Inhibition of Serum Response Factor Improves Response to Enzalutamide in Prostate Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
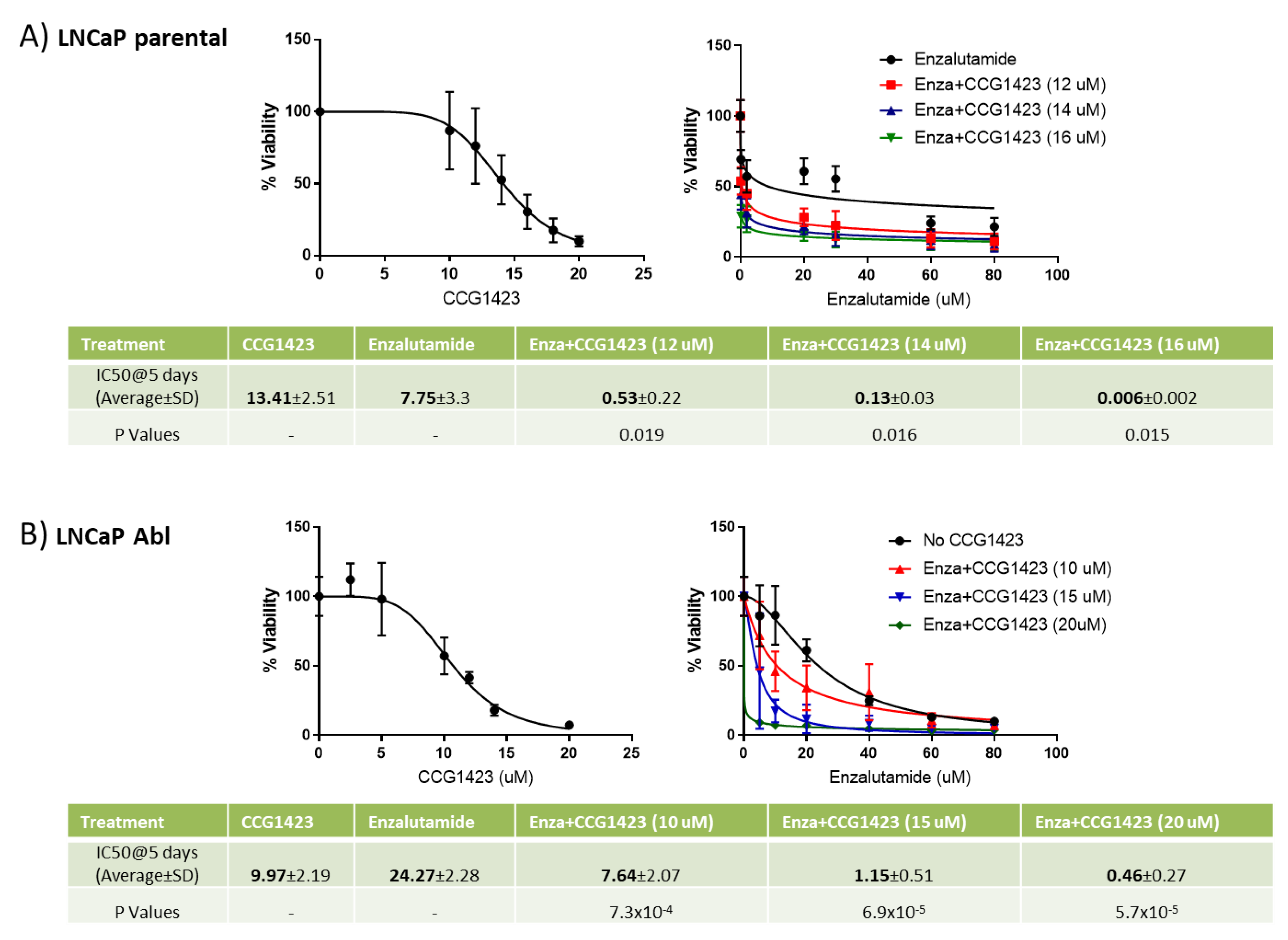
2.1. Combined Effect of Enzalutamide and CCG1423 In Vitro
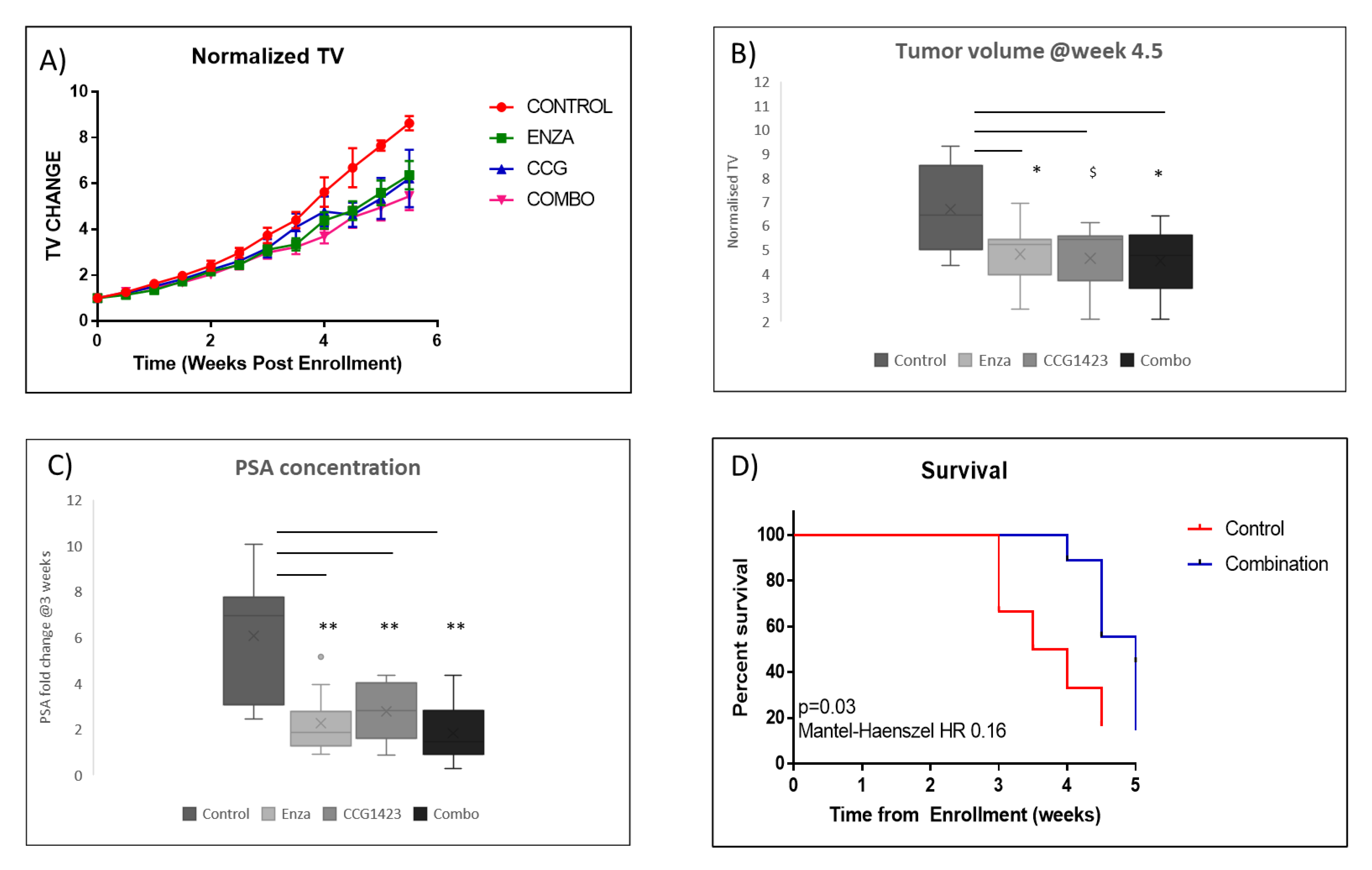
2.2. Combined Effect of Enzalutamide and CCG1423 in LuCaP 35CR Patient-Derived Xenograft
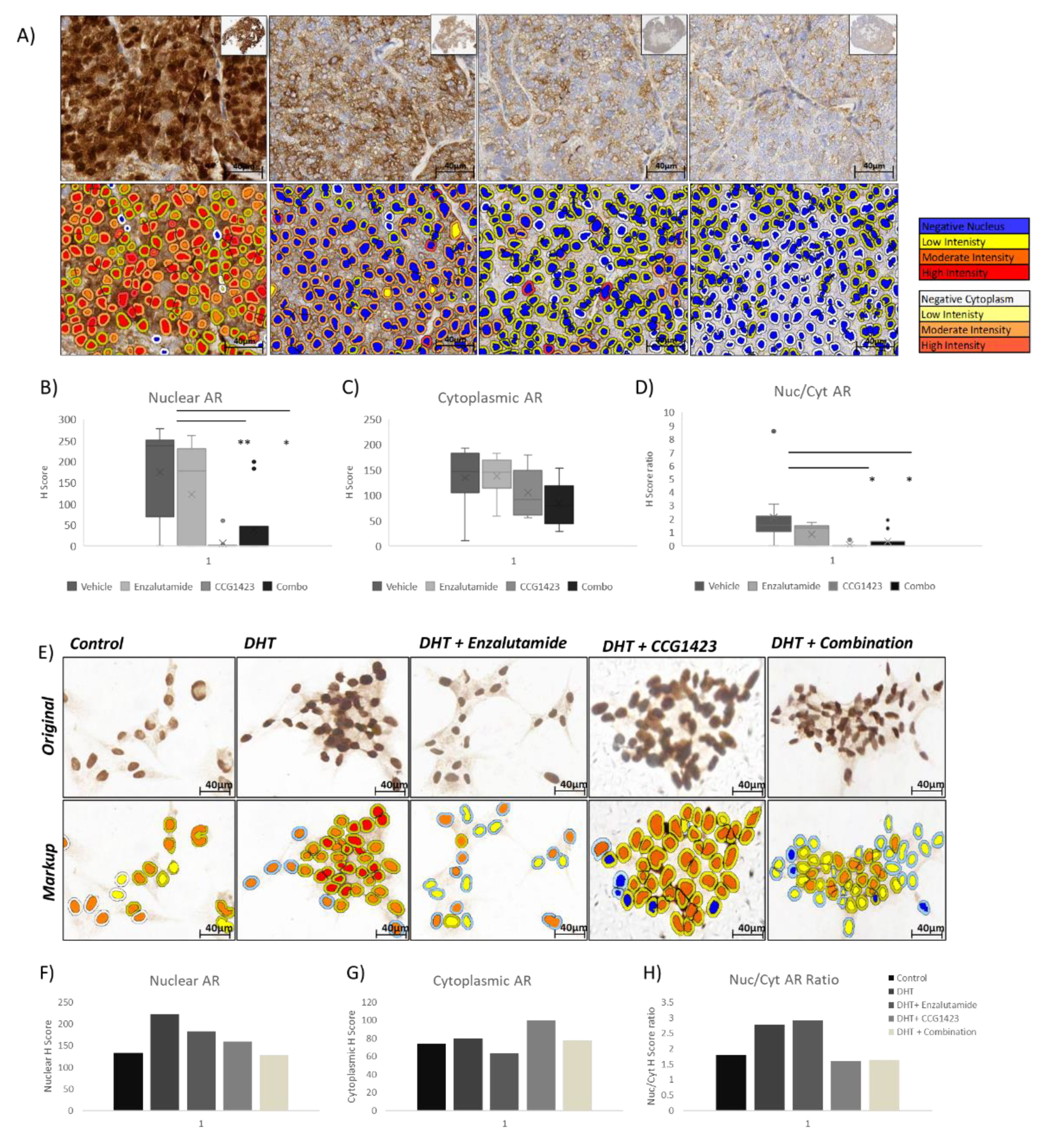
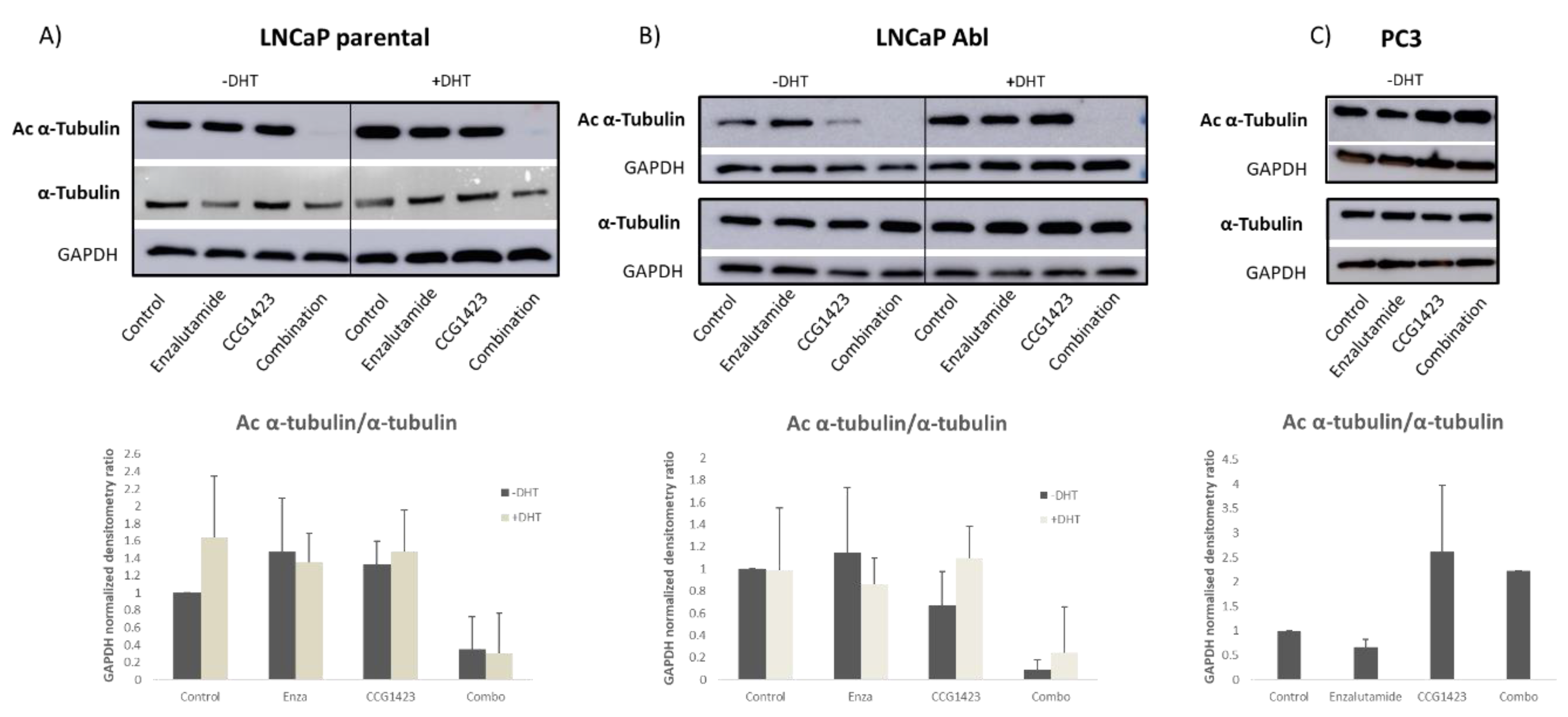
2.3. Androgen Receptor Nuclear Translocation Is Inhibited by CCG1423 through Inhibition of α-Tubulin Acetylation
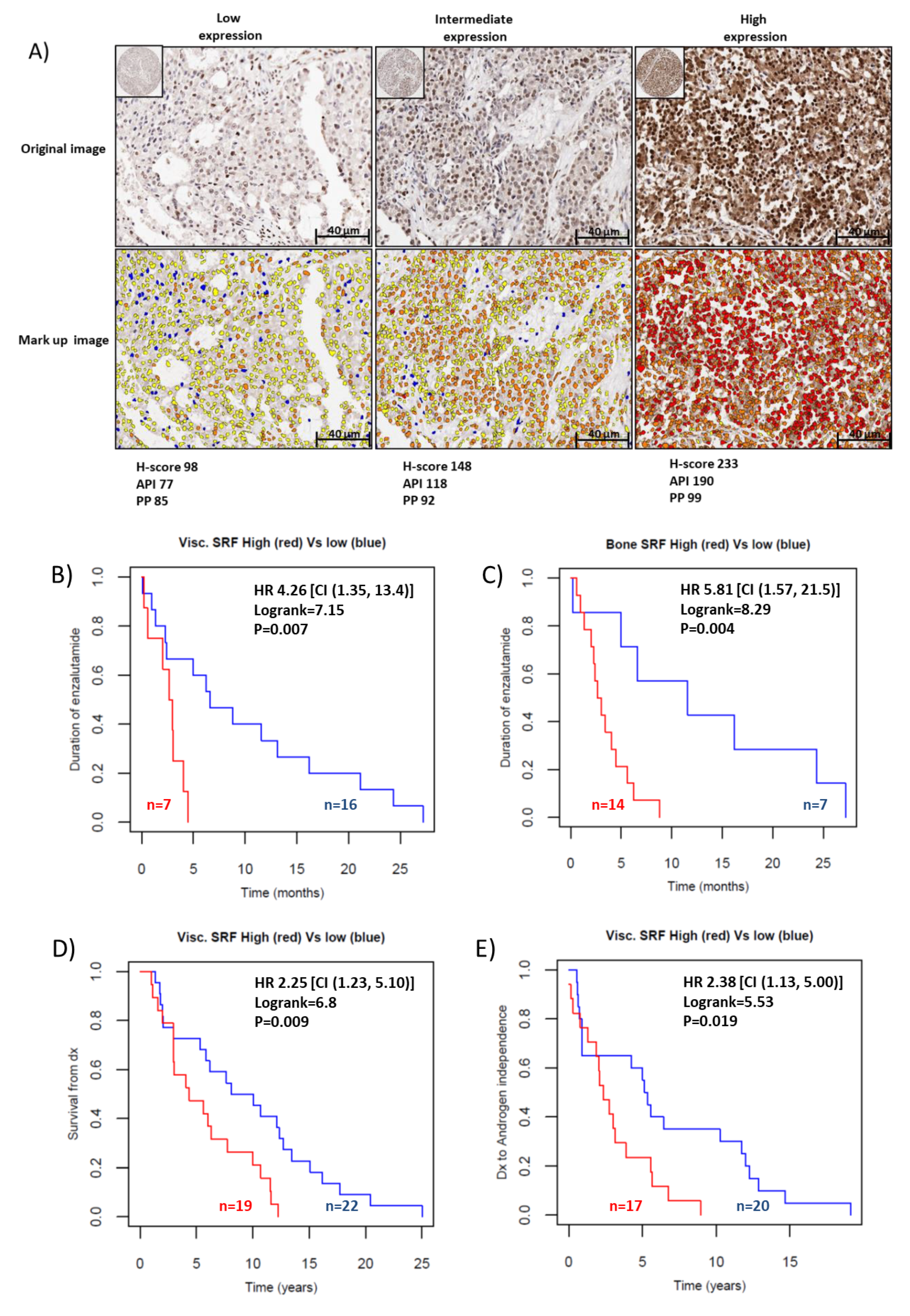
2.4. SRF Expression Is Associated with Enzalutamide Resistance in Patients with CRPC
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. 3-(4,5)-dimethylthiazol-2-yl-2,5-diphenyltetrazolium Bromide (MTT) Cell Viability Assay
4.3. PDX Study and Statistical Analysis
4.4. Immunohistochemical (IHC) and Immunocytochemical (ICC) Staining
4.5. Scoring of AR Protein Immunoreactivity and Statistical Analysis
4.6. Western Blotting
4.7. Sample Collection/Tissue Microarray Description
4.8. Scoring of SRF Protein Expression and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Claessens, F.; Helsen, C.; Prekovic, S.; Broeck, T.V.D.; Spans, L.; Van Poppel, H.; Joniau, S. Emerging mechanisms of enzalutamide resistance in prostate cancer. Nat. Rev. Urol. 2014, 11, 712–716. [Google Scholar] [CrossRef]
