Transcriptional Control of Regulatory T Cells in Cancer: Toward Therapeutic Targeting?
Simple Summary
Abstract
1. Introduction
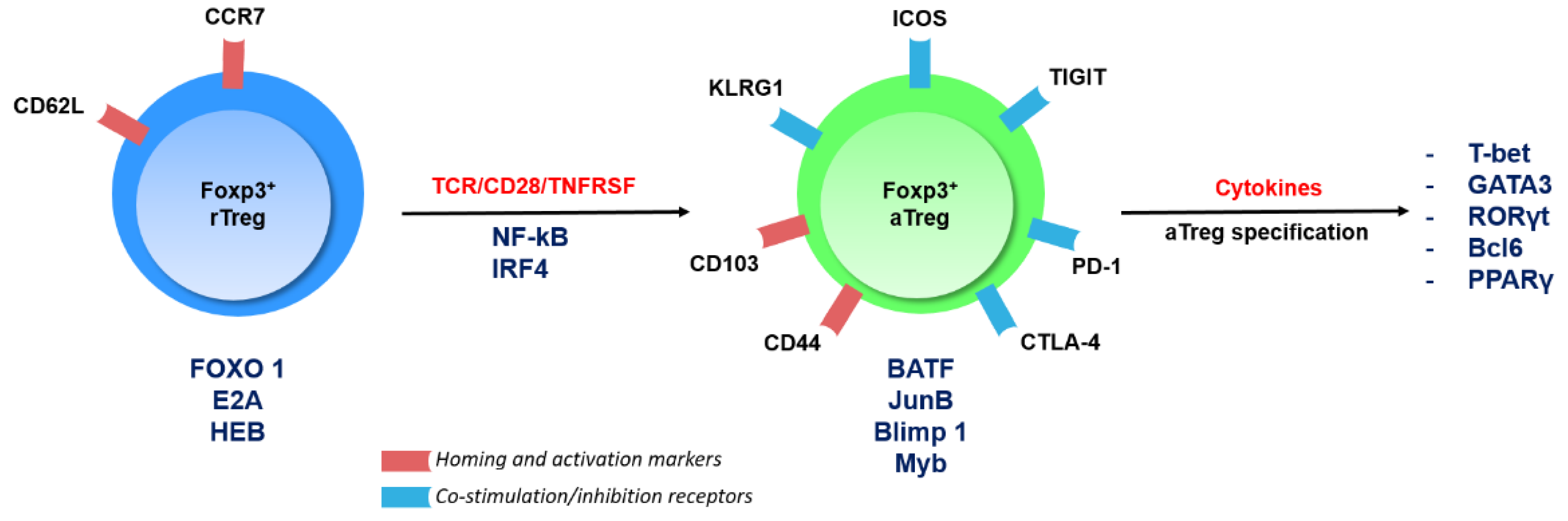
2. An Overview of Treg Cell Subsets and Their Transcriptional Regulation
3. Signaling Pathways and Transcription Factors in Treg Cells in Cancer
3.1. Foxp3
3.2. IRF4
3.3. The Complex Functions of the PI3K/AKT/Foxo Axis
3.4. Helios
3.5. Eos
3.6. The Nr4a Family
3.7. Bach2
3.8. E-Proteins and Their Inhibitor of DNA Binding (Id) Counterparts
3.9. STAT3
3.10. The NF-κB Pathway
3.10.1. The Many Roles of NF-κB in Treg Cells
3.10.2. NF-κB Is a Nexus Regulator of the Treg Cell Transcriptional Program in Cancer
4. Epigenetic Programing of Treg Cells in Cancer
5. Shared Mechanisms of Treg Cell Reprogramming in Cancer
6. Challenges on the Path toward Cancer Therapy
6.1. Missing Pieces of the Puzzle
6.2. Avoiding Autoimmunity
6.3. The Issue of Multiple Cell Type Targeting with a Single Agent
6.4. Mouse and Human Treg Cells: Commonalities and Divergences
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sakaguchi, S.; Yamaguchi, T.; Nomura, T.; Ono, M. Regulatory T cells and immune tolerance. Cell 2008, 133, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, V.L.; Wilkinson, J.E.; Russell, L.B. X-linked lymphoreticular disease in the scurfy (sf) mutant mouse. Am. J. Pathol. 1991, 138, 1379–1387. [Google Scholar] [PubMed]
- Bennett, C.L.; Christie, J.; Ramsdell, F.; Brunkow, M.E.; Ferguson, P.J.; Whitesell, L.; Kelly, T.E.; Saulsbury, F.T.; Chance, P.F.; Ochs, H.D. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 2001, 27, 20–21. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Rasmussen, J.P.; Rudensky, A.Y. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 2007, 8, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Lahl, K.; Loddenkemper, C.; Drouin, C.; Freyer, J.; Arnason, J.; Eberl, G.; Hamann, A.; Wagner, H.; Huehn, J.; Sparwasser, T. Selective depletion of Foxp3+ regulatory T cells induces a scurfy-like disease. J. Exp. Med. 2007, 204, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Vignali, D.A.; Collison, L.W.; Workman, C.J. How regulatory T cells work. Nat. Rev. Immunol. 2008, 8, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Onizuka, S.; Tawara, I.; Shimizu, J.; Sakaguchi, S.; Fujita, T.; Nakayama, E. Tumor rejection by in vivo administration of anti-CD25 (interleukin-2 receptor alpha) monoclonal antibody. Cancer Res. 1999, 59, 3128–3133. [Google Scholar]
- Nishikawa, H.; Sakaguchi, S. Regulatory T cells in tumor immunity. Int. J. Cancer 2010, 127, 759–767. [Google Scholar] [CrossRef]
- Whiteside, T.L. FOXP3+ Treg as a therapeutic target for promoting anti-tumor immunity. Expert Opin. Ther. Targets 2018, 22, 353–363. [Google Scholar] [CrossRef]
- Simpson, T.R.; Li, F.; Montalvo-Ortiz, W.; Sepulveda, M.A.; Bergerhoff, K.; Arce, F.; Roddie, C.; Henry, J.Y.; Yagita, H.; Wolchok, J.D.; et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J. Exp. Med. 2013, 210, 1695–1710. [Google Scholar] [CrossRef]
- Arce Vargas, F.; Furness, A.J.S.; Litchfield, K.; Joshi, K.; Rosenthal, R.; Ghorani, E.; Solomon, I.; Lesko, M.H.; Ruef, N.; Roddie, C.; et al. Fc Effector Function Contributes to the Activity of Human Anti-CTLA-4 Antibodies. Cancer Cell 2018, 33, 649–663. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Subudhi, S.K.; Blando, J.; Vence, L.; Wargo, J.; Allison, J.P.; Ribas, A.; Sharma, P. Anti-CTLA-4 Immunotherapy Does Not Deplete FOXP3(+) Regulatory T Cells (Tregs) in Human Cancers-Response. Clin. Cancer Res. 2019, 25, 3469–3470. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Wang, D.; Zhang, G.; Guo, X. The Role Of PD-1/PD-L1 Axis in Treg Development and Function: Implications for Cancer Immunotherapy. Onco. Targets Ther. 2019, 12, 8437–8445. [Google Scholar] [CrossRef]
- Kamada, T.; Togashi, Y.; Tay, C.; Ha, D.; Sasaki, A.; Nakamura, Y.; Sato, E.; Fukuoka, S.; Tada, Y.; Tanaka, A.; et al. PD-1(+) regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 9999–10008. [Google Scholar] [CrossRef]
- Lee, W.; Lee, G.R. Transcriptional regulation and development of regulatory T cells. Exp. Mol. Med. 2018, 50, e456. [Google Scholar] [CrossRef]
- Savage, P.A.; Klawon, D.E.J.; Miller, C.H. Regulatory T Cell Development. Annu. Rev. Immunol. 2020, 38, 421–453. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, S.I.; Ishikawa, H. Transcriptional Regulation of Differentiation and Functions of Effector T Regulatory Cells. Cells 2019, 8, 939. [Google Scholar] [CrossRef]
- Plitas, G.; Konopacki, C.; Wu, K.; Bos, P.D.; Morrow, M.; Putintseva, E.V.; Chudakov, D.M.; Rudensky, A.Y. Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer. Immunity 2016, 45, 1122–1134. [Google Scholar] [CrossRef] [PubMed]
- De Simone, M.; Arrigoni, A.; Rossetti, G.; Gruarin, P.; Ranzani, V.; Politano, C.; Bonnal, R.J.P.; Provasi, E.; Sarnicola, M.L.; Panzeri, I.; et al. Transcriptional Landscape of Human Tissue Lymphocytes Unveils Uniqueness of Tumor-Infiltrating T Regulatory Cells. Immunity 2016, 45, 1135–1147. [Google Scholar] [CrossRef]
