Are Adaptive Chemotherapy Schedules Robust? A Three-Strategy Stochastic Evolutionary Game Theory Model
Abstract
Simple Summary
Abstract
1. Introduction
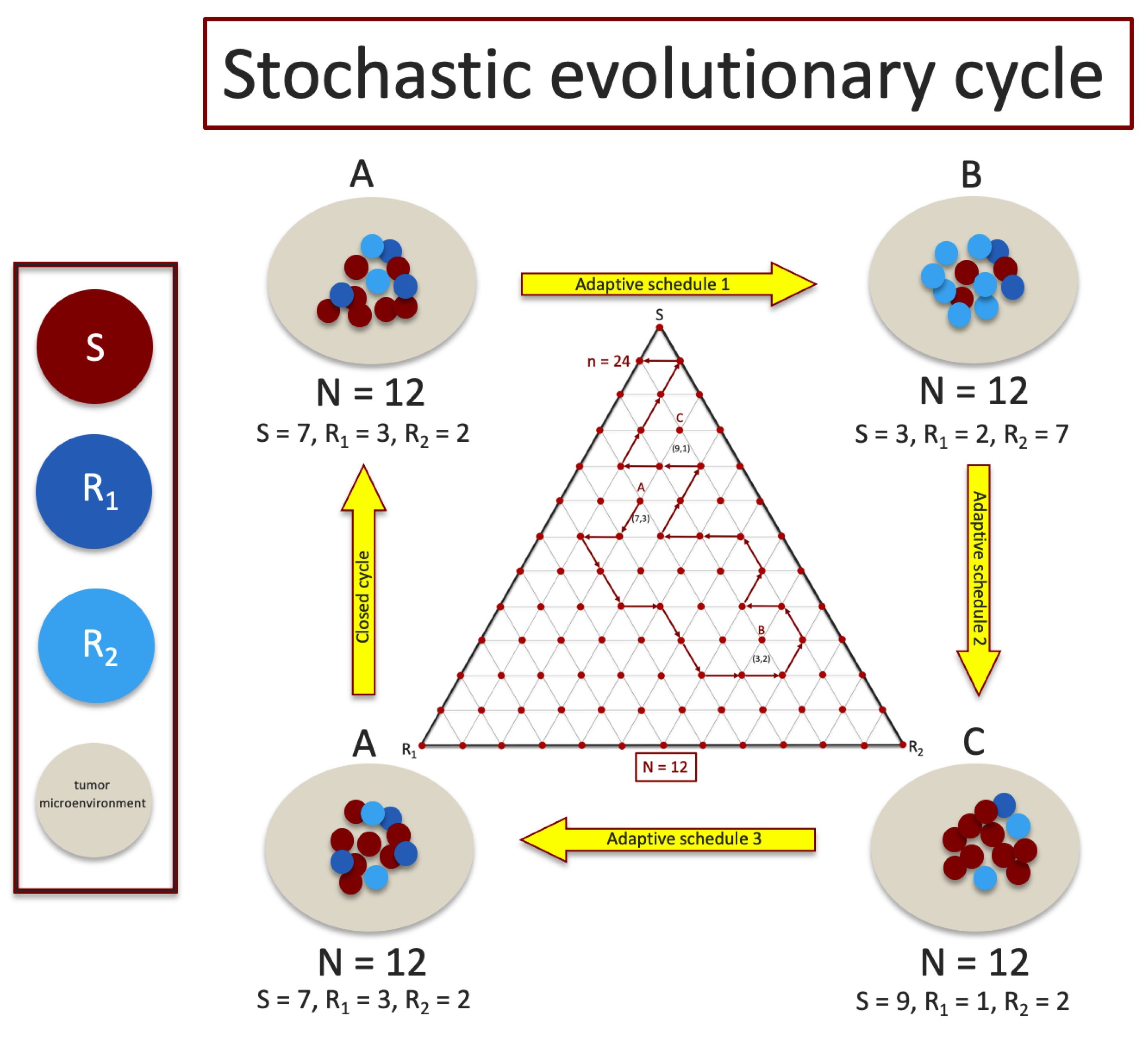
2. Model Description
- 1.
- We implement a version of the adjusted replicator dynamics equations to track the frequency evolution of the three sub-populations of cancer cells that make up the tumor. Our model consists of a chemo-sensitive population (S), a population resistant to drug 1 (), and a population resistant to drug 2 (). The competition among the three populations is determined by a payoff matrix A that builds in a fitness cost of resistance. In the absence of chemotherapy, the sensitive population is most fit and will outcompete the resistant populations, saturating the tumor.
- 2.