- Prekovic, S.; Broeck, T.V.D.; Linder, S.; Van Royen, M.E.; Houtsmuller, A.; Handle, F.; Joniau, S.; Zwart, W.; Claessens, F. Molecular underpinnings of enzalutamide resistance. Endocr. Related Cancer 2018, 25, R545–R557. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Lu, C.; Wang, H.; Luber, B.; Nakazawa, M.; Roeser, J.C.; Chen, Y.; Mohammad, T.A.; Chen, Y.; Fedor, H.L.; et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014, 371, 1028–1038. [Google Scholar] [CrossRef] [Green Version]
- Buttigliero, C.; Tucci, M.; Bertaglia, V.; Vignani, F.; Bironzo, P.; Di Maio, M.; Scagliotti, G. Understanding and overcoming the mechanisms of primary and acquired resistance to abiraterone and enzalutamide in castration resistant prostate cancer. Cancer Treat. Rev. 2015, 41, 884–892. [Google Scholar] [CrossRef]
- Foley, C.; Mitsiades, N. Moving Beyond the Androgen Receptor (AR): Targeting AR-Interacting Proteins to Treat Prostate Cancer. Horm. Cancer 2016, 7, 84–103. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Chen, G.; Streb, J.W.; Long, X.; Yang, Y.; Stoeckert, C.J.; Miano, J.M. Defining the mammalian CArGome. Genome Res. 2005, 16, 197–207. [Google Scholar] [CrossRef] [Green Version]