- Rudensky, A.Y. Regulatory T cells and Foxp3. Immunol. Rev. 2011, 241, 260–268. [Google Scholar] [CrossRef]
- Samstein, R.M.; Arvey, A.; Josefowicz, S.Z.; Peng, X.; Reynolds, A.; Sandstrom, R.; Neph, S.; Sabo, P.; Kim, J.M.; Liao, W.; et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell 2012, 151, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Rudra, D.; deRoos, P.; Chaudhry, A.; Niec, R.E.; Arvey, A.; Samstein, R.M.; Leslie, C.; Shaffer, S.A.; Goodlett, D.R.; Rudensky, A.Y. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat. Immunol. 2012, 13, 1010–1019. [Google Scholar] [CrossRef]
- Kwon, H.K.; Chen, H.M.; Mathis, D.; Benoist, C. Different molecular complexes that mediate transcriptional induction and repression by FoxP3. Nat. Immunol. 2017, 18, 1238–1248. [Google Scholar] [CrossRef]
- Cretney, E.; Xin, A.; Shi, W.; Minnich, M.; Masson, F.; Miasari, M.; Belz, G.T.; Smyth, G.K.; Busslinger, M.; Nutt, S.L.; et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat. Immunol. 2011, 12, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Dias, S.; D’Amico, A.; Cretney, E.; Liao, Y.; Tellier, J.; Bruggeman, C.; Almeida, F.F.; Leahy, J.; Belz, G.T.; Smyth, G.K.; et al. Effector Regulatory T Cell Differentiation and Immune Homeostasis Depend on the Transcription Factor Myb. Immunity 2017, 46, 78–91. [Google Scholar] [CrossRef]
- Grinberg-Bleyer, Y.; Oh, H.; Desrichard, A.; Bhatt, D.M.; Caron, R.; Chan, T.A.; Schmid, R.M.; Klein, U.; Hayden, M.S.; Ghosh, S. NF-kappaB c-Rel Is Crucial for the Regulatory T Cell Immune Checkpoint in Cancer. Cell 2017, 170, 1096–1108.e13. [Google Scholar] [CrossRef]
- Vasanthakumar, A.; Liao, Y.; Teh, P.; Pascutti, M.F.; Oja, A.E.; Garnham, A.L.; Gloury, R.; Tempany, J.C.; Sidwell, T.; Cuadrado, E.; et al. The TNF Receptor Superfamily-NF-kappaB Axis Is Critical to Maintain Effector Regulatory T Cells in Lymphoid and Non-lymphoid Tissues. Cell Rep. 2017, 20, 2906–2920. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, S.I.; Sasaki, D.; Hsieh, T.H.; Taira, N.; Arakaki, N.; Yamasaki, S.; Wang, K.; Sarkar, S.; Shirahata, H.; Miyagi, M.; et al. JunB regulates homeostasis and suppressive functions of effector regulatory T cells. Nat. Commun. 2018, 9, 5344. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Huang, H.; Gao, P.; Zhang, Q.; Liu, X.; Jia, B.; Strober, W.; Hou, B.; Zhou, X.; Gao, G.F.; et al. E-protein regulatory network links TCR signaling to effector Treg cell differentiation. Proc. Natl. Acad. Sci. USA 2019, 116, 4471–4480. [Google Scholar] [CrossRef]
- Luo, C.T.; Liao, W.; Dadi, S.; Toure, A.; Li, M.O. Graded Foxo1 activity in Treg cells differentiates tumour immunity from spontaneous autoimmunity. Nature 2016, 529, 532–536. [Google Scholar] [CrossRef]
- Wang, K.; Fu, W. Transcriptional regulation of Treg homeostasis and functional specification. Cell. Mol. Life Sci. 2020, 77, 4269–4287. [Google Scholar] [CrossRef]
- Casares, N.; Rudilla, F.; Arribillaga, L.; Llopiz, D.; Riezu-Boj, J.I.; Lozano, T.; Lopez-Sagaseta, J.; Guembe, L.; Sarobe, P.; Prieto, J.; et al. A peptide inhibitor of FOXP3 impairs regulatory T cell activity and improves vaccine efficacy in mice. J. Immunol. 2010, 185, 5150–5159. [Google Scholar] [CrossRef]
- Lozano, T.; Villanueva, L.; Durantez, M.; Gorraiz, M.; Ruiz, M.; Belsue, V.; Riezu-Boj, J.I.; Hervas-Stubbs, S.; Oyarzabal, J.; Bandukwala, H.; et al. Inhibition of FOXP3/NFAT Interaction Enhances T Cell Function after TCR Stimulation. J. Immunol. 2015, 195, 3180–3189. [Google Scholar] [CrossRef]
- Lozano, T.; Gorraiz, M.; Lasarte-Cia, A.; Ruiz, M.; Rabal, O.; Oyarzabal, J.; Hervas-Stubbs, S.; Llopiz, D.; Sarobe, P.; Prieto, J.; et al. Blockage of FOXP3 transcription factor dimerization and FOXP3/AML1 interaction inhibits T regulatory cell activity: Sequence optimization of a peptide inhibitor. Oncotarget 2017, 8, 71709–71724. [Google Scholar] [CrossRef]
- Wang, J.; Ioan-Facsinay, A.; van der Voort, E.I.; Huizinga, T.W.; Toes, R.E. Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur. J. Immunol. 2007, 37, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Miyara, M.; Yoshioka, Y.; Kitoh, A.; Shima, T.; Wing, K.; Niwa, A.; Parizot, C.; Taflin, C.; Heike, T.; Valeyre, D.; et al. Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity 2009, 30, 899–911. [Google Scholar] [CrossRef] [PubMed]
- Huber, M.; Lohoff, M. IRF4 at the crossroads of effector T-cell fate decision. Eur. J. Immunol. 2014, 44, 1886–1895. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Chaudhry, A.; Kas, A.; deRoos, P.; Kim, J.M.; Chu, T.T.; Corcoran, L.; Treuting, P.; Klein, U.; Rudensky, A.Y. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature 2009, 458, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Alvisi, G.; Brummelman, J.; Puccio, S.; Mazza, E.M.; Tomada, E.P.; Losurdo, A.; Zanon, V.; Peano, C.; Colombo, F.S.; Scarpa, A.; et al. IRF4 instructs effector Treg differentiation and immune suppression in human cancer. J. Clin. Investig. 2020, 130, 3137–3150. [Google Scholar] [CrossRef]