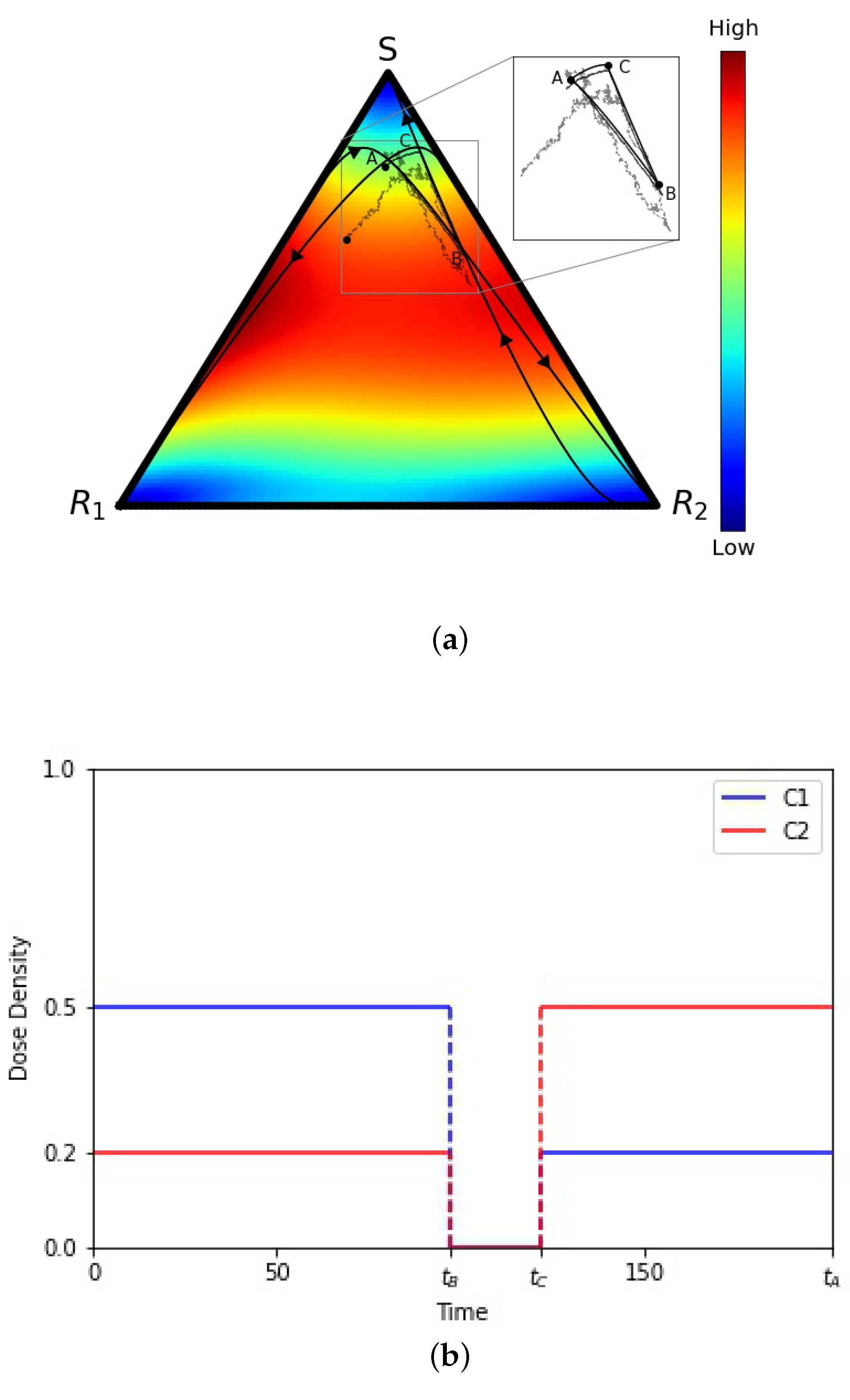
- We use two time-dependent control functions and to model the chemotherapy dosing schedules. These chemotherapy dosing functions control three selection pressure parameters , , and by altering the relative fitness values of the three sub-populations. A dose of chemotherapy () lowers the relative fitness of the targeted cell population by altering the selection pressures on the sub-populations to effectively favor ones that are not targeted. This mechanism allows us to ‘design’ favorable fitness landscapes indirectly by adaptively monitoring the sub-population frequencies and altering our dosing schedule in response. Our goals when we design chemotherapy schedules are to: (i) avoid fixation of the sensitive cell population; (ii) avoid chemoresistance (fixation of either of the resistant populations) by keeping the three populations of cells in competition without allowing any of them to saturate the tumor. We implement this by designing schedules that keep us confined to a closed ‘evolutionary cycle’ which keep the sub-populations in competition forever (ideally) for the deterministic () model.
- 3.
- We then test the performance of these designed schedules on a finite cell () stochastic Moran process model to track the sub-population frequencies during these cycles, where the limit corresponds to our adjusted replicator model from which the cycle was designed.
- 4.
