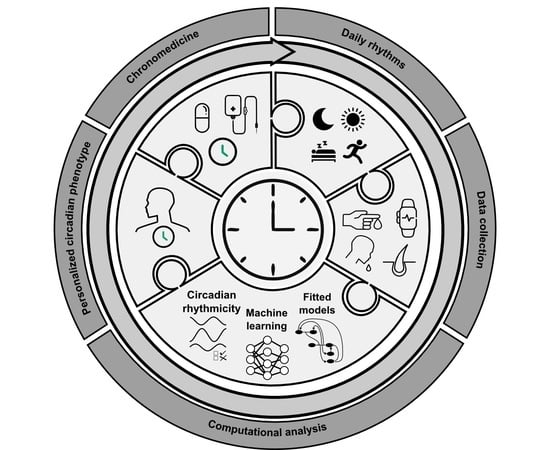
An Optimal Time for Treatment—Predicting Circadian Time by Machine Learning and Mathematical Modelling
Abstract
:Simple Summary
Abstract
1. Introduction
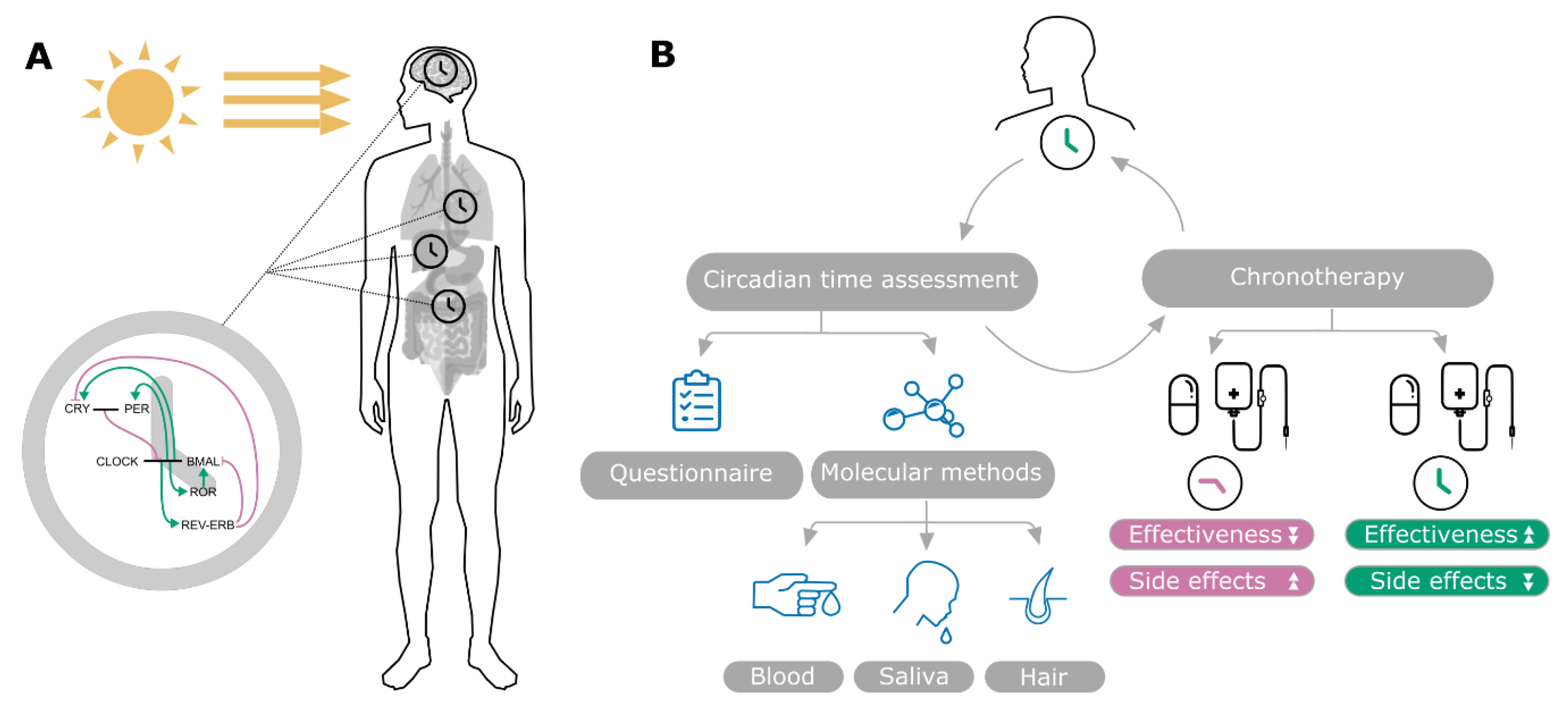
2. Clinical Overview
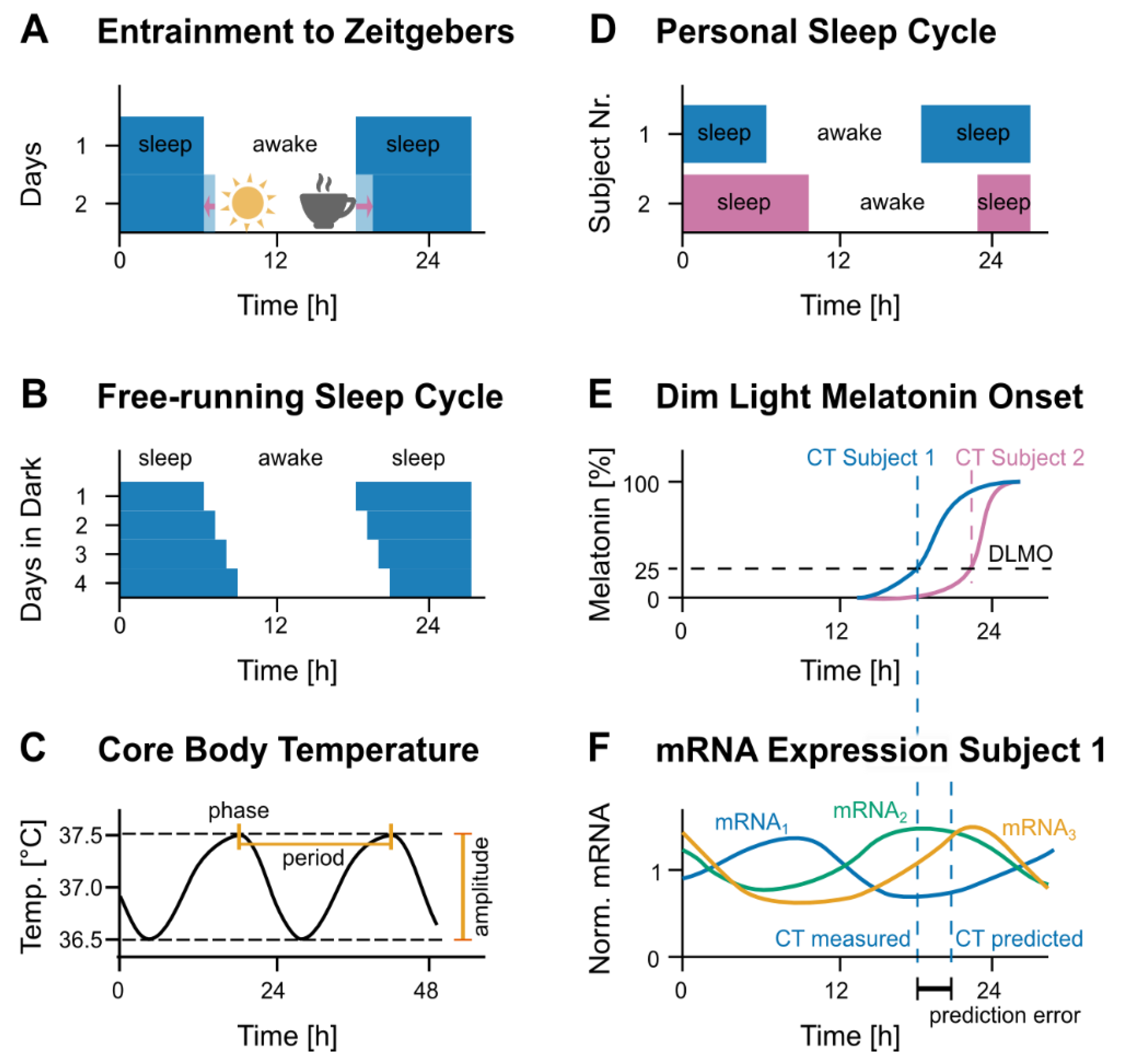
2.1. Current Methods to Assess Circadian Time
2.2. Chronotherapy and Its Importance for Cancer Treatment
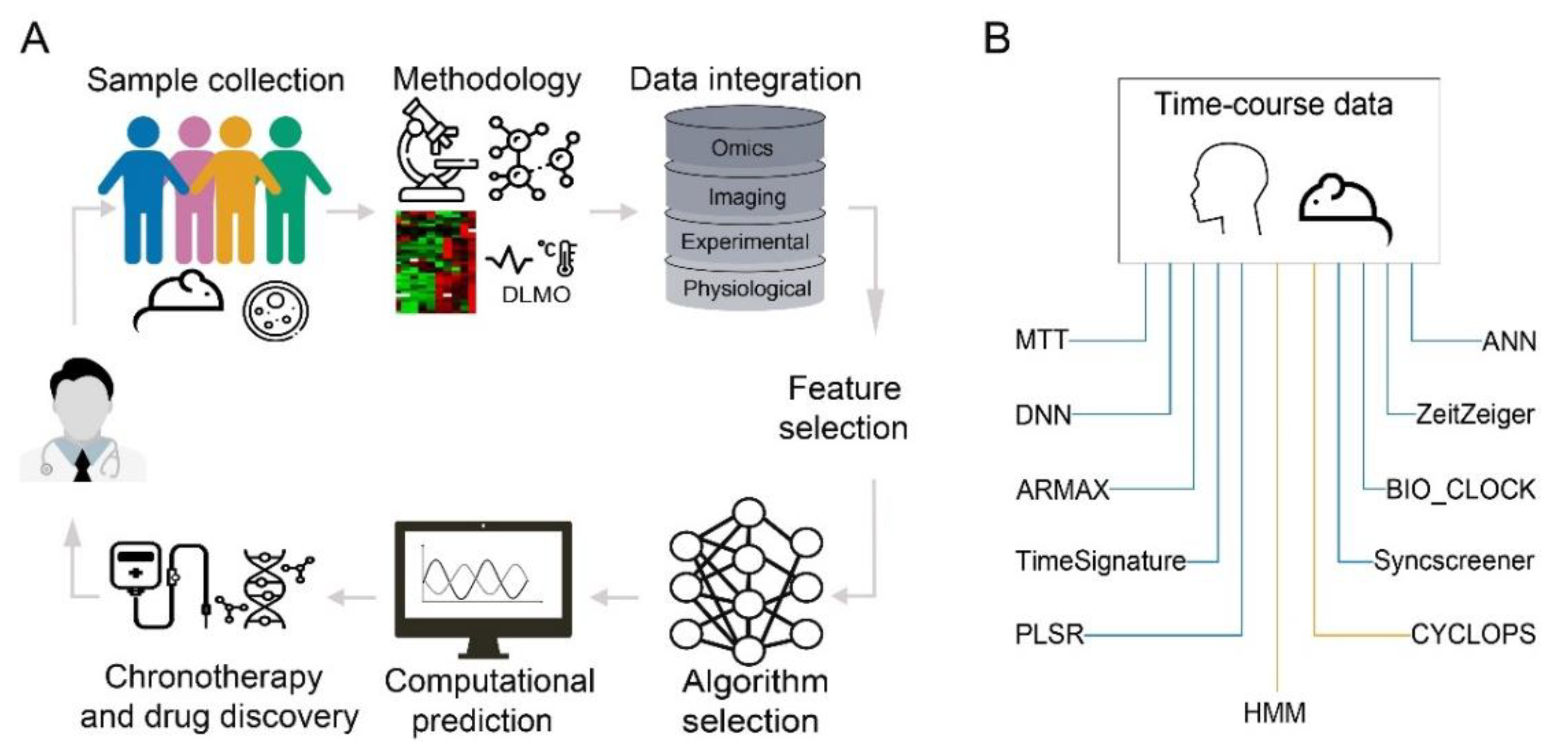
3. Computational Methods
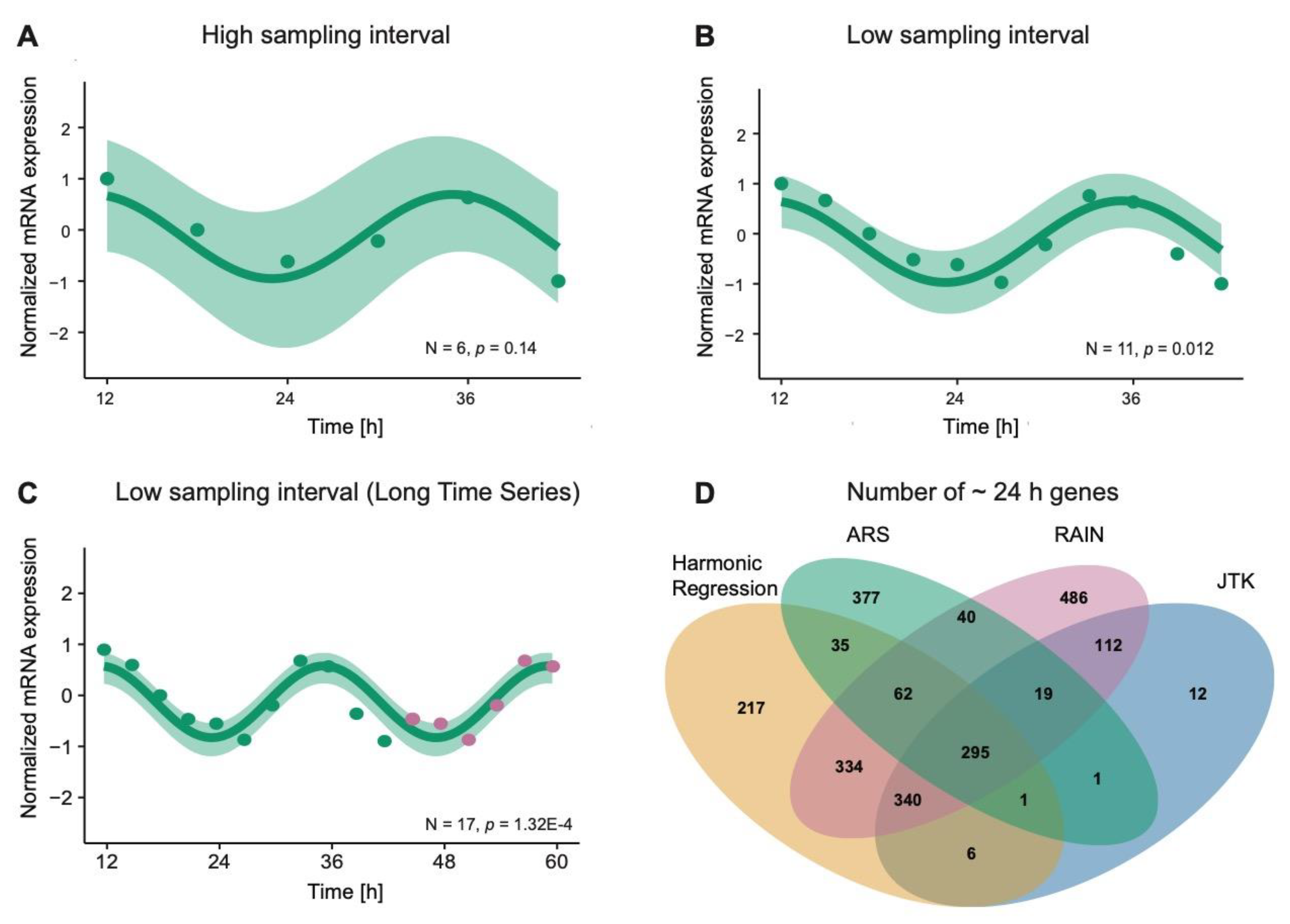
3.1. Computational Methods for Detecting Rhythmic Patterns
3.2. Machine Learning in Prediction of Circadian Time
3.2.1. Application of Machine Learning Methodologies on Gene Expression Data
3.2.2. Application of Machine Learning on Non-Invasive and Phenotypic Measurements
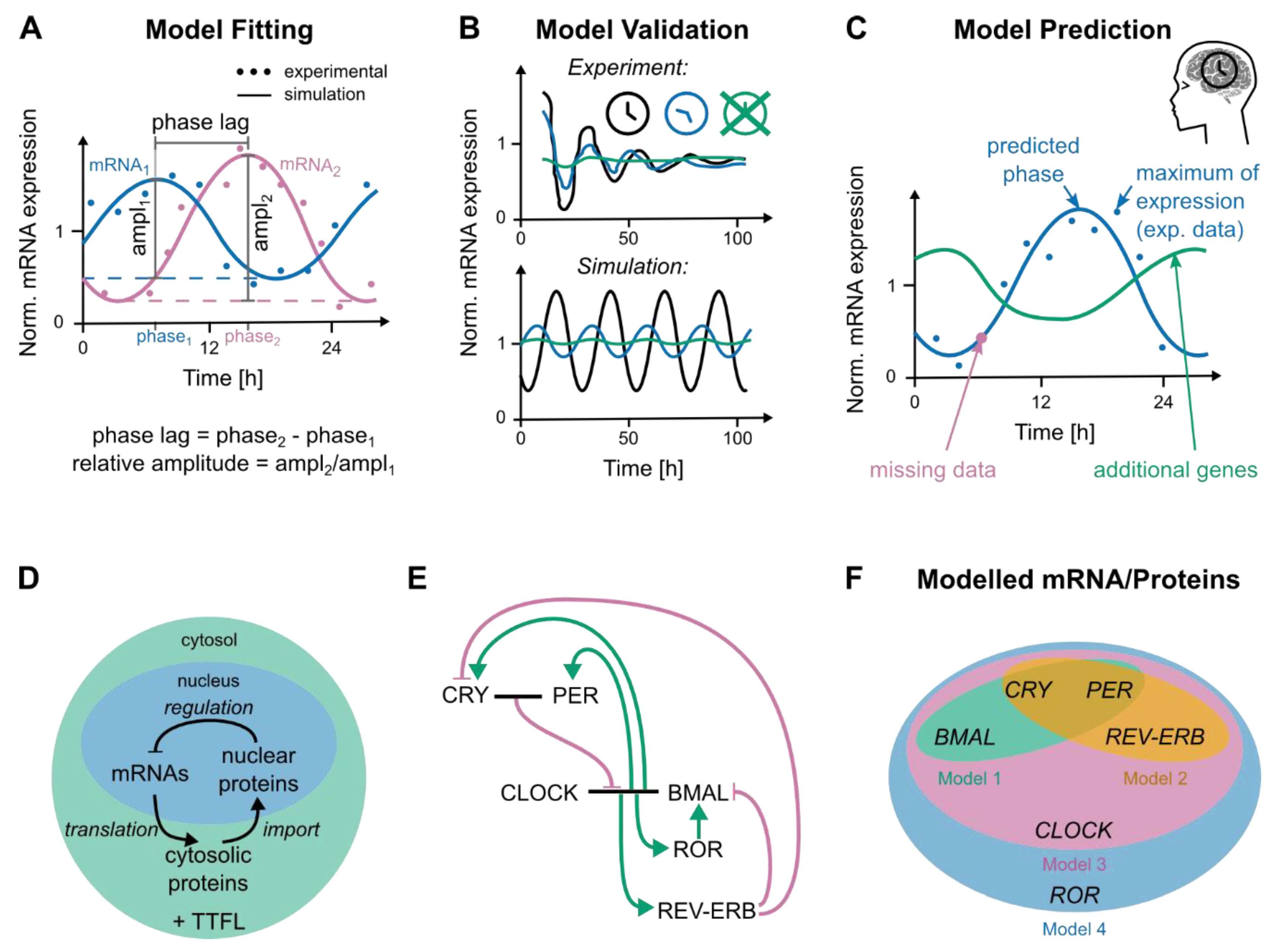
3.3. Modelling Genetic Networks Related to Cancer and Circadian Time
4. Discussion, Limitations and Conclusions
5. Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ballesta, A.; Innominato, P.F.; Dallmann, R.; Rand, D.A.; Levi, F.A. Systems Chronotherapeutics. Pharm. Rev. 2017, 69, 161–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gachon, F.; Olela, F.F.; Schaad, O.; Descombes, P.; Schibler, U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006, 4, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Bollinger, T.; Schibler, U. Circadian rhythms—From genes to physiology and disease. Swiss Med. Wkly. 2014, 144, w13984. [Google Scholar] [CrossRef] [PubMed]
- Bicker, J.; Alves, G.; Falcao, A.; Fortuna, A. Timing in drug absorption and disposition: The past, present, and future of chronopharmacokinetics. Br. J. Pharmacol. 2020, 177, 2215–2239. [Google Scholar] [CrossRef]
- Levi, F.; Zidani, R.; Misset, J.L. Randomised multicentre trial of chronotherapy with oxaliplatin, fluorouracil, and folinic acid in metastatic colorectal cancer. International Organization for Cancer Chronotherapy. Lancet 1997, 350, 681–686. [Google Scholar] [CrossRef]
- Giacchetti, S.; Bjarnason, G.; Garufi, C.; Genet, D.; Iacobelli, S.; Tampellini, M.; Smaaland, R.; Focan, C.; Coudert, B.; Humblet, Y.; et al. Phase III trial comparing 4-day chronomodulated therapy versus 2-day conventional delivery of fluorouracil, leucovorin, and oxaliplatin as first-line chemotherapy of metastatic colorectal cancer: The European Organisation for Research and Treatment of Cancer Chronotherapy Group. J. Clin. Oncol. 2006, 24, 3562–3569. [Google Scholar] [CrossRef]
- Focan, C.; Levi, F.; Kreutz, F.; Focan-Henrard, D.; Lobelle, J.P.; Adam, R.; Dallemagne, B.; Jehaes, C.; Markiewicz, S.; Weerts, J.; et al. Continuous delivery of venous 5-fluorouracil and arterial 5-fluorodeoxyuridine for hepatic metastases from colorectal cancer: Feasibility and tolerance in a randomized phase II trial comparing flat versus chronomodulated infusion. Anticancer Drugs 1999, 10, 385–392. [Google Scholar] [CrossRef]
- Fuhr, L.; Abreu, M.; Pett, P.; Relogio, A. Circadian systems biology: When time matters. Comput. Struct. Biotechnol. J. 2015, 13, 417–426. [Google Scholar] [CrossRef] [Green Version]
- Eastman, C.I.; Tomaka, V.A.; Crowley, S.J. Sex and ancestry determine the free-running circadian period. J. Sleep Res. 2017, 26, 547–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duffy, J.F.; Rimmer, D.W.; Czeisler, C.A. Association of intrinsic circadian period with morningness-eveningness, usual wake time, and circadian phase. Behav. Neurosci. 2001, 115, 895–899. [Google Scholar] [CrossRef]