- Man, K.; Gabriel, S.S.; Liao, Y.; Gloury, R.; Preston, S.; Henstridge, D.C.; Pellegrini, M.; Zehn, D.; Berberich-Siebelt, F.; Febbraio, M.A.; et al. Transcription Factor IRF4 Promotes CD8(+) T Cell Exhaustion and Limits the Development of Memory-like T Cells during Chronic Infection. Immunity 2017, 47, 1129–1141.e5. [Google Scholar] [CrossRef]
- Sharma, M.D.; Shinde, R.; McGaha, T.L.; Huang, L.; Holmgaard, R.B.; Wolchok, J.D.; Mautino, M.R.; Celis, E.; Sharpe, A.H.; Francisco, L.M.; et al. The PTEN pathway in Tregs is a critical driver of the suppressive tumor microenvironment. Sci. Adv. 2015, 1, e1500845. [Google Scholar] [CrossRef] [PubMed]
- Delgoffe, G.M.; Woo, S.R.; Turnis, M.E.; Gravano, D.M.; Guy, C.; Overacre, A.E.; Bettini, M.L.; Vogel, P.; Finkelstein, D.; Bonnevier, J.; et al. Stability and function of regulatory T cells is maintained by a neuropilin-1-semaphorin-4a axis. Nature 2013, 501, 252–256. [Google Scholar] [CrossRef]
- Overacre-Delgoffe, A.E.; Chikina, M.; Dadey, R.E.; Yano, H.; Brunazzi, E.A.; Shayan, G.; Horne, W.; Moskovitz, J.M.; Kolls, J.K.; Sander, C.; et al. Interferon-gamma Drives Treg Fragility to Promote Anti-tumor Immunity. Cell 2017, 169, 1130–1141.e11. [Google Scholar] [CrossRef] [PubMed]
- Ali, K.; Soond, D.R.; Pineiro, R.; Hagemann, T.; Pearce, W.; Lim, E.L.; Bouabe, H.; Scudamore, C.L.; Hancox, T.; Maecker, H.; et al. Inactivation of PI(3)K p110delta breaks regulatory T-cell-mediated immune tolerance to cancer. Nature 2014, 510, 407–411. [Google Scholar] [CrossRef]
- Lim, E.L.; Cugliandolo, F.M.; Rosner, D.R.; Gyori, D.; Roychoudhuri, R.; Okkenhaug, K. Phosphoinositide 3-kinase delta inhibition promotes antitumor responses but antagonizes checkpoint inhibitors. JCI Insight 2018, 3, e120626. [Google Scholar] [CrossRef] [PubMed]
- Abu-Eid, R.; Samara, R.N.; Ozbun, L.; Abdalla, M.Y.; Berzofsky, J.A.; Friedman, K.M.; Mkrtichyan, M.; Khleif, S.N. Selective inhibition of regulatory T cells by targeting the PI3K-Akt pathway. Cancer Immunol. Res. 2014, 2, 1080–1089. [Google Scholar] [CrossRef]
- Chellappa, S.; Kushekhar, K.; Munthe, L.A.; Tjonnfjord, G.E.; Aandahl, E.M.; Okkenhaug, K.; Tasken, K. The PI3K p110delta Isoform Inhibitor Idelalisib Preferentially Inhibits Human Regulatory T Cell Function. J. Immunol. 2019, 202, 1397–1405. [Google Scholar] [CrossRef]
- Ouyang, W.; Liao, W.; Luo, C.T.; Yin, N.; Huse, M.; Kim, M.V.; Peng, M.; Chan, P.; Ma, Q.; Mo, Y.; et al. Novel Foxo1-dependent transcriptional programs control T(reg) cell function. Nature 2012, 491, 554–559. [Google Scholar] [CrossRef]
- Kim, H.J.; Barnitz, R.A.; Kreslavsky, T.; Brown, F.D.; Moffett, H.; Lemieux, M.E.; Kaygusuz, Y.; Meissner, T.; Holderried, T.A.; Chan, S.; et al. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science 2015, 350, 334–339. [Google Scholar] [CrossRef]
- Thornton, A.M.; Korty, P.E.; Tran, D.Q.; Wohlfert, E.A.; Murray, P.E.; Belkaid, Y.; Shevach, E.M. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 2010, 184, 3433–3441. [Google Scholar] [CrossRef]
- Gottschalk, R.A.; Corse, E.; Allison, J.P. Expression of Helios in peripherally induced Foxp3+ regulatory T cells. J. Immunol. 2012, 188, 976–980. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, M.; Lopez-Ocasio, M.; Metidji, A.; Rieder, S.A.; Shevach, E.M.; Thornton, A.M. Helios Controls a Limited Subset of Regulatory T Cell Functions. J. Immunol. 2016, 196, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, H.; Sido, J.M.; Reyes, E.E.; Kiers, V.; Cantor, H.; Kim, H.J. Instability of Helios-deficient Tregs is associated with conversion to a T-effector phenotype and enhanced antitumor immunity. Proc. Natl. Acad. Sci. USA 2016, 113, 6248–6253. [Google Scholar] [CrossRef]
- Yates, K.; Bi, K.; Haining, W.N.; Cantor, H.; Kim, H.J. Comparative transcriptome analysis reveals distinct genetic modules associated with Helios expression in intratumoral regulatory T cells. Proc. Natl. Acad. Sci. USA 2018, 115, 2162–2167. [Google Scholar] [CrossRef]
- Li, X.; Li, D.; Huang, X.; Zhou, P.; Shi, Q.; Zhang, B.; Ju, X. Helios expression in regulatory T cells promotes immunosuppression, angiogenesis and the growth of leukemia cells in pediatric acute lymphoblastic leukemia. Leuk. Res. 2018, 67, 60–66. [Google Scholar] [CrossRef]
- Cai, Q.; Dierich, A.; Oulad-Abdelghani, M.; Chan, S.; Kastner, P. Helios deficiency has minimal impact on T cell development and function. J. Immunol. 2009, 183, 2303–2311. [Google Scholar] [CrossRef]
- Pan, F.; Yu, H.; Dang, E.V.; Barbi, J.; Pan, X.; Grosso, J.F.; Jinasena, D.; Sharma, S.M.; McCadden, E.M.; Getnet, D.; et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science 2009, 325, 1142–1146. [Google Scholar] [CrossRef]
