Impact of Neoantigen Expression and T-Cell Activation on Breast Cancer Survival
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects and Data Sources
2.2. Statistical Analysis
3. Results
3.1. Clinical and Pathologic Characteristics of Patients
3.2. Correlation between Neoantigen Expression and Clinical Pathological Variables
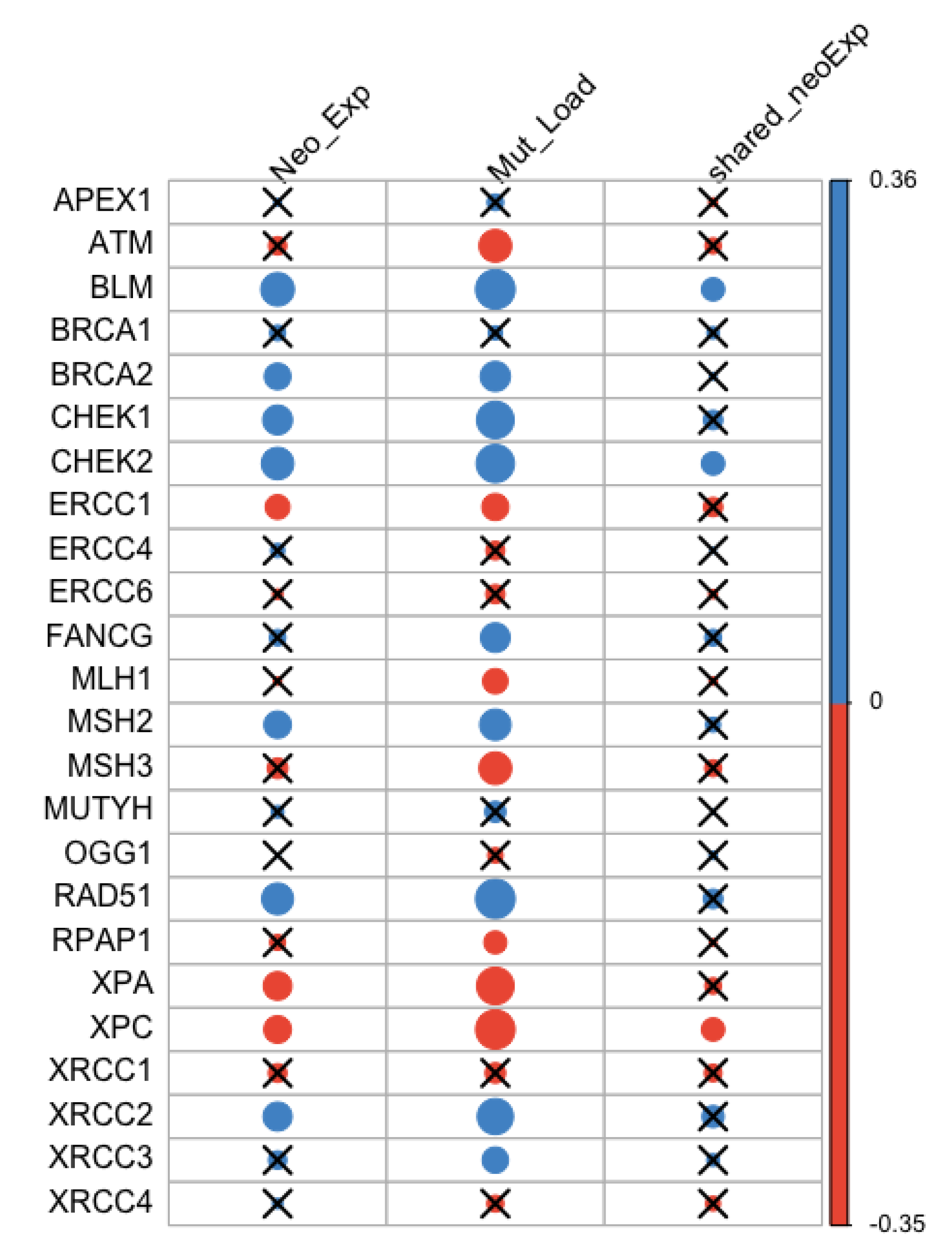
3.3. Correlation between Neoantigen Expression, Mutation Load, and DNA Repair Genes
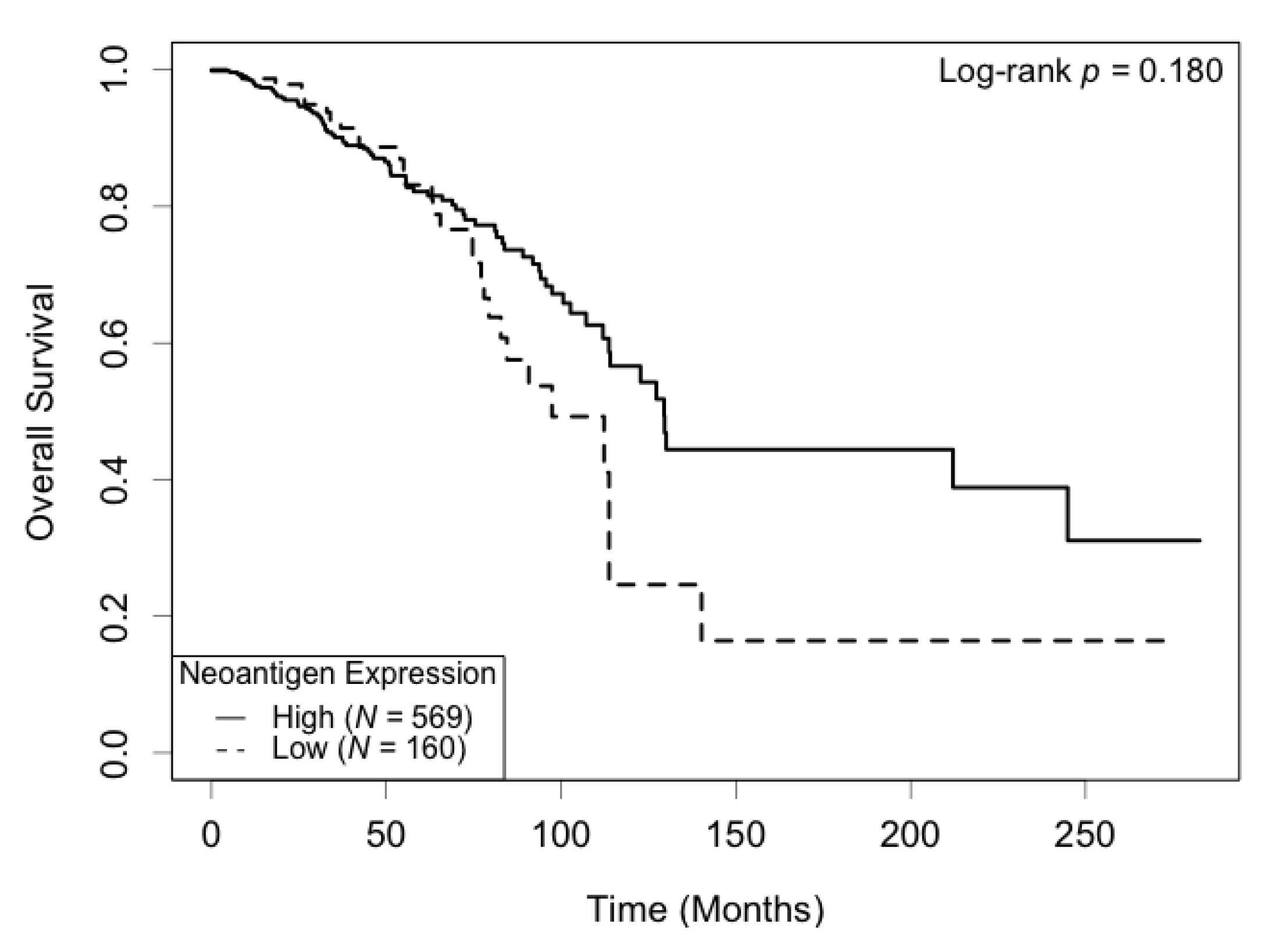
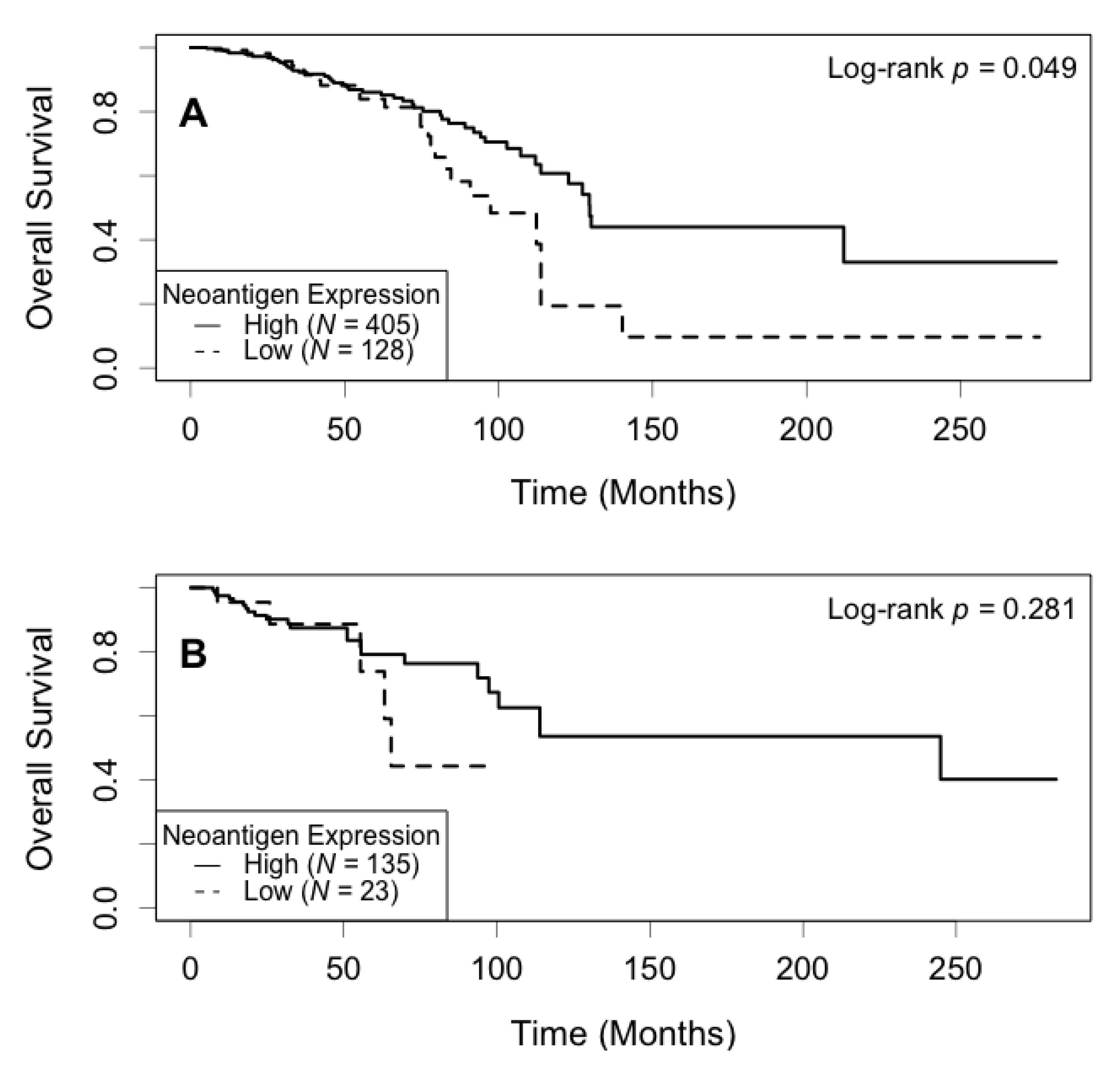
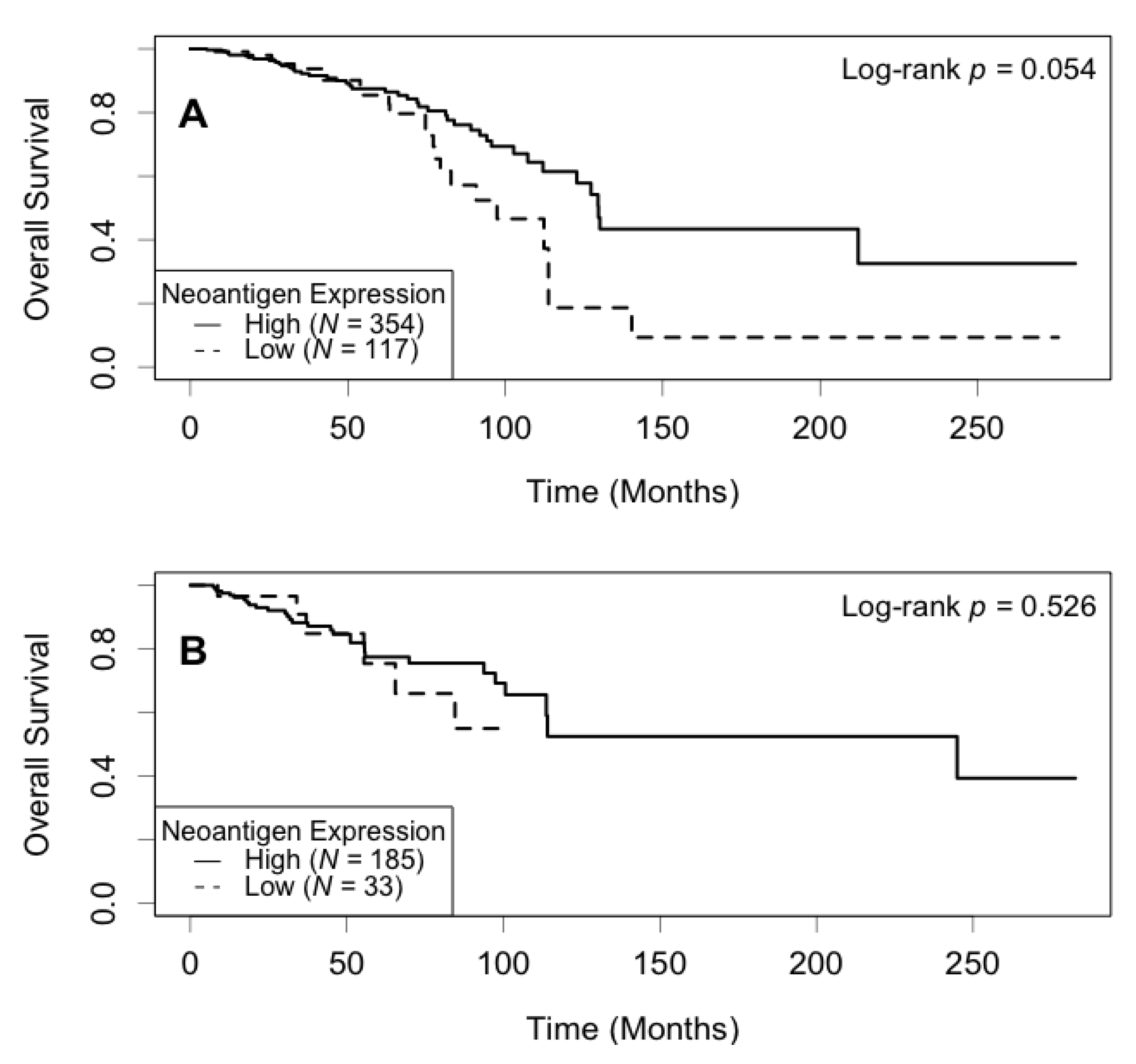
3.4. Association of Neoantigen Expression with Patient Survival
3.5. Association of Expression of the Most Shared Neoantigens with Patient Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sugie, T. Immunotherapy for metastatic breast cancer. Chin. Clin. Oncol. 2018, 7, 28. [Google Scholar] [CrossRef]
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in cancer immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Carreno, B.M.; Magrini, V.; Becker-Hapak, M.; Kaabinejadian, S.; Hundal, J.; Petti, A.A.; Ly, A.; Lie, W.R.; Hildebrand, W.H.; Mardis, E.R.; et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 2015, 348, 803–808. [Google Scholar] [CrossRef]
- Peng, M.; Mo, Y.; Wang, Y.; Wu, P.; Zhang, Y.; Xiong, F.; Guo, C.; Wu, X.; Li, Y.; Li, X.; et al. Neoantigen vaccine: An emerging tumor immunotherapy. Mol. Cancer 2019, 18, 128. [Google Scholar] [CrossRef] [PubMed]
- De Mattos-Arruda, L.; Blanco-Heredia, J.; Aguilar-Gurrieri, C.; Carrillo, J.; Blanco, J. New emerging targets in cancer immunotherapy: The role of neoantigens. ESMO Open 2020, 4 (Suppl. 3), e000684. [Google Scholar] [CrossRef]
- Ren, Y.; Cherukuri, Y.; Wickland, D.P.; Sarangi, V.; Tian, S.; Carter, J.M.; Mansfield, A.S.; Block, M.S.; Sherman, M.E.; Knutson, K.L.; et al. HLA class-I and class-II restricted neoantigen loads predict overall survival in breast cancer. Oncoimmunology 2020, 9, 1744947. [Google Scholar] [CrossRef]
- Jiang, T.; Shi, T.; Zhang, H.; Hu, J.; Song, Y.; Wei, J.; Ren, S.; Zhou, C. Tumor neoantigens: From basic research to clinical applications. J. Hematol. Oncol. 2019, 12, 93. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.Y.; Chung, W.H.; Chu, M.T.; Chen, S.J.; Chen, H.C.; Zheng, L.; Hung, S.I. Recent Development and Clinical Application of Cancer Vaccine: Targeting Neoantigens. J. Immunol. Res. 2018, 2018, 4325874. [Google Scholar] [CrossRef]