- Wright, K.P., Jr.; Gronfier, C.; Duffy, J.F.; Czeisler, C.A. Intrinsic period and light intensity determine the phase relationship between melatonin and sleep in humans. J. Biol. Rhythm. 2005, 20, 168–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hida, A.; Kitamura, S.; Ohsawa, Y.; Enomoto, M.; Katayose, Y.; Motomura, Y.; Moriguchi, Y.; Nozaki, K.; Watanabe, M.; Aritake, S.; et al. In vitro circadian period is associated with circadian/sleep preference. Sci. Rep. 2013, 3, 2074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandi-Perumal, S.R.; Smits, M.; Spence, W.; Srinivasan, V.; Cardinali, D.P.; Lowe, A.D.; Kayumov, L. Dim light melatonin onset (DLMO): A tool for the analysis of circadian phase in human sleep and chronobiological disorders. Prog. Neuropsychopharmacol. Biol. Psychiatry 2007, 31, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Astaburuaga, R.; Basti, A.; Li, Y.; Herms, D.; Relógio, A. Circadian regulation of physiology: Relevance for space medicine. Reach 2019, 14–15, 100029. [Google Scholar] [CrossRef]
- Lewy, A.J.; Cutler, N.L.; Sack, R.L. The endogenous melatonin profile as a marker for circadian phase position. J. Biol. Rhythm. 1999, 14, 227–236. [Google Scholar] [CrossRef]
- Lehmann, R.; Childs, L.; Thomas, P.; Abreu, M.; Fuhr, L.; Herzel, H.; Leser, U.; Relogio, A. Assembly of a comprehensive regulatory network for the mammalian circadian clock: A bioinformatics approach. PLoS ONE 2015, 10, e0126283. [Google Scholar] [CrossRef]
- Moore, R.Y.; Eichler, V.B. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 1972, 42, 201–206. [Google Scholar] [CrossRef]
- Levi, F.; Schibler, U. Circadian rhythms: Mechanisms and therapeutic implications. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 593–628. [Google Scholar] [CrossRef]
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef] [Green Version]
- Zehring, W.A.; Wheeler, D.A.; Reddy, P.; Konopka, R.J.; Kyriacou, C.P.; Rosbash, M.; Hall, J.C. P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster. Cell 1984, 39, 369–376. [Google Scholar] [CrossRef]
- Vosshall, L.B.; Price, J.L.; Sehgal, A.; Saez, L.; Young, M.W. Block in nuclear localization of period protein by a second clock mutation, timeless. Science 1994, 263, 1606–1609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, J.L.; Blau, J.; Rothenfluh, A.; Abodeely, M.; Kloss, B.; Young, M.W. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 1998, 94, 83–95. [Google Scholar] [CrossRef] [Green Version]
- Relogio, A.; Westermark, P.O.; Wallach, T.; Schellenberg, K.; Kramer, A.; Herzel, H. Tuning the mammalian circadian clock: Robust synergy of two loops. PLoS Comput. Biol. 2011, 7, e1002309. [Google Scholar] [CrossRef] [Green Version]
- Zhang, E.E.; Kay, S.A. Clocks not winding down: Unravelling circadian networks. Nat. Rev. Mol. Cell Biol. 2010, 11, 764–776. [Google Scholar] [CrossRef]
- El-Athman, R.; Genov, N.N.; Mazuch, J.; Zhang, K.; Yu, Y.; Fuhr, L.; Abreu, M.; Li, Y.; Wallach, T.; Kramer, A.; et al. The Ink4a/Arf locus operates as a regulator of the circadian clock modulating RAS activity. PLoS Biol. 2017, 15, e2002940. [Google Scholar] [CrossRef] [Green Version]