- Rieder, S.A.; Metidji, A.; Glass, D.D.; Thornton, A.M.; Ikeda, T.; Morgan, B.A.; Shevach, E.M. Eos Is Redundant for Regulatory T Cell Function but Plays an Important Role in IL-2 and Th17 Production by CD4+ Conventional T Cells. J. Immunol. 2015, 195, 553–563. [Google Scholar] [CrossRef]
- Gokhale, A.S.; Gangaplara, A.; Lopez-Occasio, M.; Thornton, A.M.; Shevach, E.M. Selective deletion of Eos (Ikzf4) in T-regulatory cells leads to loss of suppressive function and development of systemic autoimmunity. J. Autoimmun. 2019, 105, 102300. [Google Scholar] [CrossRef] [PubMed]
- Moran, A.E.; Holzapfel, K.L.; Xing, Y.; Cunningham, N.R.; Maltzman, J.S.; Punt, J.; Hogquist, K.A. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 2011, 208, 1279–1289. [Google Scholar] [CrossRef]
- Sekiya, T.; Kashiwagi, I.; Inoue, N.; Morita, R.; Hori, S.; Waldmann, H.; Rudensky, A.Y.; Ichinose, H.; Metzger, D.; Chambon, P.; et al. The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat. Commun. 2011, 2, 269. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, T.; Kashiwagi, I.; Yoshida, R.; Fukaya, T.; Morita, R.; Kimura, A.; Ichinose, H.; Metzger, D.; Chambon, P.; Yoshimura, A. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat. Immunol. 2013, 14, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, T.; Kondo, T.; Shichita, T.; Morita, R.; Ichinose, H.; Yoshimura, A. Suppression of Th2 and Tfh immune reactions by Nr4a receptors in mature T reg cells. J. Exp. Med. 2015, 212, 1623–1640. [Google Scholar] [CrossRef]
- Hibino, S.; Chikuma, S.; Kondo, T.; Ito, M.; Nakatsukasa, H.; Omata-Mise, S.; Yoshimura, A. Inhibition of Nr4a Receptors Enhances Antitumor Immunity by Breaking Treg-Mediated Immune Tolerance. Cancer Res. 2018, 78, 3027–3040. [Google Scholar] [CrossRef]
- Venditto, V.J.; Simanek, E.E. Cancer therapies utilizing the camptothecins: A review of the in vivo literature. Mol. Pharm. 2010, 7, 307–349. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lopez-Moyado, I.F.; Seo, H.; Lio, C.J.; Hempleman, L.J.; Sekiya, T.; Yoshimura, A.; Scott-Browne, J.P.; Rao, A. NR4A transcription factors limit CAR T cell function in solid tumours. Nature 2019, 567, 530–534. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Y.; Lu, H.; Li, J.; Yan, X.; Xiao, M.; Hao, J.; Alekseev, A.; Khong, H.; Chen, T.; et al. Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature 2019, 567, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Roychoudhuri, R.; Clever, D.; Li, P.; Wakabayashi, Y.; Quinn, K.M.; Klebanoff, C.A.; Ji, Y.; Sukumar, M.; Eil, R.L.; Yu, Z.; et al. BACH2 regulates CD8(+) T cell differentiation by controlling access of AP-1 factors to enhancers. Nat. Immunol. 2016, 17, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Jang, E.; Lee, H.R.; Lee, G.H.; Oh, A.R.; Cha, J.Y.; Igarashi, K.; Youn, J. Bach2 represses the AP-1-driven induction of interleukin-2 gene transcription in CD4 T cells. BMB Rep. 2017, 50, 472–477. [Google Scholar] [CrossRef]
- Roychoudhuri, R.; Hirahara, K.; Mousavi, K.; Clever, D.; Klebanoff, C.A.; Bonelli, M.; Sciume, G.; Zare, H.; Vahedi, G.; Dema, B.; et al. BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature 2013, 498, 506–510. [Google Scholar] [CrossRef]
- Kim, E.H.; Gasper, D.J.; Lee, S.H.; Plisch, E.H.; Svaren, J.; Suresh, M. Bach2 regulates homeostasis of Foxp3+ regulatory T cells and protects against fatal lung disease in mice. J. Immunol. 2014, 192, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Sidwell, T.; Liao, Y.; Garnham, A.L.; Vasanthakumar, A.; Gloury, R.; Blume, J.; Teh, P.P.; Chisanga, D.; Thelemann, C.; de Labastida Rivera, F.; et al. Attenuation of TCR-induced transcription by Bach2 controls regulatory T cell differentiation and homeostasis. Nat. Commun. 2020, 11, 252. [Google Scholar] [CrossRef]
- Yu, X.; Lao, Y.; Teng, X.L.; Li, S.; Zhou, Y.; Wang, F.; Guo, X.; Deng, S.; Chang, Y.; Wu, X.; et al. SENP3 maintains the stability and function of regulatory T cells via BACH2 deSUMOylation. Nat. Commun. 2018, 9, 3157. [Google Scholar] [CrossRef] [PubMed]
- Roychoudhuri, R.; Eil, R.L.; Clever, D.; Klebanoff, C.A.; Sukumar, M.; Grant, F.M.; Yu, Z.; Mehta, G.; Liu, H.; Jin, P.; et al. The transcription factor BACH2 promotes tumor immunosuppression. J. Clin. Investig. 2016, 126, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Grant, F.M.; Yang, J.; Nasrallah, R.; Clarke, J.; Sadiyah, F.; Whiteside, S.K.; Imianowski, C.J.; Kuo, P.; Vardaka, P.; Todorov, T.; et al. BACH2 drives quiescence and maintenance of resting Treg cells to promote homeostasis and cancer immunosuppression. J. Exp. Med. 2020, 217, e20190711. [Google Scholar] [CrossRef]
- Miyazaki, M.; Miyazaki, K.; Chen, S.; Itoi, M.; Miller, M.; Lu, L.F.; Varki, N.; Chang, A.N.; Broide, D.H.; Murre, C. Id2 and Id3 maintain the regulatory T cell pool to suppress inflammatory disease. Nat. Immunol. 2014, 15, 767–776. [Google Scholar] [CrossRef]
- Hwang, S.M.; Sharma, G.; Verma, R.; Byun, S.; Rudra, D.; Im, S.H. Inflammation-induced Id2 promotes plasticity in regulatory T cells. Nat. Commun. 2018, 9, 4736. [Google Scholar] [CrossRef]