- Since the Moran process model uses a fixed value of N (hence, it cannot be used directly to determine tumor growth where the total cancer cell population increases), we add a tumor growth equation with growth rate determined as a function of the average fitness of the three cancer cell sub-population frequencies comprising the tumor. When the average fitness of the cancer cell populations is above a fixed microenvironmental average, the tumor grows (exponentially), and when it is below, it shrinks (exponentially).
2.1. Three-Component Two-Drug Adjusted Replicator Dynamics Model
2.2. Three-Component Two-Drug Discrete Stochastic Moran Process
2.3. Continuous Limit Which Relates the Moran Process to the Adjusted Replicator System
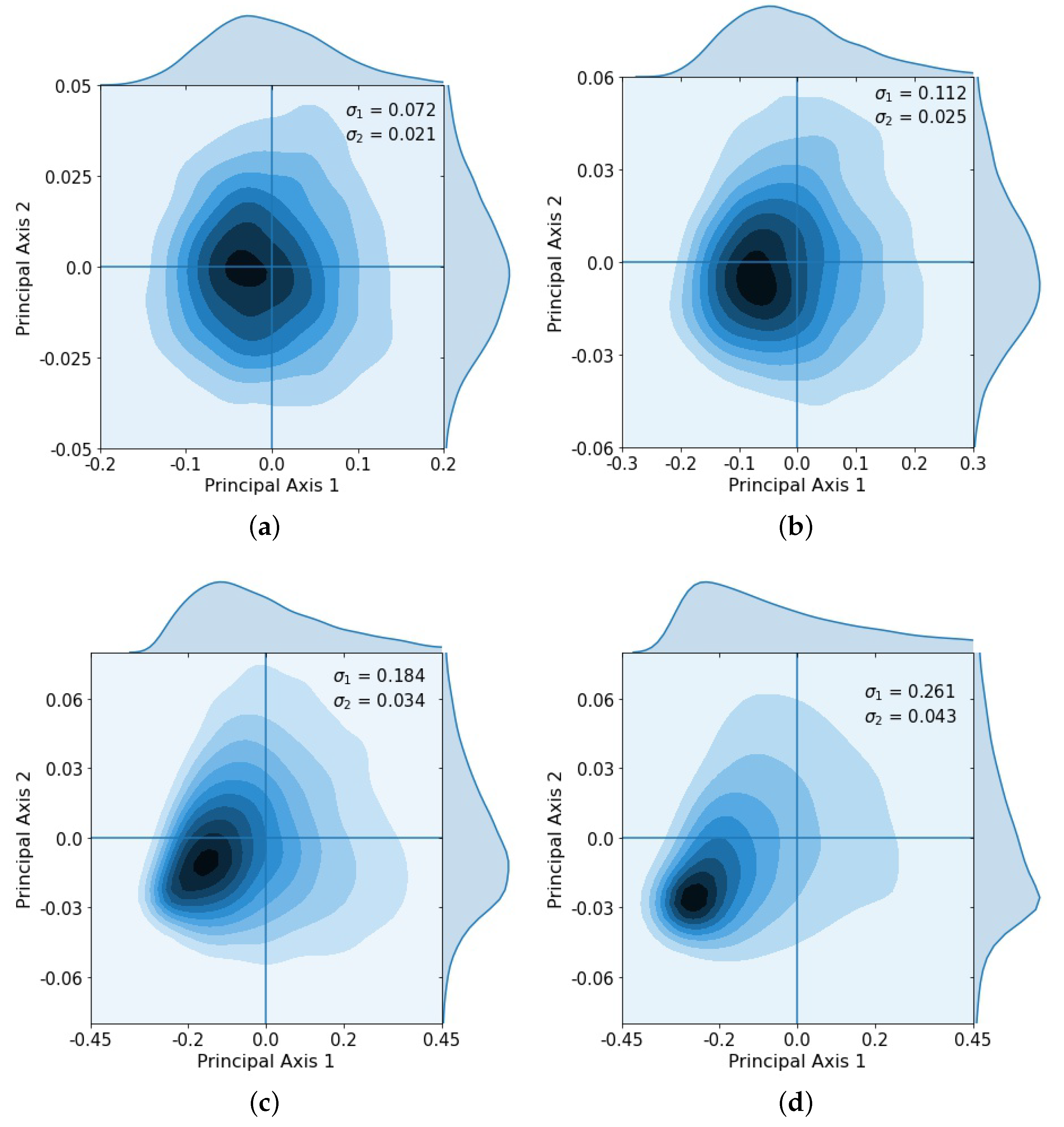
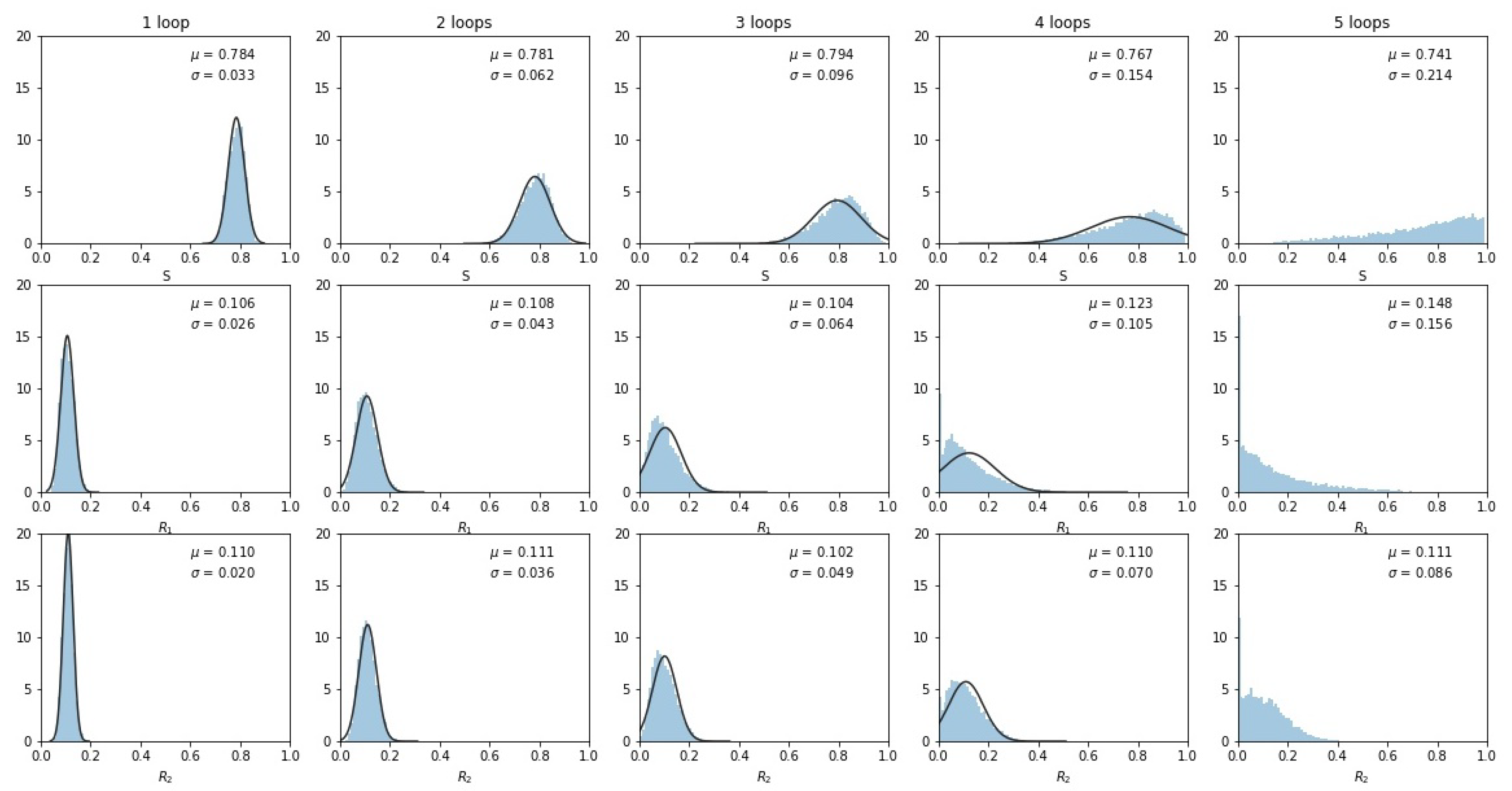
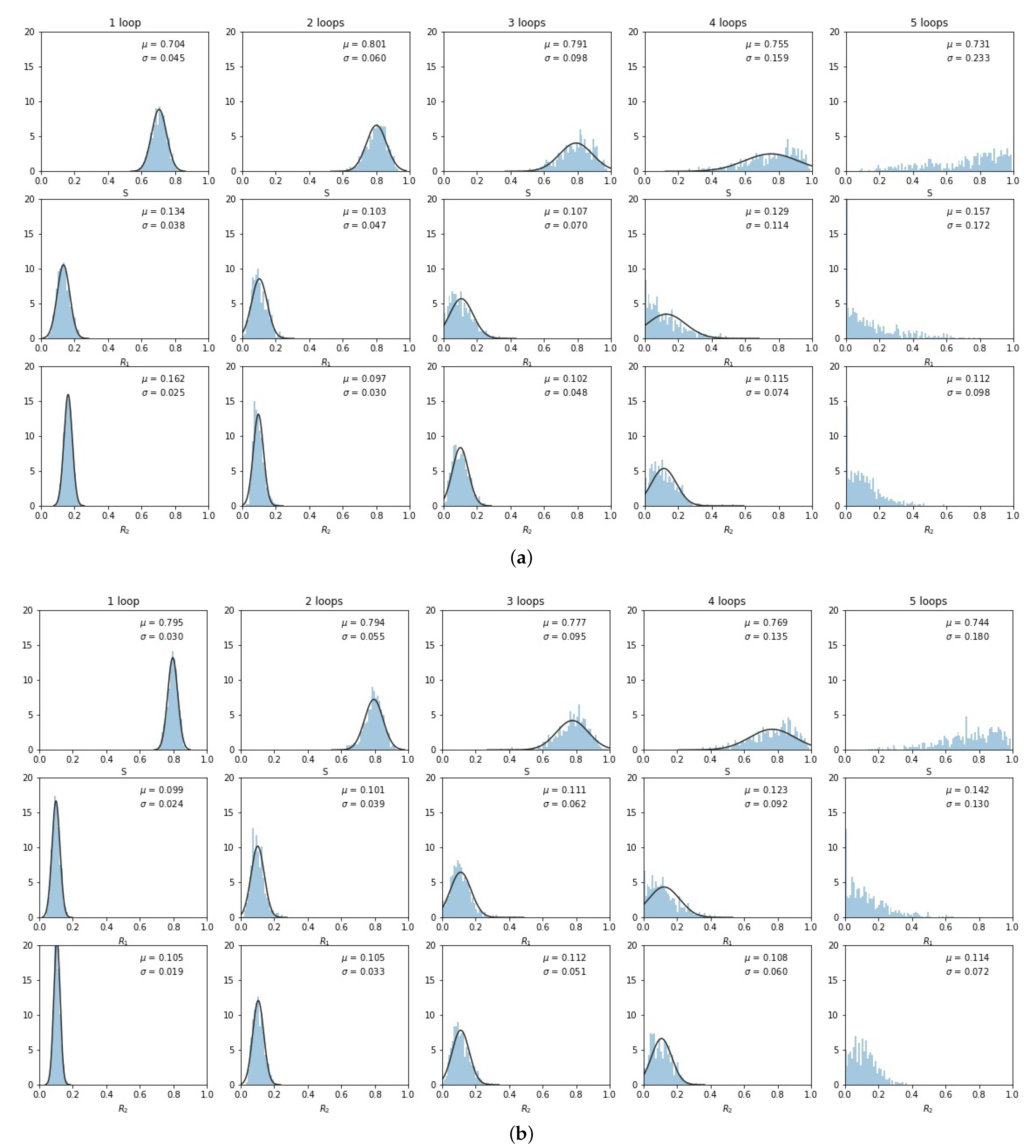
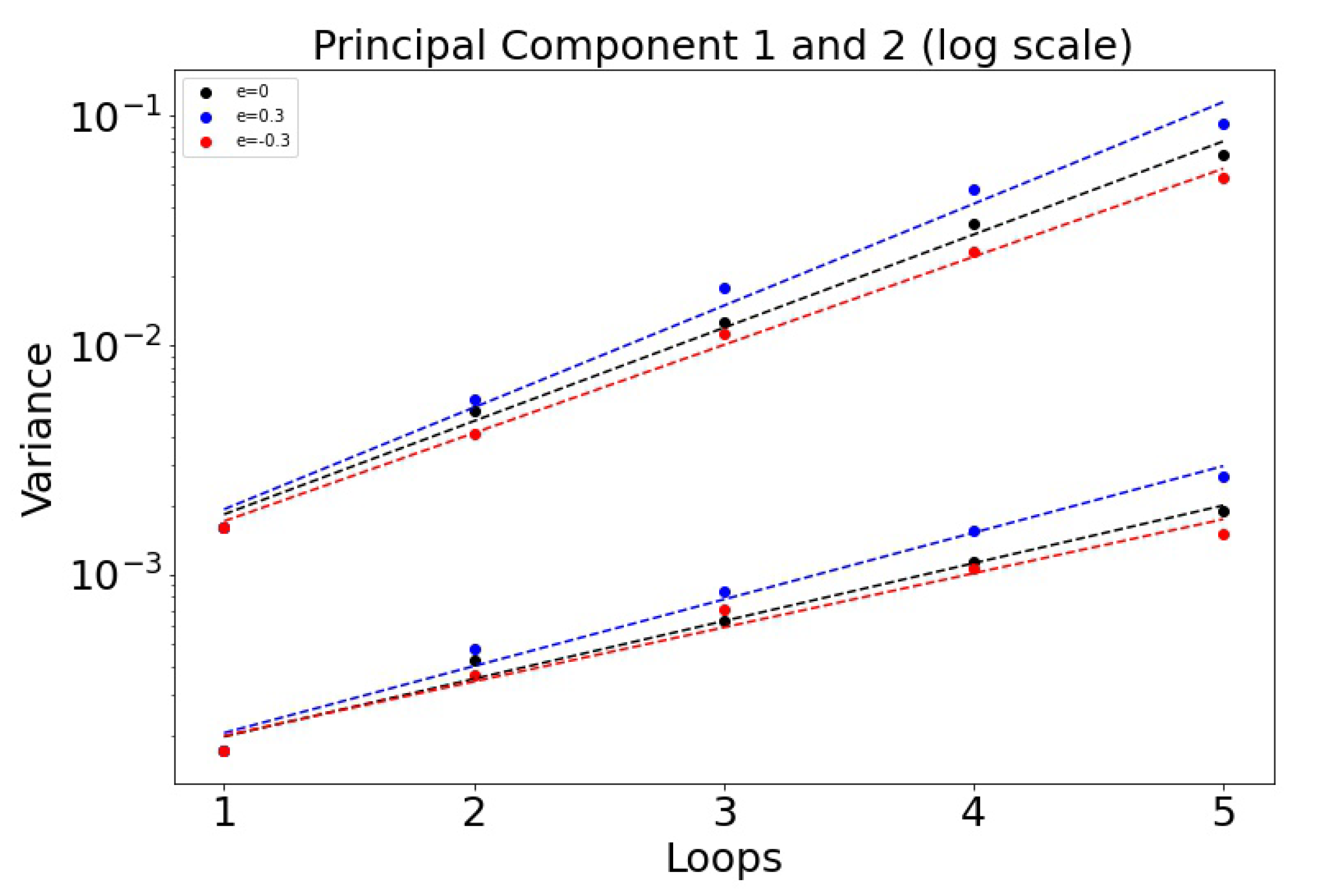
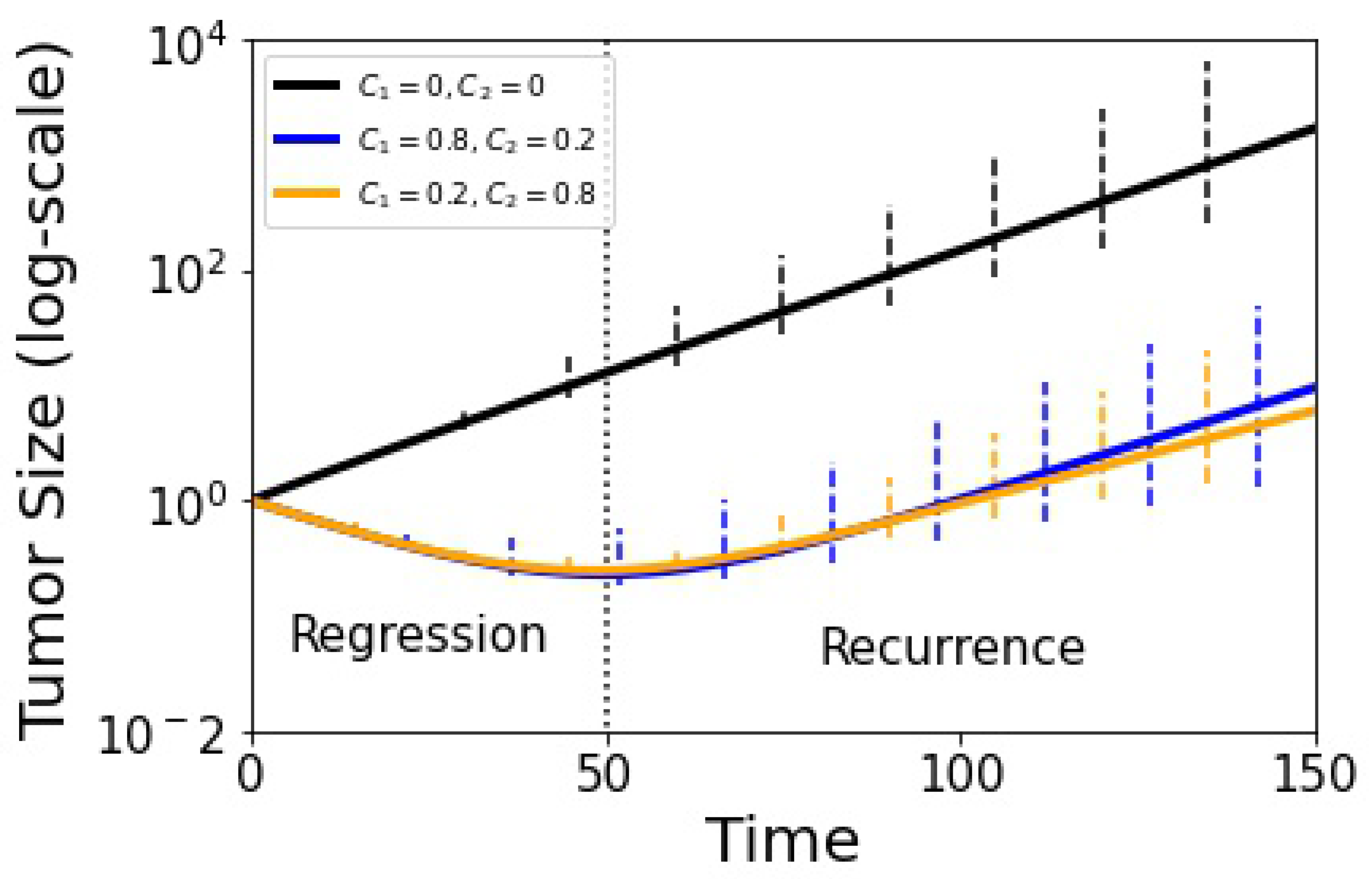
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gatenby, R.; Silva, A.; Gillies, R.; Frieden, B. Adaptive therapy. Cancer Res. 2009, 69, 4894–4903. [Google Scholar] [CrossRef] [PubMed]
- West, J.; You, L.; Zhang, J.; Gatenby, R.; Brown, J.; Newton, P.K.; Anderson, A. Towards multidrug adaptive therapy. Cancer Res. 2020, 80, 1578–1589. [Google Scholar] [CrossRef] [PubMed]