- Abreu, M.; Basti, A.; Genov, N.; Mazzoccoli, G.; Relogio, A. The reciprocal interplay between TNFalpha and the circadian clock impacts on cell proliferation and migration in Hodgkin lymphoma cells. Sci. Rep. 2018, 8, 11474. [Google Scholar] [CrossRef] [Green Version]
- El-Athman, R.; Fuhr, L.; Relogio, A. A Systems-Level Analysis Reveals Circadian Regulation of Splicing in Colorectal Cancer. EBioMedicine 2018, 33, 68–81. [Google Scholar] [CrossRef]
- Fuhr, L.; El-Athman, R.; Scrima, R.; Cela, O.; Carbone, A.; Knoop, H.; Li, Y.; Hoffmann, K.; Laukkanen, M.O.; Corcione, F.; et al. The Circadian Clock Regulates Metabolic Phenotype Rewiring Via HKDC1 and Modulates Tumor Progression and Drug Response in Colorectal Cancer. EBioMedicine 2018, 33, 105–121. [Google Scholar] [CrossRef] [Green Version]
- El-Athman, R.; Knezevic, D.; Fuhr, L.; Relogio, A. A Computational Analysis of Alternative Splicing across Mammalian Tissues Reveals Circadian and Ultradian Rhythms in Splicing Events. Int. J. Mol. Sci. 2019, 20, 3977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genov, N.; Basti, A.; Abreu, M.; Relogio, A. Temporal Splicing Switches in Elements of the TNF-Pathway Identified by Computational Analysis of Transcriptome Data for Human Cell Lines. Int. J. Mol. Sci. 2019, 20, 1182. [Google Scholar] [CrossRef] [Green Version]
- Mazzoccoli, G.; Vinciguerra, M.; Carbone, A.; Relogio, A. The Circadian Clock, the Immune System, and Viral Infections: The Intricate Relationship Between Biological Time and Host-Virus Interaction. Pathogens 2020, 9, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pourcet, B.; Duez, H. Circadian Control of Inflammasome Pathways: Implications for Circadian Medicine. Front. Immunol. 2020, 11, 1630. [Google Scholar] [CrossRef] [PubMed]
- Reinke, H.; Asher, G. Crosstalk between metabolism and circadian clocks. Nat. Rev. Mol. Cell Biol. 2019, 20, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Gaucher, J.; Montellier, E.; Sassone-Corsi, P. Molecular Cogs: Interplay between Circadian Clock and Cell Cycle. Trends Cell Biol. 2018, 28, 368–379. [Google Scholar] [CrossRef]
- El-Athman, R.; Relogio, A. Escaping Circadian Regulation: An Emerging Hallmark of Cancer? Cell Syst. 2018, 6, 266–267. [Google Scholar] [CrossRef] [Green Version]
- Fuhr, L.; Abreu, M.; Carbone, A.; El-Athman, R.; Bianchi, F.; Laukkanen, M.O.; Mazzoccoli, G.; Relogio, A. The Interplay between Colon Cancer Cells and Tumour-Associated Stromal Cells Impacts the Biological Clock and Enhances Malignant Phenotypes. Cancers 2019, 11, 988. [Google Scholar] [CrossRef] [Green Version]
- Levi, F.; Okyar, A.; Dulong, S.; Innominato, P.F.; Clairambault, J. Circadian timing in cancer treatments. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 377–421. [Google Scholar] [CrossRef] [Green Version]
- Panda, S. The arrival of circadian medicine. Nat. Rev. Endocrinol. 2019, 15, 67–69. [Google Scholar] [CrossRef]
- Finger, A.M.; Dibner, C.; Kramer, A. Coupled network of the circadian clocks: A driving force of rhythmic physiology. FEBS Lett. 2020, 594, 2734–2769. [Google Scholar] [CrossRef]
- Dong, D.; Yang, D.; Lin, L.; Wang, S.; Wu, B. Circadian rhythm in pharmacokinetics and its relevance to chronotherapy. Biochem. Pharmacol. 2020, 178, 114045. [Google Scholar] [CrossRef]
- Klerman, E.B.; Rahman, S.A.; St Hilaire, M.A. What time is it? A tale of three clocks, with implications for personalized medicine. J. Pineal. Res. 2020, 68, e12646. [Google Scholar] [CrossRef]