- Zorn, E.; Nelson, E.A.; Mohseni, M.; Porcheray, F.; Kim, H.; Litsa, D.; Bellucci, R.; Raderschall, E.; Canning, C.; Soiffer, R.J.; et al. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood 2006, 108, 1571–1579. [Google Scholar] [CrossRef]
- Kortylewski, M.; Xin, H.; Kujawski, M.; Lee, H.; Liu, Y.; Harris, T.; Drake, C.; Pardoll, D.; Yu, H. Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell 2009, 15, 114–123. [Google Scholar] [CrossRef]
- Chaudhry, A.; Rudra, D.; Treuting, P.; Samstein, R.M.; Liang, Y.; Kas, A.; Rudensky, A.Y. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science 2009, 326, 986–991. [Google Scholar] [CrossRef]
- Oweida, A.J.; Darragh, L.; Phan, A.; Binder, D.; Bhatia, S.; Mueller, A.; Court, B.V.; Milner, D.; Raben, D.; Woessner, R.; et al. STAT3 Modulation of Regulatory T Cells in Response to Radiation Therapy in Head and Neck Cancer. J. Natl. Cancer Inst. 2019, 111, 1339–1349. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Zhu, Y.; Mu, R.; Xu, J.; Zhang, X.; Wang, C.; Li, Q.; Huang, Z.; Zhang, J.; Pan, Y.; et al. A Jak2-selective inhibitor potently reverses the immune suppression by modulating the tumor microenvironment for cancer immunotherapy. Biochem. Pharmacol. 2017, 145, 132–146. [Google Scholar] [CrossRef] [PubMed]
- Rebe, C.; Ghiringhelli, F. STAT3, a Master Regulator of Anti-Tumor Immune Response. Cancers (Basel) 2019, 11, 1280. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. Shared principles in NF-kappaB signaling. Cell 2008, 132, 344–362. [Google Scholar] [CrossRef] [PubMed]
- Grinberg-Bleyer, Y.; Caron, R.; Seeley, J.J.; De Silva, N.S.; Schindler, C.W.; Hayden, M.S.; Klein, U.; Ghosh, S. The Alternative NF-kappaB Pathway in Regulatory T Cell Homeostasis and Suppressive Function. J. Immunol. 2018, 200, 2362–2371. [Google Scholar] [CrossRef]
- Di Pilato, M.; Kim, E.Y.; Cadilha, B.L.; Prussmann, J.N.; Nasrallah, M.N.; Seruggia, D.; Usmani, S.M.; Misale, S.; Zappulli, V.; Carrizosa, E.; et al. Targeting the CBM complex causes Treg cells to prime tumours for immune checkpoint therapy. Nature 2019, 570, 112–116. [Google Scholar] [CrossRef]
- Heuser, C.; Gotot, J.; Piotrowski, E.C.; Philipp, M.S.; Courreges, C.J.F.; Otte, M.S.; Guo, L.; Schmid-Burgk, J.L.; Hornung, V.; Heine, A.; et al. Prolonged IKKbeta Inhibition Improves Ongoing CTL Antitumor Responses by Incapacitating Regulatory T Cells. Cell Rep. 2017, 21, 578–586. [Google Scholar] [CrossRef]
- Cheng, L.; Deng, N.; Yang, N.; Zhao, X.; Lin, X. Malt1 Protease Is Critical in Maintaining Function of Regulatory T Cells and May Be a Therapeutic Target for Antitumor Immunity. J. Immunol. 2019, 202, 3008–3019. [Google Scholar] [CrossRef]
- Rosenbaum, M.; Gewies, A.; Pechloff, K.; Heuser, C.; Engleitner, T.; Gehring, T.; Hartjes, L.; Krebs, S.; Krappmann, D.; Kriegsmann, M.; et al. Bcl10-controlled Malt1 paracaspase activity is key for the immune suppressive function of regulatory T cells. Nat. Commun. 2019, 10, 2352. [Google Scholar] [CrossRef]
- Oh, H.; Grinberg-Bleyer, Y.; Liao, W.; Maloney, D.; Wang, P.; Wu, Z.; Wang, J.; Bhatt, D.M.; Heise, N.; Schmid, R.M.; et al. An NF-kappaB Transcription-Factor-Dependent Lineage-Specific Transcriptional Program Promotes Regulatory T Cell Identity and Function. Immunity 2017, 47, 450–465.e5. [Google Scholar] [CrossRef]
- Konczalla, L.; Perez, D.R.; Wenzel, N.; Wolters-Eisfeld, G.; Klemp, C.; Luddeke, J.; Wolski, A.; Landschulze, D.; Meier, C.; Buchholz, A.; et al. Biperiden and mepazine effectively inhibit MALT1 activity and tumor growth in pancreatic cancer. Int. J. Cancer 2020, 146, 1618–1630. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, J.; Jiang, B.; Guo, J. The roles of curcumin in regulating the tumor immunosuppressive microenvironment. Oncol. Lett. 2020, 19, 3059–3070. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Su, Z.; Han, S.; Huang, J.; Lin, L.; Shuai, X. Dual pH-sensitive nanodrug blocks PD-1 immune checkpoint and uses T cells to deliver NF-kappaB inhibitor for antitumor immunotherapy. Sci. Adv. 2020, 6, eaay7785. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Md Sakib Hossain, D.; Mohanty, S.; Sankar Sen, G.; Chattopadhyay, S.; Banerjee, S.; Chakraborty, J.; Das, K.; Sarkar, D.; Das, T.; et al. Curcumin reverses T cell-mediated adaptive immune dysfunctions in tumor-bearing hosts. Cell. Mol. Immunol. 2010, 7, 306–315. [Google Scholar] [CrossRef]
- Zou, J.Y.; Su, C.H.; Luo, H.H.; Lei, Y.Y.; Zeng, B.; Zhu, H.S.; Chen, Z.G. Curcumin converts Foxp3+ regulatory T cells to T helper 1 cells in patients with lung cancer. J. Cell. Biochem. 2018, 119, 1420–1428. [Google Scholar] [CrossRef]
- Xu, B.; Yu, L.; Zhao, L.Z. Curcumin up regulates T helper 1 cells in patients with colon cancer. Am. J. Transl. Res. 2017, 9, 1866–1875. [Google Scholar] [PubMed]
- Li, T.; Li, X.; Zamani, A.; Wang, W.; Lee, C.N.; Li, M.; Jin, J.; Ramachandran, M.; Qingguo, R.; Weiyun, S.; et al. c-Rel is a myeloid checkpoint for cancer immunotherapy. Nat. Cancer 2020, 1, 507–517. [Google Scholar] [CrossRef]
