- Lindén, M.; Lind, S.B.; Mayrhofer, C.; Segersten, U.; Wester, K.; Lyutvinskiy, Y.; Zubarev, R.; Malmström, P.U.; Pettersson, U. Proteomic analysis of urinary biomarker candidates for nonmuscle invasive bladder cancer. Proteomics 2012, 12, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Luo, X.; Hu, H.; Wang, R.; Sun, Y.; Zeng, R.; Chen, H. Integrative proteomics and tissue microarray profiling indicate the association between overexpressed serum proteins and non-small cell lung cancer. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [Green Version]
- Furukawa, K.; Kawamoto, K.; Eguchi, H.; Tanemura, M.; Tanida, T.; Tomimaru, Y.; Akita, H.; Hama, N.; Wada, H.; Kobayashi, S.; et al. Clinicopathological Significance of Leucine-Rich α2-Glycoprotein-1 in Sera of Patients with Pancreatic Cancer. Pancreas 2015, 44, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Han, L.; Yang, C.; Liu, Y.J.; Zhang, X.M. Prognostic Value of LRG1 in Breast Cancer: A Retrospective Study. Oncol. Res. Treat. 2021, 44, 36–41. [Google Scholar] [CrossRef]
- Guldvik, I.J.; Zuber, V.; Braadland, P.R.; Grytli, H.H.; Ramberg, H.; Lilleby, W.; Thiede, B.; Zucknick, M.; Saatcioglu, F.; Gislefoss, R.; et al. Identification and Validation of Leucine-rich α-2-glycoprotein 1 as a Noninvasive Biomarker for Improved Precision in Prostate Cancer Risk Stratification. Eur. Urol. Open Sci. 2020, 21, 51–60. [Google Scholar] [CrossRef]
- Sandanayake, N.; Sinclair, J.; Andreola, F.; Chapman, M.H.; Xue, A.; Webster, G.J.; Clarkson, A.; Gill, A.; Norton, I.D.; Smith, R.C.; et al. A combination of serum leucine-rich α-2-glycoprotein 1, CA19-9 and interleukin-6 differentiate biliary tract cancer from benign biliary strictures. Br. J. Cancer. 2011, 105, 1370–1378. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, L.; Fang, J.; Ge, Z.; Li, X. LRG1 modulates epithelial-mesenchymal transition and angiogenesis in colorectal cancer via HIF-1α activation. J. Exp. Clin. Cancer Res. 2016, 35, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.; Yang, Z.; Yu, D.; Lin, J.; Cai, W. RUNX1 regulates TGF-β induced migration and EMT in colorectal cancer. Pathol. Res. Pract. 2020, 216, 153142. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Ren, Y.; Wang, Y.; Zhao, L.; Wang, B.; Ma, N.; Gao, Z.; Cao, B. LRG1 Suppresses migration and invasion of esophageal squamous cell carcinoma by modulating epithelial to mesenchymal transition. J. Cancer 2020, 11, 1486–1494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honda, H.; Fujimoto, M.; Serada, S.; Urushima, H.; Mishima, T.; Lee, H.; Ohkawara, T.; Kohno, N.; Hattori, N.; Yokoyama, A. Leucine-rich α-2 glycoprotein promotes lung fibrosis by modulating TGF-β signaling in fibroblasts. Physiol. Rep. 2017, 5, e13556. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Lim, S.T.; Teo, M.H.Y.; Tan, M.S.Y.; Kulkarni, M.D.; Qiu, B.; Li, A.; Lal, S.; dos Remedios, C.G.; Tan, N.S.; et al. Collaborative Regulation of LRG1 by TGF-β1 and PPAR-β/δ Modulates Chronic Pressure Overload–Induced Cardiac Fibrosis. Circ. Heart Fail. 2019, 12, e005962. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, V.; Le Mercier, M.; De Neve, N.; Sauvage, S.; Gras, T.; Roland, I.; Lefranc., F.; Kiss, R. Galectin-1 Knockdown Increases Sensitivity to Temozolomide in a B16F10 Mouse Metastatic Melanoma Model. J. Invest. Dermatol. 2007, 127, 2399–2410. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, V.; de Lassalle, E.M.; Toelen, J.; Mohr, T.; Bellahcene, A.; Goietsenoven, G.V.; Verschuere, T.; Bouzin, C.; Debyser, Z.; De Vleeschouwer, S.; et al. Galectin-1 in Melanoma Biology and Related Neo-Angiogenesis Processes. J. Invest. Dermatol. 2012, 132, 2245–2254. [Google Scholar] [CrossRef] [Green Version]