- Enriquez-Navas, P.; Kam, Y.; Das, T.; Hassan, S.; Silva, A.; Foroutan, P.; Ruiz, E.; Martiniez, G.; Minton, S.; Gillies, R.; et al. Exploiting evolutionary principles to prolong tumor control in preclinical models of breast cancer. Sci. Trans. Med. 2016, 8, 1–8. [Google Scholar] [CrossRef]
- Gasparini, G. Metronomic scheduling: The future of chemotherapy. Lancet 2001, 2, 733–738. [Google Scholar] [CrossRef]
- West, J.; Newton, P.K. Chemotherapeutic dose scheduling based on tumor growth rates provides a case for low-dose metronomic high-entropy therapies. Cancer Res. 2017, 77, 6717–6728. [Google Scholar] [CrossRef]
- Cunningham, J.; Gatenby, R.; Brown, J. Evolutionary dynamics in cancer therapy. Mol. Pharma. 2011, 8, 2094–2100. [Google Scholar] [CrossRef]
- Fisher, A.; Vazquez-Garcia, I.; Mustonen, V. The value of monitoring to control evolving populations. Proc. Natl Acad. Sci. USA 2015, 112, 1007–1012. [Google Scholar] [CrossRef] [PubMed]
- Dagogo-Jack, I.; Shaw, A. Tumor heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef] [PubMed]
- West, J.; Dinh, M.; Brown, J.; Zhang, J.; Anderson, A.; Gatenby, R. Multidrug cancer therapy in metastatic castrate-resistant prostate cancer: An evolution-based strategy. Clin. Cancer Res. 2019, 25, 4413–4421. [Google Scholar] [CrossRef]
- Zhang, J.; Cunningham, J.; Brown, J.; Gatenby, R. Integrating evolutionary dynamics into treatment of metastatic castrate-resistant prostate cancer. Nat. Commun. 2017, 8, 2094–2100. [Google Scholar] [CrossRef] [PubMed]