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mure, L.S.; Le, H.D.; Benegiamo, G.; Chang, M.W.; Rios, L.; Jillani, N.; Ngotho, M.; Kariuki, T.; Dkhissi-Benyahya, O.; Cooper, H.M.; et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 2018, 359, eaao0318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yalcin, M.; El-Athman, R.; Ouk, K.; Priller, J.; Relogio, A. Analysis of the Circadian Regulation of Cancer Hallmarks by a Cross-Platform Study of Colorectal Cancer Time-Series Data Reveals an Association with Genes Involved in Huntington’s Disease. Cancers 2020, 12, 963. [Google Scholar] [CrossRef] [Green Version]
- Robles, M.S.; Cox, J.; Mann, M. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 2014, 10, e1004047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basti, A.; Fior, R.; Yalin, M.; Povoa, V.; Astaburuaga, R.; Li, Y.; Naderi, J.; Godinho Ferreira, M.; Relogio, A. The Core-Clock Gene NR1D1 Impacts Cell Motility In Vitro and Invasiveness in A Zebrafish Xenograft Colon Cancer Model. Cancers 2020, 12, 853. [Google Scholar] [CrossRef] [Green Version]
- Robles, M.S.; Humphrey, S.J.; Mann, M. Phosphorylation Is a Central Mechanism for Circadian Control of Metabolism and Physiology. Cell Metab. 2017, 25, 118–127. [Google Scholar] [CrossRef]
- Relogio, A.; Thomas, P.; Medina-Perez, P.; Reischl, S.; Bervoets, S.; Gloc, E.; Riemer, P.; Mang-Fatehi, S.; Maier, B.; Schafer, R.; et al. Ras-mediated deregulation of the circadian clock in cancer. PLoS Genet. 2014, 10, e1004338. [Google Scholar] [CrossRef]
- Gaspar, L.S.; Alvaro, A.R.; Carmo-Silva, S.; Mendes, A.F.; Relogio, A.; Cavadas, C. The importance of determining circadian parameters in pharmacological studies. Br. J. Pharmacol. 2019, 176, 2827–2847. [Google Scholar] [CrossRef]
- Horne, J.A.; Ostberg, O. A self-assessment questionnaire to determine morningness-eveningness in human circadian rhythms. Int. J. Chronobiol. 1976, 4, 97–110. [Google Scholar]
- Solomon, N.L.; Zeitzer, J.M. The impact of chronotype on prosocial behavior. PLoS ONE 2019, 14, e0216309. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.S.; Reilly, C.; Midkiff, K. Evaluation of three circadian rhythm questionnaires with suggestions for an improved measure of morningness. J. Appl. Psychol. 1989, 74, 728–738. [Google Scholar] [CrossRef] [PubMed]
- Roenneberg, T.; Wirz-Justice, A.; Merrow, M. Life between clocks: Daily temporal patterns of human chronotypes. J. Biol. Rhythm. 2003, 18, 80–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghotbi, N.; Pilz, L.K.; Winnebeck, E.C.; Vetter, C.; Zerbini, G.; Lenssen, D.; Frighetto, G.; Salamanca, M.; Costa, R.; Montagnese, S.; et al. The microMCTQ: An Ultra-Short Version of the Munich ChronoType Questionnaire. J. Biol. Rhythm. 2020, 35, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Danilenko, K.V.; Verevkin, E.G.; Antyufeev, V.S.; Wirz-Justice, A.; Cajochen, C. The hockey-stick method to estimate evening dim light melatonin onset (DLMO) in humans. Chronobiol. Int. 2014, 31, 349–355. [Google Scholar] [CrossRef]
- Roenneberg, T.; Pilz, L.K.; Zerbini, G.; Winnebeck, E.C. Chronotype and Social Jetlag: A (Self-) Critical Review. Biology 2019, 8, 54. [Google Scholar] [CrossRef] [Green Version]
- Bhake, R.C.; Kluckner, V.; Stassen, H.; Russell, G.M.; Leendertz, J.; Stevens, K.; Linthorst, A.C.E.; Lightman, S.L. Continuous Free Cortisol Profiles-Circadian Rhythms in Healthy Men. J. Clin. Endocrinol. Metab. 2019, 104, 5935–5947. [Google Scholar] [CrossRef]