- Fidler, I.J.; Gersten, D.M.; Budmen, M.B. Characterization in vivo and in vitro of tumor cells selected for resistance to syngeneic lymphocyte-mediated cytotoxicity. Cancer Res. 1976, 36, 3160–3165. [Google Scholar]
- Overwijk, W.W.; Restifo, N.P. B16 as a mouse model for human melanoma. Curr. Protoc. Immunol. 2001, 20. [Google Scholar] [CrossRef]
- Nishida, N.; Yano, H.; Nishida, T.; Kamura, T.; Kojiro, M. Angiogenesis in cancer. Vasc. Health Risk Manag. 2006, 2, 213–219. [Google Scholar] [CrossRef]
- Azevedo, A.S.; Follain, G.; Patthabhiraman, S.; Harlepp, S.; Goetz, J.G. Metastasis of circulating tumor cells: Favorable soil or suitable biomechanics, or both? Cell Adh. Migr. 2015, 9, 345–356. [Google Scholar] [CrossRef]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhan, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Sig. Transduct. Target Ther. 2020, 5, 28. [Google Scholar] [CrossRef]
- Spada, S.; Tocci, A.; Di Modugno, F.; Nistico, P. Fibronectin as a multiregulatory molecule crucial in tumor matrisome: From structural and functional features to clinical practice in oncology. J. Exp. Clin. Cancer Res. 2021, 40, 102. [Google Scholar] [CrossRef]
- Li, W.; Wang, X.; Cheng, J.; Li, J.; Wang, Q.; Zhou, Q.; Li, H.; Xue, J.; Zhang, Y.; Yang, L. Leucine-rich α-2-glycoprotein-1 promotes diabetic corneal epithelial wound healing and nerve regeneration via regulation of matrix metalloproteinases. Exp. Eye Res. 2020, 196, 108060. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer. 2014, 14, 736–746. [Google Scholar] [CrossRef]
- Irwin, M.E.; Bohin, N.; Boerner, J.L. Src family kinases mediate epidermal growth factor receptor signaling from lipid rafts in breast cancer cells. Cancer Biol. Ther. 2011, 12, 718–726. [Google Scholar] [CrossRef] [Green Version]
- Silva, C.M. Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis. Oncogene 2004, 23, 8017–8023. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, M.; Takahashi, T.; Serada, S.; Sugase, T.; Tanaka, K.; Miyazaki, Y.; Makino, T.; Kurokawa, Y.; Yamasaki, M.; Nakajima, K.; et al. Overexpression of leucine-rich alpha2-glycoprotein-1 is a prognostic marker and enhances tumor migration in gastric cancer. Cancer Sci. 2017, 108, 2052–2060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ladd, J.J.; Busald, T.; Johnson, M.M.; Zhang, Q.; Pitteri, S.J.; Wang, H.; Brenner, D.E.; Lampe, P.D.; Kucherlapati, R.; Feng, Z.; et al. Increased plasma levels of the APC-interacting protein MAPRE1, LRG1, and IGFBP2 preceding a diagnosis of colorectal cancer in women. Cancer Prev. Res. 2012, 5, 655–664. [Google Scholar] [CrossRef] [Green Version]
- Wen, S.Y.; Zhang, L.N.; Yang, X.M.; Zhang, Y.L.; Ma, L.; Ge, Q.L.; Jiang, S.H.; Zhu, X.L.; Xu, W.; Ding, W.J. LRG1 is an independent prognostic factor for endometrial carcinoma. Tumor Biol. 2014, 35, 7125–7133. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.B.; Zhang, Y.F.; Jin, C.; Mao, Y.S.; Fu, D.L. LRG-1 promotes pancreatic cancer growth and metastasis via modulation of the EGFR/p38 signaling. J. Exp. Clin. Cancer Res. 2019, 38, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, D.; He, G.; Zhao, S.; Li, J.; Lang, Y.; Ye, W.; Li, Y.; Jiang, C.; Li, X. LRG1 modulates invasion and migration of glioma cell lines through TGF-β signaling pathway. Acta Histochem. 2015, 117, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Ban, Z.; He, J.; Tang, Z.; Zhang, L.; Xu, Z. LRG-1 enhances the migration of thyroid carcinoma cells through promotion of the epithelial-mesenchymal transition by activating MAPK/p38 signaling. Oncol. Rep. 2019, 41, 3270–3280. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Jove, R. The stats of cancer—New molecular targets come of age. Nat. Rev. Cancer. 2004, 4, 97–105. [Google Scholar] [CrossRef]