- Nowak, M.A. Evolutionary Dynamics: Exploring the Equations of Life; Harvard University Press: Cambridge, MA, USA, 2006. [Google Scholar]
- West, J.; Hasnain, Z.; Macklin, P.; Newton, P.K. An evolutionary model of tumor cell kinetics and the emergence of molecular heterogeneity driving gompertzian growth. SIAM Rev. 2016, 58, 716–736. [Google Scholar] [CrossRef]
- Komarova, N. Stochastic modeling of drug resistance in cancer. J. Theor. Biol. 2006, 239, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The different mechanisms of cancer drug resistance: A brief review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Khatami, M. Analyses of repeated failures in cancer therapy for solid tumors: Poor tumor-selective drug delivery, low therapeutic efficacy and unsustainable costs. Clin. Trans. Med. 2018, 7, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Kaznatchaeev, A.; Peacock, J.; Basanta, D.; Marusyk, A.; Scott, J. Fibroblasts and alectinib switch the evolutionary games being played by non-small cell lung cancer. Nat. Ecol. Evol. 2019, 3, 450–456. [Google Scholar] [CrossRef]
- Enriquez-Navas, M.; Wojtkowiak, J.; Gatenby, R. Application of evolutionary principles to cancer therapy. Cancer Res. 2015, 75, 4675–4680. [Google Scholar] [CrossRef]