- Ohdo, S. Circadian rhythms in the CNS and peripheral clock disorders: Chronopharmacological findings on antitumor drugs. J. Pharmacol. Sci. 2007, 103, 155–158. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Ruben, M.D.; Francey, L.J.; Smith, D.F.; Sherrill, J.D.; Oblong, J.E.; Mills, K.J.; Hogenesch, J.B. A population-based gene expression signature of molecular clock phase from a single epidermal sample. Genome Med. 2020, 12, 73. [Google Scholar] [CrossRef]
- Wittenbrink, N.; Ananthasubramaniam, B.; Munch, M.; Koller, B.; Maier, B.; Weschke, C.; Bes, F.; de Zeeuw, J.; Nowozin, C.; Wahnschaffe, A.; et al. High-accuracy determination of internal circadian time from a single blood sample. J. Clin. Investig. 2018, 128, 3826–3839. [Google Scholar] [CrossRef]
- Braun, R.; Kath, W.L.; Iwanaszko, M.; Kula-Eversole, E.; Abbott, S.M.; Reid, K.J.; Zee, P.C.; Allada, R. Universal method for robust detection of circadian state from gene expression. Proc. Natl. Acad. Sci. USA 2018, 115, E9247–E9256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughey, J.J. Machine learning identifies a compact gene set for monitoring the circadian clock in human blood. Genome Med. 2017, 9, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laing, E.E.; Moller-Levet, C.S.; Poh, N.; Santhi, N.; Archer, S.N.; Dijk, D.J. Blood transcriptome based biomarkers for human circadian phase. eLife 2017, 6, e20214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, M.; Tahara, Y.; Tsubosaka, M.; Fukazawa, M.; Ozaki, M.; Iwakami, T.; Nakaoka, T.; Shibata, S. Chronotype and social jetlag influence human circadian clock gene expression. Sci. Rep. 2018, 8, 10152. [Google Scholar] [CrossRef] [Green Version]
- Koshy, A.; Cuesta, M.; Boudreau, P.; Cermakian, N.; Boivin, D.B. Disruption of central and peripheral circadian clocks in police officers working at night. FASEB J. 2019, 33, 6789–6800. [Google Scholar] [CrossRef]
- Bjarnason, G.A.; Jordan, R.C.; Wood, P.A.; Li, Q.; Lincoln, D.W.; Sothern, R.B.; Hrushesky, W.J.; Ben-David, Y. Circadian expression of clock genes in human oral mucosa and skin: Association with specific cell-cycle phases. Am. J. Pathol. 2001, 158, 1793–1801. [Google Scholar] [CrossRef]
- Kim, D.W.; Zavala, E.; Kim, J.K. Wearable technology and systems modeling for personalized chronotherapy. Curr. Opin. Syst. Biol. 2020, 21, 9–15. [Google Scholar] [CrossRef]
- Hasselberg, M.J.; McMahon, J.; Parker, K. The validity, reliability, and utility of the iButton(R) for measurement of body temperature circadian rhythms in sleep/wake research. Sleep Med. 2013, 14, 5–11. [Google Scholar] [CrossRef]
- Walch, O.; Huang, Y.; Forger, D.; Goldstein, C. Sleep stage prediction with raw acceleration and photoplethysmography heart rate data derived from a consumer wearable device. Sleep 2019, 42, zsz180. [Google Scholar] [CrossRef]
- Roberts, D.M.; Schade, M.M.; Mathew, G.M.; Gartenberg, D.; Buxton, O.M. Detecting sleep using heart rate and motion data from multisensor consumer-grade wearables, relative to wrist actigraphy and polysomnography. Sleep 2020, 43, zsaa045. [Google Scholar] [CrossRef]
- Smets, E.; Rios Velazquez, E.; Schiavone, G.; Chakroun, I.; D’Hondt, E.; De Raedt, W.; Cornelis, J.; Janssens, O.; Van Hoecke, S.; Claes, S.; et al. Large-scale wearable data reveal digital phenotypes for daily-life stress detection. NPJ Digit. Med. 2018, 1, 67. [Google Scholar] [CrossRef] [PubMed]