- Gao, F.; Cheng, Q.; Liu, M.D.; Rong, L.; Liu, C.J.; Zhang, X.Z. Local T regulatory cells depletion by an integrated nanodrug system for efficient chem-immunotherapy of tumor. Sci. China Chem. 2019, 62, 1230–1244. [Google Scholar] [CrossRef]
- Shono, Y.; Tuckett, A.Z.; Liou, H.C.; Doubrovina, E.; Derenzini, E.; Ouk, S.; Tsai, J.J.; Smith, O.M.; Levy, E.R.; Kreines, F.M.; et al. Characterization of a c-Rel Inhibitor That Mediates Anticancer Properties in Hematologic Malignancies by Blocking NF-kappaB-Controlled Oxidative Stress Responses. Cancer Res. 2016, 76, 377–389. [Google Scholar] [CrossRef]
- Neo, W.H.; Lim, J.F.; Grumont, R.; Gerondakis, S.; Su, I.H. c-Rel regulates Ezh2 expression in activated lymphocytes and malignant lymphoid cells. J. Biol. Chem. 2014, 289, 31693–31707. [Google Scholar] [CrossRef]
- Hunter, J.E.; Butterworth, J.A.; Zhao, B.; Sellier, H.; Campbell, K.J.; Thomas, H.D.; Bacon, C.M.; Cockell, S.J.; Gewurz, B.E.; Perkins, N.D. The NF-kappaB subunit c-Rel regulates Bach2 tumour suppressor expression in B-cell lymphoma. Oncogene 2016, 35, 3476–3484. [Google Scholar] [CrossRef] [PubMed]
- Grumont, R.J.; Gerondakis, S. Rel induces interferon regulatory factor 4 (IRF-4) expression in lymphocytes: Modulation of interferon-regulated gene expression by rel/nuclear factor kappaB. J. Exp. Med. 2000, 191, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Shen, S.; Verma, I.M. NF-kappaB, an active player in human cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, T.D.; Herscovitch, M. Inhibitors of NF-kappaB signaling: 785 and counting. Oncogene 2006, 25, 6887–6899. [Google Scholar] [CrossRef] [PubMed]
- Baud, V.; Karin, M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat. Rev. Drug Discov. 2009, 8, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Leporini, C.; Pisano, A.; Russo, E.; D’Arrigo, G.; de Sarro, G.; Coppolino, G.; Bolignano, D. Effect of pentoxifylline on renal outcomes in chronic kidney disease patients: A systematic review and meta-analysis. Pharm. Res. 2016, 107, 315–332. [Google Scholar] [CrossRef] [PubMed]
- Arvey, A.; van der Veeken, J.; Samstein, R.M.; Feng, Y.; Stamatoyannopoulos, J.A.; Rudensky, A.Y. Inflammation-induced repression of chromatin bound by the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 2014, 15, 580–587. [Google Scholar] [CrossRef]
- DuPage, M.; Chopra, G.; Quiros, J.; Rosenthal, W.L.; Morar, M.M.; Holohan, D.; Zhang, R.; Turka, L.; Marson, A.; Bluestone, J.A. The chromatin-modifying enzyme Ezh2 is critical for the maintenance of regulatory T cell identity after activation. Immunity 2015, 42, 227–238. [Google Scholar] [CrossRef]
- Yang, X.P.; Jiang, K.; Hirahara, K.; Vahedi, G.; Afzali, B.; Sciume, G.; Bonelli, M.; Sun, H.W.; Jankovic, D.; Kanno, Y.; et al. EZH2 is crucial for both differentiation of regulatory T cells and T effector cell expansion. Sci. Rep. 2015, 5, 10643. [Google Scholar] [CrossRef]
- Wang, D.; Quiros, J.; Mahuron, K.; Pai, C.C.; Ranzani, V.; Young, A.; Silveria, S.; Harwin, T.; Abnousian, A.; Pagani, M.; et al. Targeting EZH2 Reprograms Intratumoral Regulatory T Cells to Enhance Cancer Immunity. Cell Rep. 2018, 23, 3262–3274. [Google Scholar] [CrossRef]
- Goswami, S.; Apostolou, I.; Zhang, J.; Skepner, J.; Anandhan, S.; Zhang, X.; Xiong, L.; Trojer, P.; Aparicio, A.; Subudhi, S.K.; et al. Modulation of EZH2 expression in T cells improves efficacy of anti-CTLA-4 therapy. J. Clin. Investig. 2018, 128, 3813–3818. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Roberts, C.W. Targeting EZH2 in cancer. Nat. Med. 2016, 22, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Zingg, D.; Arenas-Ramirez, N.; Sahin, D.; Rosalia, R.A.; Antunes, A.T.; Haeusel, J.; Sommer, L.; Boyman, O. The Histone Methyltransferase Ezh2 Controls Mechanisms of Adaptive Resistance to Tumor Immunotherapy. Cell Rep. 2017, 20, 854–867. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Beier, U.H.; Akimova, T.; Dahiya, S.; Han, R.; Samanta, A.; Levine, M.H.; Hancock, W.W. Histone/protein deacetylase inhibitor therapy for enhancement of Foxp3+ T-regulatory cell function posttransplantation. Am. J. Transpl. 2018, 18, 1596–1603. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Ciesielski, M.; Ramakrishnan, S.; Miles, K.M.; Ellis, L.; Sotomayor, P.; Shrikant, P.; Fenstermaker, R.; Pili, R. Class I histone deacetylase inhibitor entinostat suppresses regulatory T cells and enhances immunotherapies in renal and prostate cancer models. PLoS ONE 2012, 7, e30815. [Google Scholar] [CrossRef]
- Xiong, Y.; Wang, L.; Di Giorgio, E.; Akimova, T.; Beier, U.H.; Han, R.; Trevisanut, M.; Kalin, J.H.; Cole, P.A.; Hancock, W.W. Inhibiting the coregulator CoREST impairs Foxp3+ Treg function and promotes antitumor immunity. J. Clin. Investig. 2020, 130, 1830–1842. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Predina, J.; Han, R.; Beier, U.H.; Wang, L.C.; Kapoor, V.; Bhatti, T.R.; Akimova, T.; Singhal, S.; et al. Inhibition of p300 impairs Foxp3(+) T regulatory cell function and promotes antitumor immunity. Nat. Med. 2013, 19, 1173–1177. [Google Scholar] [CrossRef]