- Prencipe, M.; Madden, S.F.; O’Neill, A.; O’hurley, G.; Culhane, A.C.; O’Connor, D.P.; Klocker, H.; Kay, E.W.; Gallagher, W.M.; Watson, W.R. Identification of transcription factors associated with castration-resistance: Is the serum responsive factor a potential therapeutic target? Prostate 2013, 73, 743–753. [Google Scholar] [CrossRef]
- Culig, Z.; Hoffmann, J.; Erdel, M.; E Eder, I.; Hobisch, A.; Hittmair, A.; Bartsch, G.; Utermann, G.; Schneider, M.R.; Parczyk, K.; et al. Switch from antagonist to agonist of the androgen receptor blocker bicalutamide is associated with prostate tumour progression in a new model system. Br. J. Cancer 1999, 81, 242–251. [Google Scholar] [CrossRef]
- Prencipe, M.; O’Neill, A.; O’Hurley, G.; Nguyen, L.K.; Fabre, A.; Bjartell, A.; Gallagher, W.M.; Morrissey, C.; Kay, E.W.; Watson, W.R. Relationship between serum response factor and androgen receptor in prostate cancer. Prostate 2015, 75, 1704–1717. [Google Scholar] [CrossRef] [Green Version]
- Heemers, H.V.; Regan, K.M.; Dehm, S.M.; Tindall, D.J. Androgen Induction of the Androgen Receptor Coactivator Four and a Half LIM Domain Protein-2: Evidence for a Role for Serum Response Factor in Prostate Cancer. Cancer Res. 2007, 67, 10592–10599. [Google Scholar] [CrossRef] [Green Version]
- Heemers, H.V.; Schmidt, L.J.; Sun, Z.; Regan, K.M.; Anderson, S.K.; Duncan, K.; Wang, D.; Liu, S.; Ballman, K.V.; Tindall, N.J. Identification of a clinically relevant androgen-dependent gene signature in prostate cancer. Cancer Res. 2011, 71, 1978–1988. [Google Scholar] [CrossRef] [Green Version]