- Porumb, M.; Stranges, S.; Pescape, A.; Pecchia, L. Precision Medicine and Artificial Intelligence: A Pilot Study on Deep Learning for Hypoglycemic Events Detection based on ECG. Sci. Rep. 2020, 10, 170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Innominato, P.; Komarzynski, S.; Karaboue, A.; Ulusakarya, A.; Bouchahda, M.; Haydar, M.; Bossevot-Desmaris, R.; Mocquery, M.; Plessis, V.; Levi, F. Home-Based e-Health Platform for Multidimensional Telemonitoring of Symptoms, Body Weight, Sleep, and Circadian Activity: Relevance for Chronomodulated Administration of Irinotecan, Fluorouracil-Leucovorin, and Oxaliplatin at Home-Results from a Pilot Study. JCO Clin. Cancer Inform. 2018, 2, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.F.; Ruben, M.D.; Francey, L.J.; Walch, O.J.; Hogenesch, J.B. When Should You Take Your Medicines? J. Biol. Rhythm. 2019, 34, 582–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levi, F. From circadian rhythms to cancer chronotherapeutics. Chronobiol. Int. 2002, 19, 1–19. [Google Scholar] [CrossRef]
- Reinberg, A.; Sidi, E. Circadian changes in the inhibitory effects of an antihistaminic drug in man. J. Investig. Derm. 1966, 46, 415–419. [Google Scholar] [CrossRef] [Green Version]
- Haye, R.; Hoye, K.; Berg, O.; Frones, S.; Odegard, T. Morning versus evening dosing of desloratadine in seasonal allergic rhinitis: A randomized controlled study [ISRCTN23032971]. Clin. Mol. Allergy 2005, 3, 3. [Google Scholar] [CrossRef] [Green Version]
- Buttgereit, F.; Doering, G.; Schaeffler, A.; Witte, S.; Sierakowski, S.; Gromnica-Ihle, E.; Jeka, S.; Krueger, K.; Szechinski, J.; Alten, R. Efficacy of modified-release versus standard prednisone to reduce duration of morning stiffness of the joints in rheumatoid arthritis (CAPRA-1): A double-blind, randomised controlled trial. Lancet 2008, 371, 205–214. [Google Scholar] [CrossRef]
- To, H.; Yoshimatsu, H.; Tomonari, M.; Ida, H.; Tsurumoto, T.; Tsuji, Y.; Sonemoto, E.; Shimasaki, N.; Koyanagi, S.; Sasaki, H.; et al. Methotrexate chronotherapy is effective against rheumatoid arthritis. Chronobiol. Int. 2011, 28, 267–274. [Google Scholar] [CrossRef]
- Faurschou, P.; Engel, A.M.; Haanaes, O.C. Salmeterol in two different doses in the treatment of nocturnal bronchial asthma poorly controlled by other therapies. Allergy 1994, 49, 827–832. [Google Scholar] [CrossRef]
- Burioka, N.; Miyata, M.; Endo, M.; Fukuoka, Y.; Suyama, H.; Nakazaki, H.; Igawa, K.; Shimizu, E. Alteration of the circadian rhythm in peak expiratory flow of nocturnal asthma following nighttime transdermal beta2-adrenoceptor agonist tulobuterol chronotherapy. Chronobiol. Int. 2005, 22, 383–390. [Google Scholar] [CrossRef]
- Yi, Y.J.; Kim, H.J.; Jo, S.K.; Kim, S.G.; Song, Y.R.; Chung, W.; Han, K.H.; Lee, C.H.; Hwang, Y.H.; Oh, K.H. Comparison of the efficacy and safety profile of morning administration of controlled-release simvastatin versus evening administration of immediate-release simvastatin in chronic kidney disease patients with dyslipidemia. Clin. Ther. 2014, 36, 1182–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, H.S.; Kim, S.H.; Kim, J.K.; Ko, S.H.; Ko, J.E.; Park, S.J.; Park, M.G.; Lee, J.H.; Hyon, M.S. Comparison of effects of morning versus evening administration of ezetimibe/simvastatin on serum cholesterol in patients with primary hypercholesterolemia. Ann. Pharm. 2011, 45, 841–849. [Google Scholar] [CrossRef]