- Yarchoan, M.; Johnson, B.A., III; Lutz, E.R.; Laheru, D.A.; Jaffee, E.M. Targeting neoantigens to augment antitumour immunity. Nat. Rev. Cancer 2017, 17, 569. [Google Scholar] [CrossRef]
- Lu, L.; Bai, Y.; Wang, Z. Elevated T cell activation score is associated with improved survival of breast cancer. Breast Cancer Res. Treat. 2017, 164, 689–696. [Google Scholar] [CrossRef]
- Lu, L.; Huang, H.; Zhou, J.; Ma, W.; Mackay, S.; Wang, Z. BRCA1 mRNA expression modifies the effect of T cell activation score on patient survival in breast cancer. BMC Cancer 2019, 19, 387. [Google Scholar] [CrossRef] [PubMed]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H., II; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Han, X. Anti-PD-1/PD-L1 therapy of human cancer: Past, present, and future. J. Clin. Investig. 2015, 125, 3384–3391. [Google Scholar] [CrossRef]
- Kim, T.; Amaria, R.N.; Spencer, C.; Reuben, A.; Cooper, Z.A.; Wargo, J.A. Combining targeted therapy and immune checkpoint inhibitors in the treatment of metastatic melanoma. Cancer Biol. Med. 2014, 11, 237–246. [Google Scholar] [PubMed]
- Stronen, E.; Toebes, M.; Kelderman, S.; van Buuren, M.M.; Yang, W.; van Rooij, N.; Donia, M.; Boschen, M.L.; Lund-Johansen, F.; Olweus, J.; et al. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science 2016, 352, 1337–1341. [Google Scholar] [CrossRef] [PubMed]
- Chae, Y.K.; Anker, J.F.; Oh, M.S.; Bais, P.; Namburi, S.; Agte, S.; Giles, F.J.; Chuang, J.H. Mutations in DNA repair genes are associated with increased neoantigen burden and a distinct immunophenotype in lung squamous cell carcinoma. Sci. Rep. 2019, 9, 3235. [Google Scholar] [CrossRef]
- Zhang, J.; Caruso, F.P.; Sa, J.K.; Justesen, S.; Nam, D.H.; Sims, P.; Ceccarelli, M.; Lasorella, A.; Iavarone, A. The combination of neoantigen quality and T lymphocyte infiltrates identifies glioblastomas with the longest survival. Commun. Biol. 2019, 2, 135. [Google Scholar] [CrossRef]
- Miller, A.; Asmann, Y.; Cattaneo, L.; Braggio, E.; Keats, J.; Auclair, D.; Lonial, S.; Network, M.C.; Russell, S.J.; Stewart, A.K. High somatic mutation and neoantigen burden are correlated with decreased progression-free survival in multiple myeloma. Blood Cancer J. 2017, 7, e612. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef]