- West, J.; Ma, Y.; Newton, P.K. Capitalizing on competition: An evolutionary model of competitive release in metastatic castration resistant prostate cancer treatment. J. Theor. Biol. 2018, 455, 249–260. [Google Scholar] [CrossRef]
- Newton, P.K.; Ma, Y. Nonlinear adaptive control of competitive release and chemotherapeutic resistance. Phys. Rev. E 2019, 99, 022404. [Google Scholar] [CrossRef]
- Hansen, E.; Karslake, J.; Woods, R.; Read, A.; Wood, K. Antibiotics can be used to contain drug-resistant bacteria by maintaining sufficiently large sensitive populations. PLoS Biol. 2020, 18. [Google Scholar] [CrossRef] [PubMed]
- Pastan, I.; Gottesman, M. Multidrug resistance. Annu. Rev. Med. 1991, 42, 277–286. [Google Scholar] [CrossRef]
- Hoffmann, A.; Scherrer, A.; Kufer, K.H. Analyzing the quality robustness of chemotherapy plans with respect to model uncertainties. Math. Biosci. 2015, 259, 55–61. [Google Scholar] [CrossRef]
- Smith, R.C. Uncertainty Quantification: Theory, Implementation, and Applications; SIAM: Philadelphia, PA, USA, 2014. [Google Scholar]
- Kostelich, E.; Kuang, Y.; McDaniel, J.; Moore, N.; Martirosyan, N.; Preul, M. Accurate state estimation from uncertain data and models: An application of data assimilation to mathematical models of human brain tumors. Biol. Direct 2011, 6, 1–20. [Google Scholar] [CrossRef]