- Wang, X.; Crowe, P.J.; Goldstein, D.; Yang, J.L. STAT3 inhibition, a novel approach to enhancing targeted therapy in human cancers (review). Int. J. Oncol. 2012, 41, 1181–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, P.A.; Grandis, J.R. STAT3 signaling: Anticancer strategies and challenges. Mol. Interv. 2011, 11, 18–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Huang, J.; Li, W.; Chen, Y.; Liu, X.; Wang, J. Meta-analysis of STAT3 and phospho-STAT3 expression and survival of patients with breast cancer. Oncotarget 2018, 9, 13060–13067. [Google Scholar] [CrossRef] [Green Version]
- Zou, S.; Tong, Q.; Liu, B.; Huang, W.; Tian, Y. Targeting STAT3 in Cancer Immunotherapy. Mol. Cancer 2020, 19, 145. [Google Scholar] [CrossRef] [PubMed]
- Laudisi, F.; Cherubini, F.; Monteleone, G.; Stolfi, C. STAT3 Interactors as potential therapeutic targets for cancer treatment. Int. J. Mol. Sci. 2018, 19, 1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, G.; Bowman, T.; Huang, M.; Shivers, S.; Reintgen, D.; Daud, A.; Chang, A.; Kraker, A.; Jove, R.; Yu, H. Roles of activated Src and Stat3 signaling in melanoma tumor cell growth. Oncogene 2002, 21, 7001–7010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Jeong, A.J.; Ye, S.K. Highlighted STAT3 as a potential drug target for cancer therapy. BMB Rep. 2019, 52, 415–423. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Lin, S.; Xu, L.; Lin, J.; Zhao, C.; Huang, X. Novel activators and small-molecule inhibitors of STAT3 in cancer. Cytokine Growth Factor Rev. 2019, 49, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Huynh, J.; Chand, A.; Gough, D.; Ernst, M. Therapeutically exploiting STAT3 activity in cancer—using tissue repair as a road map. Nat. Rev. Cancer 2019, 19, 82–96. [Google Scholar] [CrossRef] [PubMed]

| Target (Mouse) | Forward Sequence (5’–3’) | Reverse Sequence (5’–3’) |
|---|---|---|
| Lrg1 | TGCACCTCTCGAGCAATCG | AGAGCATTGCGGGTCAGATC |
| Endoglin | CGATAGCAGCACTGGATGAC | AGAATGGTGCCTTTGGGTCT |
| Alk1 | CTTGGGGAGCTTCAGAAGGGG | GGTGGCCTCCAGCATCAGAGA |
| Alk5 | AAATTGCTCGACGCTGTTCT | GGTACAAGATCATAATAAGGCAACTG |
| Gapdh | ACTGAGGACCAGGTTGTCTCC | CTGTAGCCGTATTCATTGTCATACC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwan, Y.P.; Teo, M.H.Y.; Lim, J.C.W.; Tan, M.S.; Rosellinny, G.; Wahli, W.; Wang, X. LRG1 Promotes Metastatic Dissemination of Melanoma through Regulating EGFR/STAT3 Signalling. Cancers 2021, 13, 3279. https://doi.org/10.3390/cancers13133279
Kwan YP, Teo MHY, Lim JCW, Tan MS, Rosellinny G, Wahli W, Wang X. LRG1 Promotes Metastatic Dissemination of Melanoma through Regulating EGFR/STAT3 Signalling. Cancers. 2021; 13(13):3279. https://doi.org/10.3390/cancers13133279
Chicago/Turabian StyleKwan, Yuet Ping, Melissa Hui Yen Teo, Jonathan Chee Woei Lim, Michelle Siying Tan, Graciella Rosellinny, Walter Wahli, and Xiaomeng Wang. 2021. "LRG1 Promotes Metastatic Dissemination of Melanoma through Regulating EGFR/STAT3 Signalling" Cancers 13, no. 13: 3279. https://doi.org/10.3390/cancers13133279
APA StyleKwan, Y. P., Teo, M. H. Y., Lim, J. C. W., Tan, M. S., Rosellinny, G., Wahli, W., & Wang, X. (2021). LRG1 Promotes Metastatic Dissemination of Melanoma through Regulating EGFR/STAT3 Signalling. Cancers, 13(13), 3279. https://doi.org/10.3390/cancers13133279