- Garcia-Garijo, A.; Fajardo, C.A.; Gros, A. Determinants for Neoantigen Identification. Front. Immunol. 2019, 10, 1392. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, R.; Cadieux, E.L.; Salgado, R.; Bakir, M.A.; Moore, D.A.; Hiley, C.T.; Lund, T.; Tanic, M.; Reading, J.L.; Joshi, K.; et al. Neoantigen-directed immune escape in lung cancer evolution. Nature 2019, 567, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Jurtz, V.; Paul, S.; Andreatta, M.; Marcatili, P.; Peters, B.; Nielsen, M. NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J. Immunol. 2017, 199, 3360–3368. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, D.G.; Manolio, T.A.; Dimmock, D.P.; Rehm, H.L.; Shendure, J.; Abecasis, G.R.; Adams, D.R.; Altman, R.B.; Antonarakis, S.E.; Ashley, E.A.; et al. Guidelines for investigating causality of sequence variants in human disease. Nature 2014, 508, 469–476. [Google Scholar] [CrossRef]
- Hountis, P.; Dedeilias, P.; Douzinas, M. The management of Castleman’s disease of the mediastinum: A case report. Cases J. 2008, 1, 330. [Google Scholar] [CrossRef]
- Kappil, M.A.; Liao, Y.; Terry, M.B.; Santella, R.M. DNA Repair Gene Expression Levels as Indicators of Breast Cancer in the Breast Cancer Family Registry. Anticancer Res. 2016, 36, 4039–4044. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.D.; Lin, S.Y. DNA damage and breast cancer. World J. Clin. Oncol. 2011, 2, 329–338. [Google Scholar] [CrossRef]
- Zhu, G.; Su, H.; Lu, L.; Guo, H.; Chen, Z.; Sun, Z.; Song, R.; Wang, X.; Li, H.; Wang, Z. Association of nineteen polymorphisms from seven DNA repair genes and the risk for bladder cancer in Gansu province of China. Oncotarget 2016, 7, 31372–31383. [Google Scholar] [CrossRef]
- Zhao, J.; Huang, J. Breast cancer immunology and immunotherapy: Targeting the programmed cell death protein-1/programmed cell death protein ligand-1. Chin. Med. J. 2020, 133, 853–862. [Google Scholar] [CrossRef]
- Strickland, K.C.; Howitt, B.E.; Shukla, S.A.; Rodig, S.; Ritterhouse, L.L.; Liu, J.F.; Garber, J.E.; Chowdhury, D.; Wu, C.J.; D’Andrea, A.D.; et al. Association and prognostic significance of BRCA1/2-mutation status with neoantigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget 2016, 7, 13587–13598. [Google Scholar] [CrossRef]
- Lauss, M.; Donia, M.; Harbst, K.; Andersen, R.; Mitra, S.; Rosengren, F.; Salim, M.; Vallon-Christersson, J.; Torngren, T.; Kvist, A.; et al. Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma. Nat. Commun. 2017, 8, 1738. [Google Scholar] [CrossRef]