- O’hurley, G.; Prencipe, M.; Lundon, D.; O’Neill, A.; Boyce, S.; O’Grady, A.; Gallagher, W.M.; Morrissey, C.; Kay, E.W.; Watson, W.R. The analysis of serum response factor expression in bone and soft tissue prostate cancer metastases. Prostate 2014, 74, 306–313. [Google Scholar] [CrossRef]
- Prencipe, M.; Fabre, A.; Murphy, T.B.; Vargyas, E.; O’Neill, A.; Bjartell, A.; Taskén, K.A.; Grytli, H.H.; Svindland, A.; Berge, V.; et al. Role of serum response factor expression in prostate cancer biochemical recurrence. Prostate 2018, 78, 724–730. [Google Scholar] [CrossRef]
- Lundon, D.J.; Boland, A.; Prencipe, M.; Hurley, G.; O’Neill, A.; Kay, E.; Aherne, S.T.; Doolan, P.; Madden, S.F.; Clynes, M.; et al. The prognostic utility of the transcription factor SRF in docetaxel-resistant prostate cancer: In-vitro discovery and in-vivo validation. BMC Cancer 2017, 17, 163. [Google Scholar] [CrossRef] [Green Version]
- Schalken, J.A.; Fitzpatrick, J.M. Enzalutamide: Targeting the androgen signalling pathway in metastatic castration-resistant prostate cancer. BJU Int. 2015, 117, 215–225. [Google Scholar] [CrossRef]