- Yeh, P.; Hegrennes, M.; Aiden, A.; Kishony, R. Drug interactions and the evolution of antibiotic resistance. Nat. Rev. Micro. 2009, 7, 460–466. [Google Scholar] [CrossRef]
- Ma, Y.; Newton, P.K. The role of synergy and antagonism in designing multidrug adaptive chemotherapy. Phys. Rev. E 2021, 103, 032408. [Google Scholar] [CrossRef]
- Cressman, R.; Tao, Y. The replicator equation and other game dynamics. Proc. Natl. Acad. Sci. USA 2014, 111, 10810–10817. [Google Scholar] [CrossRef] [PubMed]
- Traulsen, A.; Claussen, J.C.; Hauert, C. Coevolutionary dynamics: From finite to infinite populations. Phys. Rev. Lett. 2005, 95, 238701. [Google Scholar] [CrossRef] [PubMed]
- Traulsen, A.; Claussen, J.C.; Hauert, C. Coevolutionary dynamics in large, but finite populations. Phys. Rev. E 2006, 74, 011901. [Google Scholar] [CrossRef] [PubMed]
- West, J.; Hasnain, Z.; Mason, J.; Newton, P.K. The prisoner’s dilemma as a cancer model. Converg. Sci. Phys. Oncol. 2016, 2, 035002. [Google Scholar] [CrossRef]
- Lenski, R. Bacterial evolution and the cost of antibiotic resistance. Internatl. Microbiol. 1998, 1, 265–270. [Google Scholar]
- Ferreira, E.; Neves, A. Fixation probabilities for the Moran process with three or more strategies: General and coupling results. J. Math. Bio. 2020, 81, 277–314. [Google Scholar] [CrossRef]
- Rodriquez, I.; Neves, A. Evolution of cooperation in a particular case of the infintely repeated prisoner’s dilemma with three strategies. J. Math. Bio. 2016, 73, 1665–1690. [Google Scholar] [CrossRef] [PubMed]