- Ott, P.A.; Hu, Z.; Keskin, D.B.; Shukla, S.A.; Sun, J.; Bozym, D.J.; Zhang, W.; Luoma, A.; Giobbie-Hurder, A.; Peter, L.; et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017, 547, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Keskin, D.B.; Anandappa, A.J.; Sun, J.; Tirosh, I.; Mathewson, N.D.; Li, S.; Oliveira, G.; Giobbie-Hurder, A.; Felt, K.; Gjini, E.; et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 2019, 565, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Terranova-Barberio, M.; Pawlowska, N.; Dhawan, M.; Moasser, M.; Chien, A.J.; Melisko, M.E.; Rugo, H.; Rahimi, R.; Deal, T.; Daud, A.; et al. Exhausted T cell signature predicts immunotherapy response in ER-positive breast cancer. Nat. Commun. 2020, 11, 3584. [Google Scholar] [CrossRef]
- Andor, N.; Maley, C.C.; Ji, H.P. Genomic Instability in Cancer: Teetering on the Limit of Tolerance. Cancer Res. 2017, 77, 2179–2185. [Google Scholar] [CrossRef]
- Burgess, J.T.; Rose, M.; Boucher, D.; Plowman, J.; Molloy, C.; Fisher, M.; O’Leary, C.; Richard, D.J.; O’Byrne, K.J.; Bolderson, E. The Therapeutic Potential of DNA Damage Repair Pathways and Genomic Stability in Lung Cancer. Front. Oncol. 2020, 10, 1256. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, X.; Hao, L.; Jiang, D.; Wu, B.; He, T.; Tang, Y. The diagnostic value of DNA repair gene in breast cancer metastasis. Sci. Rep. 2020, 10, 19626. [Google Scholar] [CrossRef]
- Crew, K.D.; Gammon, M.D.; Terry, M.B.; Zhang, F.F.; Zablotska, L.B.; Agrawal, M.; Shen, J.; Long, C.M.; Eng, S.M.; Sagiv, S.K.; et al. Polymorphisms in nucleotide excision repair genes, polycyclic aromatic hydrocarbon-DNA adducts, and breast cancer risk. Cancer Epidemiol. Biomark. Prev. 2007, 16, 2033–2041. [Google Scholar] [CrossRef]
- Malik, S.S.; Zia, A.; Rashid, S.; Mubarik, S.; Masood, N.; Hussain, M.; Yasmin, A.; Bano, R. XPC as breast cancer susceptibility gene: Evidence from genetic profiling, statistical inferences and protein structural analysis. Breast Cancer 2020, 27, 1168–1176. [Google Scholar] [CrossRef] [PubMed]
- Anglian Breast Cancer Study Group. Prevalence and penetrance of BRCA1 and BRCA2 mutations in a population-based series of breast cancer cases. Br. J. Cancer 2000, 83, 1301–1308. [Google Scholar] [CrossRef]
- Stratton, M.R.; Rahman, N. The emerging landscape of breast cancer susceptibility. Nat. Genet. 2008, 40, 17–22. [Google Scholar] [CrossRef]
- Melchor, L.; Benitez, J. The complex genetic landscape of familial breast cancer. Hum. Genet. 2013, 132, 845–863. [Google Scholar] [CrossRef] [PubMed]
- Claus, E.B.; Schildkraut, J.M.; Thompson, W.D.; Risch, N.J. The genetic attributable risk of breast and ovarian cancer. Cancer 1996, 77, 2318–2324. [Google Scholar] [CrossRef]