- Bates, S.E. Epigenetic Therapies for Cancer. New Engl. J. Med. 2020, 383, 650–663. [Google Scholar] [CrossRef]
- Moreno Ayala, M.A.; Li, Z.; DuPage, M. Treg programming and therapeutic reprogramming in cancer. Immunology 2019, 157, 198–209. [Google Scholar] [CrossRef]
- Lio, C.W.; Hsieh, C.S. A two-step process for thymic regulatory T cell development. Immunity 2008, 28, 100–111. [Google Scholar] [CrossRef]
- Chinen, T.; Kannan, A.K.; Levine, A.G.; Fan, X.; Klein, U.; Zheng, Y.; Gasteiger, G.; Feng, Y.; Fontenot, J.D.; Rudensky, A.Y. An essential role for the IL-2 receptor in Treg cell function. Nat. Immunol. 2016, 17, 1322–1333. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Borde, M.; Heissmeyer, V.; Feuerer, M.; Lapan, A.D.; Stroud, J.C.; Bates, D.L.; Guo, L.; Han, A.; Ziegler, S.F.; et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell 2006, 126, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Garg, G.; Muschaweckh, A.; Moreno, H.; Vasanthakumar, A.; Floess, S.; Lepennetier, G.; Oellinger, R.; Zhan, Y.; Regen, T.; Hiltensperger, M.; et al. Blimp1 Prevents Methylation of Foxp3 and Loss of Regulatory T Cell Identity at Sites of Inflammation. Cell Rep. 2019, 26, 1854–1868.e5. [Google Scholar] [CrossRef] [PubMed]
- Franckaert, D.; Dooley, J.; Roos, E.; Floess, S.; Huehn, J.; Luche, H.; Fehling, H.J.; Liston, A.; Linterman, M.A.; Schlenner, S.M. Promiscuous Foxp3-cre activity reveals a differential requirement for CD28 in Foxp3(+) and Foxp3(-) T cells. Immunol. Cell Biol. 2015, 93, 417–423. [Google Scholar] [CrossRef]
- Bittner-Eddy, P.D.; Fischer, L.A.; Costalonga, M. Cre-loxP Reporter Mouse Reveals Stochastic Activity of the Foxp3 Promoter. Front. Immunol. 2019, 10, 2228. [Google Scholar] [CrossRef]
- Wu, D.; Huang, Q.; Orban, P.C.; Levings, M.K. Ectopic germline recombination activity of the widely used Foxp3-YFP-Cre mouse: A case report. Immunology 2020, 159, 231–241. [Google Scholar] [CrossRef]
- Rubtsov, Y.P.; Niec, R.E.; Josefowicz, S.; Li, L.; Darce, J.; Mathis, D.; Benoist, C.; Rudensky, A.Y. Stability of the regulatory T cell lineage in vivo. Science 2010, 329, 1667–1671. [Google Scholar] [CrossRef]
- Cortez, J.T.; Montauti, E.; Shifrut, E.; Gatchalian, J.; Zhang, Y.; Shaked, O.; Xu, Y.; Roth, T.L.; Simeonov, D.R.; Zhang, Y.; et al. CRISPR screen in regulatory T cells reveals modulators of Foxp3. Nature 2020, 582, 416–420. [Google Scholar] [CrossRef]
- Loo, C.S.; Gatchalian, J.; Liang, Y.; Leblanc, M.; Xie, M.; Ho, J.; Venkatraghavan, B.; Hargreaves, D.C.; Zheng, Y. A Genome-wide CRISPR Screen Reveals a Role for the Non-canonical Nucleosome-Remodeling BAF Complex in Foxp3 Expression and Regulatory T Cell Function. Immunity 2020, 53, 143–157.e8. [Google Scholar] [CrossRef]
- Schumann, K.; Raju, S.S.; Lauber, M.; Kolb, S.; Shifrut, E.; Cortez, J.T.; Skartsis, N.; Nguyen, V.Q.; Woo, J.M.; Roth, T.L.; et al. Functional CRISPR dissection of gene networks controlling human regulatory T cell identity. Nat. Immunol. 2020, 21, 1456–1466. [Google Scholar] [CrossRef]
- Postow, M.A.; Sidlow, R.; Hellmann, M.D. Immune-Related Adverse Events Associated with Immune Checkpoint Blockade. New Engl. J. Med. 2018, 378, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.H.; Rincon, M.; McCoy, K.D.; Le Gros, G. CTLA4 ligation attenuates AP-1, NFAT and NF-kappaB activity in activated T cells. Eur. J. Immunol. 1999, 29, 838–844. [Google Scholar] [CrossRef]
- Walker, L.S.K. PD-1 and CTLA4: Two checkpoints, one pathway? Sci. Immunol. 2017, 2, eaan3864. [Google Scholar] [CrossRef]
- Fenwick, C.; Loredo-Varela, J.L.; Joo, V.; Pellaton, C.; Farina, A.; Rajah, N.; Esteves-Leuenberger, L.; Decaillon, T.; Suffiotti, M.; Noto, A.; et al. Tumor suppression of novel anti-PD-1 antibodies mediated through CD28 costimulatory pathway. J. Exp. Med. 2019, 216, 1525–1541. [Google Scholar] [CrossRef]
- Jutz, S.; Hennig, A.; Paster, W.; Asrak, O.; Dijanovic, D.; Kellner, F.; Pickl, W.F.; Huppa, J.B.; Leitner, J.; Steinberger, P. A cellular platform for the evaluation of immune checkpoint molecules. Oncotarget 2017, 8, 64892–64906. [Google Scholar] [CrossRef]
- Huynh, A.; DuPage, M.; Priyadharshini, B.; Sage, P.T.; Quiros, J.; Borges, C.M.; Townamchai, N.; Gerriets, V.A.; Rathmell, J.C.; Sharpe, A.H.; et al. Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat. Immunol. 2015, 16, 188–196. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, G.; Jin, J.; Degan, S.; Tameze, Y.; Zhang, J.Y. MALT1 promotes melanoma progression through JNK/c-Jun signaling. Oncogenesis 2017, 6, e365. [Google Scholar] [CrossRef]