- Hammond, J.W.; Cai, D.; Verhey, K.J. Tubulin modifications and their cellular functions. Curr. Opin. Cell Biol. 2008, 20, 71–76. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Barrera, J.; Bernabé-Rubio, M.; Casares-Arias, J.; Rangel, L.; Fernández-Martín, L.; Correas, I.; Alonso, M.A. The actin-MRTF-SRF transcriptional circuit controls tubulin acetylation via α-TAT1 gene expression. J. Cell Biol. 2018, 217, 929–944. [Google Scholar] [CrossRef] [Green Version]
- Evelyn, C.R.; Wade, S.M.; Wang, Q.; Wu, M.; Merajver, S.D.; Neubig, R.R.; Iñiguez-Lluhí, J.A. CCG-1423: A small-molecule inhibitor of RhoA transcriptional signaling. Mol. Cancer Ther. 2007, 6, 2249–2260. [Google Scholar] [CrossRef] [Green Version]
- Jin, W.; Goldfine, A.B.; Boes, T.; Henry, R.R.; Ciaraldi, T.P.; Kim, E.-Y.; Emecan, M.; Fitzpatrick, C.; Sen, A.; Shah, A.; et al. Increased SRF transcriptional activity in human and mouse skeletal muscle is a signature of insulin resistance. J. Clin. Investig. 2011, 121, 918–929. [Google Scholar] [CrossRef] [Green Version]
- Souček, K.; Kamaid, A.; Phung, A.D.; Kubala, L.; Bulinski, J.C.; Harper, R.; Eiserich, J.P. Normal and prostate cancer cells display distinct molecular profiles of α-tubulin posttranslational modifications. Prostate 2006, 66, 954–965. [Google Scholar] [CrossRef]
- Boggs, A.E.; Vitolo, M.I.; Whipple, R.A.; Charpentier, M.S.; Goloubeva, O.G.; Ioffe, O.B.; Tuttle, K.C.; Slovic, J.; Lu, Y.; Mills, G.B.; et al. α-Tubulin acetylation elevated in metastatic and basal-like breast cancer cells