- Harkness, E.F.; Barrow, E.; Newton, K.; Green, K.; Clancy, T.; Lalloo, F.; Hill, J.; Evans, D.G. Lynch syndrome caused by MLH1 mutations is associated with an increased risk of breast cancer: A cohort study. J. Med. Genet. 2015, 52, 553–556. [Google Scholar] [CrossRef] [PubMed]
- Malik, S.S.; Masood, N.; Asif, M.; Ahmed, P.; Shah, Z.U.; Khan, J.S. Expressional analysis of MLH1 and MSH2 in breast cancer. Curr. Probl. Cancer 2019, 43, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.Y.; Camp, N.J.; Cannon-Albright, L.A.; Allen-Brady, K.; Balasubramanian, S.; Reed, M.W.; Hopper, J.L.; Apicella, C.; Giles, G.G.; Southey, M.C.; et al. A role for XRCC2 gene polymorphisms in breast cancer risk and survival. J. Med. Genet. 2011, 48, 477–484. [Google Scholar] [CrossRef]
- Jara, L.; Morales, S.; de Mayo, T.; Gonzalez-Hormazabal, P.; Carrasco, V.; Godoy, R. Mutations in BRCA1, BRCA2 and other breast and ovarian cancer susceptibility genes in Central and South American populations. Biol. Res. 2017, 50, 35. [Google Scholar] [CrossRef]
- Klein, H.L. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair 2008, 7, 686–693. [Google Scholar] [CrossRef]
- Wiegmans, A.P.; Al-Ejeh, F.; Chee, N.; Yap, P.Y.; Gorski, J.J.; Da Silva, L.; Bolderson, E.; Chenevix-Trench, G.; Anderson, R.; Simpson, P.T.; et al. Rad51 supports triple negative breast cancer metastasis. Oncotarget 2014, 5, 3261–3272. [Google Scholar] [CrossRef]
- Stopper, H.; Schmitt, E.; Gregor, C.; Mueller, S.O.; Fischer, W.H. Increased cell proliferation is associated with genomic instability: Elevated micronuclei frequencies in estradiol-treated human ovarian cancer cells. Mutagenesis 2003, 18, 243–247. [Google Scholar] [CrossRef][Green Version]
- Wu, M.; Pang, J.S.; Sun, Q.; Huang, Y.; Hou, J.Y.; Chen, G.; Zeng, J.J.; Feng, Z.B. The clinical significance of CHEK1 in breast cancer: A high-throughput data analysis and immunohistochemical study. Int. J. Clin. Exp. Pathol. 2019, 12, 1–20. [Google Scholar] [PubMed]
- Tort, F.; Hernandez, S.; Bea, S.; Camacho, E.; Fernandez, V.; Esteller, M.; Fraga, M.F.; Burek, C.; Rosenwald, A.; Hernandez, L.; et al. Checkpoint kinase 1 (CHK1) protein and mRNA expression is downregulated in aggressive variants of human lymphoid neoplasms. Leukemia 2005, 19, 112–117. [Google Scholar] [CrossRef]
- Ansari, N.; Shahrabi, S.; Khosravi, A.; Shirzad, R.; Rezaeean, H. Prognostic Significance of CHEK2 Mutation in Progression of Breast Cancer. Lab. Med. 2019, 50, e36–e41. [Google Scholar] [CrossRef] [PubMed]