- Nagel, D.; Spranger, S.; Vincendeau, M.; Grau, M.; Raffegerst, S.; Kloo, B.; Hlahla, D.; Neuenschwander, M.; Peter von Kries, J.; Hadian, K.; et al. Pharmacologic inhibition of MALT1 protease by phenothiazines as a therapeutic approach for the treatment of aggressive ABC-DLBCL. Cancer Cell 2012, 22, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Schon, M.; Wienrich, B.G.; Kneitz, S.; Sennefelder, H.; Amschler, K.; Vohringer, V.; Weber, O.; Stiewe, T.; Ziegelbauer, K.; Schon, M.P. KINK-1, a novel small-molecule inhibitor of IKKbeta, and the susceptibility of melanoma cells to antitumoral treatment. J. Natl. Cancer Inst. 2008, 100, 862–875. [Google Scholar] [CrossRef]
- Edward, M.; MacKie, R.M. Pentoxifylline enhances lung colonization and alters cell adhesion and glycosaminoglycan synthesis by metastatic B16 melanoma cells. Int. J. Cancer 1991, 49, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Ratheesh, A.; Ingle, A.; Gude, R.P. Pentoxifylline modulates cell surface integrin expression and integrin mediated adhesion of B16F10 cells to extracellular matrix components. Cancer Biol. Ther. 2007, 6, 1743–1752. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.; Ghosh, S. NF-kappaB: Roles and regulation in different CD4(+) T-cell subsets. Immunol. Rev. 2013, 252, 41–51. [Google Scholar] [CrossRef]
- Barnes, S.E.; Wang, Y.; Chen, L.; Molinero, L.L.; Gajewski, T.F.; Evaristo, C.; Alegre, M.L. T cell-NF-kappaB activation is required for tumor control in vivo. J. Immunother. Cancer 2015, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Evaristo, C.; Spranger, S.; Barnes, S.E.; Miller, M.L.; Molinero, L.L.; Locke, F.L.; Gajewski, T.F.; Alegre, M.L. Cutting Edge: Engineering Active IKKbeta in T Cells Drives Tumor Rejection. J. Immunol. 2016, 196, 2933–2938. [Google Scholar] [CrossRef] [PubMed]
- Sievers, E.L.; Senter, P.D. Antibody-drug conjugates in cancer therapy. Annu. Rev. Med. 2013, 64, 15–29. [Google Scholar] [CrossRef]
- Knochelmann, H.M.; Dwyer, C.J.; Bailey, S.R.; Amaya, S.M.; Elston, D.M.; Mazza-McCrann, J.M.; Paulos, C.M. When worlds collide: Th17 and Treg cells in cancer and autoimmunity. Cell. Mol. Immunol. 2018, 15, 458–469. [Google Scholar] [CrossRef] [PubMed]

| Protein/Family | Mouse Model | Autoimmune Symptoms | Lethality | Ref. |
|---|---|---|---|---|
| IKKβ | Foxp3creIkbkbF/F | Scurfy-like syndrome | Yes | [87] |
| Ezh2 | Foxp3creEzh2F/F | From 1 month of age, spontaneous T conv cell activation and tissue infiltration | Yes | [108] |
| IRF4 | Foxp3creIrf4F/F | From 6 to 8 weeks of age, spontaneous T conv cell activation and tissue infiltration | Yes | [38] |
| Id proteins | Foxp3creId2Tg | From 6 to 8 weeks of age, progressive T conv cell activation and tissue infiltration | Yes | [77] |
| STAT3 | Foxp3creStat3F/F | From 6 to 8 weeks of age, splenomegaly and colon inflammation | Yes | [80] |
| Bach2 | Bach2−/− | From 3 months of age, spontaneous T conv cell activation and tissue infiltration | Yes | [70] |
| P300 | Foxp3creep300F/F | From 10 weeks of age, spontaneous T conv cell activation and tissue infiltration | n.d. | [117] |
| CBM complex | Foxp3creBcl10F/F | Scurfy-like syndrome | Yes | [89] |
| Foxp3creMalt1F/C4712A | from 6 to 8 weeks of age, spontaneous T conv cell activation and tissue infiltration | n.d. | [88] | |
| Foxp3cre/+Carma1F/F | No detectable signs of autoimmunity (Foxp3cre/cre Carma1 F/F develop Scurfy-like syndrome) | No | [86] | |
| NF-κB | Foxp3creRelF/F | Mild lymphoproliferation from 20 weeks of age | No | [90] |
| Helios | Foxp3creIkzf2F/F | From 5 months of age, spontaneous T conv cell activation and tissue infiltration | No | [52] |
| Foxo1 | Foxp3creFoxo1CA/+ | No detectable signs of autoimmunity (Foxo1CA/CA develop lethal autoimmunity) | No | [30] |
| Pten | Foxp3crePTENF/F | From 12 weeks of age, spontaneous T conv cell activation, kidney damage | No | [136] |
| Foxp3crePTENF/F (2nd construct) | No detectable signs of autoimmunity | No | [41] | |
| Nr4a | Foxp3creNr4a1F/FNr4a2F/F dKO | No detectable signs of autoimmunity (Nr4a1/3 dKO and Nr4a1/2/3 tKO develop lethal autoimmunity) | No | [63] |
| PI3K | Foxp3crep110dF/F | No detectable signs of autoimmunity | No | [44] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stéphan, P.; Lautraite, R.; Voisin, A.; Grinberg-Bleyer, Y. Transcriptional Control of Regulatory T Cells in Cancer: Toward Therapeutic Targeting? Cancers 2020, 12, 3194. https://doi.org/10.3390/cancers12113194
Stéphan P, Lautraite R, Voisin A, Grinberg-Bleyer Y. Transcriptional Control of Regulatory T Cells in Cancer: Toward Therapeutic Targeting? Cancers. 2020; 12(11):3194. https://doi.org/10.3390/cancers12113194
Chicago/Turabian StyleStéphan, Pierre, Raphaëlle Lautraite, Allison Voisin, and Yenkel Grinberg-Bleyer. 2020. "Transcriptional Control of Regulatory T Cells in Cancer: Toward Therapeutic Targeting?" Cancers 12, no. 11: 3194. https://doi.org/10.3390/cancers12113194
APA StyleStéphan, P., Lautraite, R., Voisin, A., & Grinberg-Bleyer, Y. (2020). Transcriptional Control of Regulatory T Cells in Cancer: Toward Therapeutic Targeting? Cancers, 12(11), 3194. https://doi.org/10.3390/cancers12113194