- Hermida, R.C.; Ayala, D.E.; Mojon, A.; Fernandez, J.R. Ambulatory blood pressure control with bedtime aspirin administration in subjects with prehypertension. Am. J. Hypertens. 2009, 22, 896–903. [Google Scholar] [CrossRef] [PubMed]
- Hermida, R.C.; Ayala, D.E.; Chayan, L.; Mojon, A.; Fernandez, J.R. Administration-time-dependent effects of olmesartan on the ambulatory blood pressure of essential hypertension patients. Chronobiol. Int. 2009, 26, 61–79. [Google Scholar] [CrossRef]
- Lin, P.; An, F.; Xu, X.; Zhao, L.; Liu, L.; Liu, N.; Wang, P.; Liu, J.; Wang, L.; Li, M. Chronopharmacodynamics and mechanisms of antitumor effect induced by erlotinib in xenograft-bearing nude mice. Biochem. Biophys. Res. Commun. 2015, 460, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, C.Y.; Ji, S.G.; Xu, X.; Wang, P.P.; Zhang, B.; Zhao, L.Y.; Liu, L.; Lin, P.P.; Liu, L.K.; et al. Chronopharmacokinetics of Erlotinib and Circadian Rhythms of Related Metabolic Enzymes in Lewis Tumor-Bearing Mice. Eur J. Drug Metab. Pharm. 2016, 41, 627–635. [Google Scholar] [CrossRef]
- Kloth, J.S.; Binkhorst, L.; de Wit, A.S.; De Bruijn, P.; Hamberg, P.; Lam, M.H.; Burger, H.; Chaves, I.; Wiemer, E.A.; van der Horst, G.T.; et al. Relationship Between Sunitinib Pharmacokinetics and Administration Time: Preclinical and Clinical Evidence. Clin. Pharm. 2015, 54, 851–858. [Google Scholar] [CrossRef] [Green Version]
- Szalek, E.; Karbownik, A.; Sobanska, K.; Polom, W.; Grabowski, T.; Wolc, A.; Matuszewski, M.; Grzeskowiak, E. The influence of the time-of-day administration of the drug on the pharmacokinetics of sunitinib in rabbits. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 2393–2399. [Google Scholar]
- Lauriola, M.; Enuka, Y.; Zeisel, A.; D’Uva, G.; Roth, L.; Sharon-Sevilla, M.; Lindzen, M.; Sharma, K.; Nevo, N.; Feldman, M.; et al. Diurnal suppression of EGFR signalling by glucocorticoids and implications for tumour progression and treatment. Nat. Commun. 2014, 5, 5073. [Google Scholar] [CrossRef] [Green Version]
- Sallam, H.; El-Serafi, A.T.; Filipski, E.; Terelius, Y.; Levi, F.; Hassan, M. The effect of circadian rhythm on pharmacokinetics and metabolism of the Cdk inhibitor, roscovitine, in tumor mice model. Chronobiol. Int. 2015, 32, 608–614. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, H.; Matsunaga, N.; Fujioka, T.; Okazaki, F.; Akagawa, Y.; Tsurudome, Y.; Ono, M.; Kuwano, M.; Koyanagi, S.; Ohdo, S. Circadian regulation of mTOR by the ubiquitin pathway in renal cell carcinoma. Cancer Res. 2014, 74, 543–551. [Google Scholar] [CrossRef] [Green Version]
- Li, X.M.; Mohammad-Djafari, A.; Dumitru, M.; Dulong, S.; Filipski, E.; Siffroi-Fernandez, S.; Mteyrek, A.; Scaglione, F.; Guettier, C.; Delaunay, F.; et al. A circadian clock transcription model for the personalization of cancer chronotherapy. Cancer Res. 2013, 73, 7176–7188. [Google Scholar] [CrossRef] [Green Version]
- Cederroth, C.R.; Albrecht, U.; Bass, J.; Brown, S.A.; Dyhrfjeld-Johnsen, J.; Gachon, F.; Green, C.B.; Hastings, M.H.; Helfrich-Forster, C.; Hogenesch, J.B.; et al. Medicine in the Fourth Dimension. Cell Metab. 2019, 30, 238–250. [Google Scholar] [CrossRef]
- Levi, F. Circadian chronotherapy for human cancers. Lancet Oncol. 2001, 2, 307–315. [Google Scholar] [CrossRef]