- Arora, A.; Abdel-Fatah, T.M.; Agarwal, D.; Doherty, R.; Moseley, P.M.; Aleskandarany, M.A.; Green, A.R.; Ball, G.; Alshareeda, A.T.; Rakha, E.A.; et al. Transcriptomic and Protein Expression Analysis Reveals Clinicopathological Significance of Bloom Syndrome Helicase (BLM) in Breast Cancer. Mol. Cancer Ther. 2015, 14, 1057–1065. [Google Scholar] [CrossRef]
- Lakshmanan, I.; Ponnusamy, M.P.; Das, S.; Chakraborty, S.; Haridas, D.; Mukhopadhyay, P.; Lele, S.M.; Batra, S.K. MUC16 induced rapid G2/M transition via interactions with JAK2 for increased proliferation and anti-apoptosis in breast cancer cells. Oncogene 2012, 31, 805–817. [Google Scholar] [CrossRef]
- Balachandran, V.P.; Luksza, M.; Zhao, J.N.; Makarov, V.; Moral, J.A.; Remark, R.; Herbst, B.; Askan, G.; Bhanot, U.; Senbabaoglu, Y.; et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 2017, 551, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Novak, P.; Jensen, T.; Oshiro, M.M.; Watts, G.S.; Kim, C.J.; Futscher, B.W. Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer Res. 2008, 68, 8616–8625. [Google Scholar] [CrossRef]
- Sahin, U.; Derhovanessian, E.; Miller, M.; Kloke, B.P.; Simon, P.; Lower, M.; Bukur, V.; Tadmor, A.D.; Luxemburger, U.; Schrors, B.; et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017, 547, 222–226. [Google Scholar] [CrossRef]
- Sosman, J.A.; Kim, K.B.; Schuchter, L.; Gonzalez, R.; Pavlick, A.C.; Weber, J.S.; McArthur, G.A.; Hutson, T.E.; Moschos, S.J.; Flaherty, K.T.; et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N. Engl. J. Med. 2012, 366, 707–714. [Google Scholar] [CrossRef]
- Chapman, P.B.; Hauschild, A.; Robert, C.; Haanen, J.B.; Ascierto, P.; Larkin, J.; Dummer, R.; Garbe, C.; Testori, A.; Maio, M.; et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011, 364, 2507–2516. [Google Scholar] [CrossRef]
- Larkin, J.; Ascierto, P.A.; Dreno, B.; Atkinson, V.; Liszkay, G.; Maio, M.; Mandala, M.; Demidov, L.; Stroyakovskiy, D.; Thomas, L.; et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N. Engl. J. Med. 2014, 371, 1867–1876. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Stroyakovskiy, D.; Gogas, H.; Levchenko, E.; de Braud, F.; Larkin, J.; Garbe, C.; Jouary, T.; Hauschild, A.; Grob, J.J.; et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N. Engl. J. Med. 2014, 371, 1877–1888. [Google Scholar] [CrossRef] [PubMed]
| Variable | N | % |
|---|---|---|
| ER | 691 | |
| Negative | 158 | 22.9 |
| Positive | 533 | 77.1 |
| PR | 689 | |
| Negative | 218 | 31.6 |
| Positive | 471 | 68.4 |