promotes microtentacle formation, adhesion, and invasive migration. Cancer Res. 2015, 75, 203–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dompierre, J.P.; Godin, J.D.; Charrin, B.C.; Cordelières, F.P.; King, S.J.; Humbert, S.; Saudou, F. Histone Deacetylase 6 Inhibition Compensates for the Transport Deficit in Huntington’s Disease by Increasing Tubulin Acetylation. J. Neurosci. 2007, 27, 3571–3583. [Google Scholar] [CrossRef] [PubMed]
- Thadani-Mulero, M.; Nanus, D.M.; Giannakakou, P. Androgen Receptor on the Move: Boarding the Microtubule Expressway to the Nucleus. Cancer Res. 2012, 72, 4611–4615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, M.-L.; Horbinski, C.M.; Garzotto, M.; Qian, D.Z.; Beer, T.M.; Kyprianou, N. Tubulin-Targeting Chemotherapy Impairs Androgen Receptor Activity in Prostate Cancer. Cancer Res. 2010, 70, 7992–8002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calhoun, B.C.; Collins, L.C. Predictive markers in breast cancer: An update on ER and HER2 testing and reporting. Semin. Diagn. Pathol. 2015, 32, 362–369. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, A.J.; Prencipe, M.; Dowling, C.; Fan, Y.; Mulrane, L.; Gallagher, W.M.; O’Connor, D.; O’Connor, R.; Devery, A.; Corcoran, C.; et al. Characterisation and manipulation of docetaxel resistant prostate cancer cell lines. Mol. Cancer 2011, 10, 126. [Google Scholar] [CrossRef] [Green Version]