- Ross, S. Introduction to Probability Models, 10th ed.; Academic Press: Burlington, MA, USA, 2010. [Google Scholar]
- Helbing, D. A stochastic behavioral model and a microscopic foundation of evolutionary game theory. Theory Decis. 1996, 40, 149–179. [Google Scholar] [CrossRef]
- Hansen, E.; Read, A. Modifying adaptive therapy to enhance competitve suppression. Cancers 2020, 12, 3556. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dua, R.; Ma, Y.; Newton, P.K. Are Adaptive Chemotherapy Schedules Robust? A Three-Strategy Stochastic Evolutionary Game Theory Model. Cancers 2021, 13, 2880. https://doi.org/10.3390/cancers13122880
Dua R, Ma Y, Newton PK. Are Adaptive Chemotherapy Schedules Robust? A Three-Strategy Stochastic Evolutionary Game Theory Model. Cancers. 2021; 13(12):2880. https://doi.org/10.3390/cancers13122880
Chicago/Turabian StyleDua, Rajvir, Yongqian Ma, and Paul K. Newton. 2021. "Are Adaptive Chemotherapy Schedules Robust? A Three-Strategy Stochastic Evolutionary Game Theory Model" Cancers 13, no. 12: 2880. https://doi.org/10.3390/cancers13122880
APA StyleDua, R., Ma, Y., & Newton, P. K. (2021). Are Adaptive Chemotherapy Schedules Robust? A Three-Strategy Stochastic Evolutionary Game Theory Model. Cancers, 13(12), 2880. https://doi.org/10.3390/cancers13122880