| HER2 | 498 | |
| Negative | 381 | 76.5 |
| Positive | 117 | 23.5 |
| Molecular subtype | 495 | |
| Luminal | 393 | 79.4 |
| Basal-like | 71 | 14.3 |
| HER2-enrich | 31 | 6.3 |
| Stage | 709 | |
| I | 125 | 17.6 |
| II | 408 | 57.6 |
| III & IV | 176 | 24.8 |
| Histology | 728 | |
| Ductal | 604 | 83.0 |
| Lobular | 62 | 8.5 |
| Mix | 44 | 6.0 |
| Other | 18 | 2.5 |
| Death | 729 | |
| No | 617 | 84.6 |
| Yes | 112 | 15.4 |
| N | Mean (Range) | |
| Age (mean ± SD 1, years) | 729 | 57.7 ± 13.1 (26–90) |
| Follow-up (months) | 729 | 42.8 (0–282.7) |
| Variables | Death | |
|---|---|---|
| HR (95% | p-Value | |
| Neoantigen Expression | ||
| Low | 1.00 | |
| High | 0.61 (0.38–0.97) | 0.038 |
| T-cell Activation | ||
| Exhaustion | 1.00 | |
| Activation | 0.48 (0.24–0.96) | 0.038 |
| Age | 1.04 (1.03–1.06) | <0.001 |
| ER | ||
| Negative | 1.00 | |
| Positive | 0.53 (0.32–0.87) | 0.012 |
| Stage | ||
| Stage I | 1.00 | |
| Stage II–IV | 2.64 (1.39–5.04) | 0.003 |
| Histology | ||
| Ductal | 1.00 | |
| Lobular | 0.73 (0.35–1.51) | 0.395 |
| Mix or Other | 0.93 (0.48–1.77) | 0.817 |
| Stratification | Death | ||
|---|---|---|---|
| Variable | Variables | HR (95% | p-Value |
| T-cell Exhaustion | Neoantigen Expression | ||
| Low | 1.00 | ||
| High | 0.55 (0.34–0.89) | 0.016 | |
| Age | 1.04 (1.03–1.06) | <0.001 | |
| ER | |||
| Negative | 1.00 | ||
| Positive | 0.45 (0.27–0.75) | 0.002 | |
| Stage | |||
| Stage I | 1.00 | ||
| Stage II–IV | 2.80 (1.38–5.67) | 0.004 | |
| Histology | |||
| Ductal | 1.00 | ||
| Lobular | 0.58 (0.25–1.30) | 0.185 | |
| Mix or Other | 0.95 (0.47–1.93) | 0.887 | |
| T-cell Activation | Neoantigen Expression | ||
| Low | 1.00 | ||
| High | 0.76 (0.08–7.44) | 0.816 | |
| Age | 1.06 (1.00–1.13) | 0.049 | |
| ER | |||
| Negative | 1.00 | ||
| Positive | 0.89 (0.18–4.34) | 0.890 | |
| Stage | |||
| Stage I | 1.00 | ||
| Stage II–IV | 5.52 (0.71–42.90) | 0.103 | |
| Histology | |||
| Ductal | 1.00 | ||
| Lobular | 7.80 (1.00–60.59) | 0.050 | |
| Mix or Other | 0.48 (0.05–4.44) | 0.521 | |
| Stratification | Death | ||
|---|---|---|---|
| Variable | Variables | HR (95% | p-Value |
| ER Positive | Neo Expression | ||
| Low | 1.00 | ||
| High | 0.61 (0.36–1.04) | 0.067 | |
| T-cell Activation | |||
| Exhaustion | 1.00 | ||
| Activation | 0.80 (0.34–1.89) | 0.613 | |
| Age | 1.05 (1.03–1.07) | <0.001 | |
| Stage | |||
| Stage I | 1.00 | ||
| Stage II–IV | 2.56 (1.25–5.24) | 0.010 | |
| Histology | |||
| Ductal | 1.00 | ||
| Lobular | 0.47 (0.19–1.14) | 0.096 | |