- Lam, H.-M.; McMullin, R.; Nguyen, H.M.; Coleman, I.; Gormley, M.; Gulati, R.; Brown, L.G.; Holt, S.K.; Li, W.; Ricci, D.S.; et al. Characterization of an Abiraterone Ultraresponsive Phenotype in Castration-Resistant Prostate Cancer Patient-Derived Xenografts. Clin. Cancer Res. 2017, 23, 2301–2312. [Google Scholar] [CrossRef] [Green Version]
- Hirsch, F.R.; Varella-Garcia, M.; Bunn, P.A.; Di Maria, M.V.; Veve, R.; Bremnes, R.M.; Barón, A.E.; Zeng, C.; A Franklin, W. Epidermal Growth Factor Receptor in Non–Small-Cell Lung Carcinomas: Correlation Between Gene Copy Number and Protein Expression and Impact on Prognosis. J. Clin. Oncol. 2003, 21, 3798–3807. [Google Scholar] [CrossRef]
- Roudier, M.P.; True, L.D.; Higano, C.S.; Vesselle, H.; Ellis, W.; Lange, P.; Vessella, R.L. Phenotypic heterogeneity of end-stage prostate carcinoma metastatic to bone. Hum. Pathol. 2003, 34, 646–653. [Google Scholar] [CrossRef]
- Zhang, X.; Morrissey, C.; Sun, S.; Ketchandji, M.; Nelson, P.S.; True, L.D.; Vakar-Lopez, F.; Vessella, R.L.; Plymate, S.R. Androgen Receptor Variants Occur Frequently in Castration Resistant Prostate Cancer Metastases. PLoS ONE 2011, 6, e27970. [Google Scholar] [CrossRef] [Green Version]
- Therneau, T.M.; Lumley, T. Package ‘Survival’. Available online: https://cran.r-project.org/web/packages/survival/survival.pdf (accessed on 25 October 2020).