| Mix or Other | 1.01 (0.49–2.07) | 0.989 | |
| ER Negative | Neo Expression | ||
| Low | 1.00 | ||
| High | 0.76 (0.26–2.16) | 0.601 | |
| T-cell Activation | |||
| Exhaustion | 1.00 | ||
| Activation | 0.32 (0.11–0.99) | 0.048 | |
| Age | 1.03 (1.00–1.06) | 0.061 | |
| Stage | |||
| Stage I | 1.00 | ||
| Stage II–IV | 4.93 (0.65–37.26) | 0.122 | |
| Histology | |||
| Ductal | 1.00 | ||
| Lobular | 4.64 (1.17–18.45) | 0.029 | |
| Mix or Other | 0.61 (0.13–2.90) | 0.538 | |
| Stratification | Death | ||
|---|---|---|---|
| Variable | Variables | HR (95% | p-Value |
| PR Positive | Neo Expression | ||
| Low | 1.00 | ||
| High | 0.57 (0.32–0.99) | 0.046 | |
| T-cell Activation | |||
| Exhaustion | 1.00 | ||
| Activation | 0.82 (0.29–2.36) | 0.720 | |
| Age | 1.05 (1.03–1.07) | <0.001 | |
| ER | |||
| Negative | 1.00 | ||
| Positive | 0.85 (0.11–6.38) | 0.876 | |
| Stage | |||
| Stage I | 1.00 | ||
| Stage II–IV | 2.45 (1.15–5.20) | 0.020 | |
| Histology | |||
| Ductal | 1.00 | ||
| Lobular | 0.46 (0.19–1.14) | 0.095 | |
| Mix or Other | 0.97 (0.45–2.10) | 0.941 | |
| PR Negative | Neo Expression | ||
| Low | 1.00 | ||
| High | 0.67 (0.24–1.84) | 0.439 | |
| T-cell Activation | |||
| Exhaustion | 1.00 | ||
| Activation | 0.45 (0.17–1.16) | 0.096 | |
| Age | 1.03 (1.00–1.06) | 0.036 | |
| ER | |||
| Negative | 1.00 | ||
| Positive | 0.63 (0.29–1.38) | 0.247 | |
| Stage | |||
| Stage I | 1.00 | ||
| Stage II–IV | 3.73 (0.88–15.91) | 0.075 | |
| Histology | |||
| Ductal | 1.00 | ||
| Lobular | 3.25 (0.92–11.55) | 0.068 | |
| Mix or Other | 0.77 (0.22–2.69) | 0.685 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Amei, A.; Bui, F.; Norouzifar, S.; Lu, L.; Wang, Z. Impact of Neoantigen Expression and T-Cell Activation on Breast Cancer Survival. Cancers 2021, 13, 2879. https://doi.org/10.3390/cancers13122879
Li W, Amei A, Bui F, Norouzifar S, Lu L, Wang Z. Impact of Neoantigen Expression and T-Cell Activation on Breast Cancer Survival. Cancers. 2021; 13(12):2879. https://doi.org/10.3390/cancers13122879
Chicago/Turabian StyleLi, Wenjing, Amei Amei, Francis Bui, Saba Norouzifar, Lingeng Lu, and Zuoheng Wang. 2021. "Impact of Neoantigen Expression and T-Cell Activation on Breast Cancer Survival" Cancers 13, no. 12: 2879. https://doi.org/10.3390/cancers13122879
APA StyleLi, W., Amei, A., Bui, F., Norouzifar, S., Lu, L., & Wang, Z. (2021). Impact of Neoantigen Expression and T-Cell Activation on Breast Cancer Survival. Cancers, 13(12), 2879. https://doi.org/10.3390/cancers13122879