- Available online: https://www.r-project.org/ (accessed on 25 October 2020).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watson, R.W.; Azam, H.; Aura, C.; Russell, N.; McCormack, J.; Corey, E.; Morrissey, C.; Crown, J.; Gallagher, W.M.; Prencipe, M. Inhibition of Serum Response Factor Improves Response to Enzalutamide in Prostate Cancer. Cancers 2020, 12, 3540. https://doi.org/10.3390/cancers12123540
Watson RW, Azam H, Aura C, Russell N, McCormack J, Corey E, Morrissey C, Crown J, Gallagher WM, Prencipe M. Inhibition of Serum Response Factor Improves Response to Enzalutamide in Prostate Cancer. Cancers. 2020; 12(12):3540. https://doi.org/10.3390/cancers12123540
Chicago/Turabian StyleWatson, R. William, Haleema Azam, Claudia Aura, Niamh Russell, Janet McCormack, Eva Corey, Colm Morrissey, John Crown, William M Gallagher, and Maria Prencipe. 2020. "Inhibition of Serum Response Factor Improves Response to Enzalutamide in Prostate Cancer" Cancers 12, no. 12: 3540. https://doi.org/10.3390/cancers12123540
APA StyleWatson, R. W., Azam, H., Aura, C., Russell, N., McCormack, J., Corey, E., Morrissey, C., Crown, J., Gallagher, W. M., & Prencipe, M. (2020). Inhibition of Serum Response Factor Improves Response to Enzalutamide in Prostate Cancer. Cancers, 12(12), 3540. https://doi.org/10.3390/cancers12123540