- Ruben, M.D.; Smith, D.F.; FitzGerald, G.A.; Hogenesch, J.B. Dosing time matters. Science 2019, 365, 547–549. [Google Scholar] [CrossRef]
- Levi, F.; Karaboue, A.; Saffroy, R.; Desterke, C.; Boige, V.; Smith, D.; Hebbar, M.; Innominato, P.; Taieb, J.; Carvalho, C.; et al. Pharmacogenetic determinants of outcomes on triplet hepatic artery infusion and intravenous cetuximab for liver metastases from colorectal cancer (European trial OPTILIV, NCT00852228). Br. J. Cancer 2017, 117, 965–973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levi, F.; Karaboue, A.; Etienne-Grimaldi, M.C.; Paintaud, G.; Focan, C.; Innominato, P.; Bouchahda, M.; Milano, G.; Chatelut, E. Pharmacokinetics of Irinotecan, Oxaliplatin and 5-Fluorouracil During Hepatic Artery Chronomodulated Infusion: A Translational European OPTILIV Study. Clin. Pharm. 2017, 56, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, J.F.; Benedetti, F.; Geoffroy, P.A.; Henriksen, T.E.G.; Lam, R.W.; Murray, G.; Phelps, J.; Sit, D.; Swartz, H.A.; Crowe, M.; et al. The chronotherapeutic treatment of bipolar disorders: A systematic review and practice recommendations from the ISBD task force on chronotherapy and chronobiology. Bipolar Disord. 2019, 21, 741–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, M.; Silver, R. Sex differences in circadian timing systems: Implications for disease. Front. Neuroendocr. 2014, 35, 111–139. [Google Scholar] [CrossRef] [Green Version]
- Rahn, D.A., 3rd; Ray, D.K.; Schlesinger, D.J.; Steiner, L.; Sheehan, J.P.; O’Quigley, J.M.; Rich, T. Gamma knife radiosurgery for brain metastasis of nonsmall cell lung cancer: Is there a difference in outcome between morning and afternoon treatment? Cancer 2011, 117, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Badiyan, S.N.; Ferraro, D.J.; Yaddanapudi, S.; Drzymala, R.E.; Lee, A.Y.; Silver, S.A.; Dyk, P.; DeWees, T.; Simpson, J.R.; Rich, K.M.; et al. Impact of time of day on outcomes after stereotactic radiosurgery for non-small cell lung cancer brain metastases. Cancer 2013, 119, 3563–3569. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.; Rowbottom, L.; McDonald, R.; Zhang, L.; Bjarnason, G.A.; Tsao, M.; Danjoux, C.; Barnes, E.; Lam, H.; Popovic, M.; et al. Could time of whole brain radiotherapy delivery impact overall survival in patients with multiple brain metastases? Ann. Palliat. Med. 2016, 5, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.M.; Choi, D.H.; Park, H.; Huh, S.J.; Park, W.; Seol, S.W.; Jeong, B.K.; Nam, S.J.; Lee, J.E.; Kil, W.H. Comparison of acute skin reaction following morning versus late afternoon radiotherapy in patients with breast cancer who have undergone curative surgical resection. J. Radiat. Res. 2014, 55, 553–558. [Google Scholar] [CrossRef] [Green Version]
- Caussanel, J.P.; Levi, F.; Brienza, S.; Misset, J.L.; Itzhaki, M.; Adam, R.; Milano, G.; Hecquet, B.; Mathe, G. Phase I trial of 5-day continuous venous infusion of oxaliplatin at circadian rhythm-modulated rate compared with constant rate. J. Natl. Cancer Inst. 1990, 82, 1046–1050. [Google Scholar] [CrossRef]
- Chan, S.; Zhang, L.; Rowbottom, L.; McDonald, R.; Bjarnason, G.A.; Tsao, M.; Barnes, E.; Danjoux, C.; Popovic, M.; Lam, H.; et al. Effects of circadian rhythms and treatment times on the response of radiotherapy for painful bone metastases. Ann. Palliat. Med. 2017, 6, 14–25. [Google Scholar] [CrossRef]
- Shukla, P.; Gupta, D.; Bisht, S.S.; Pant, M.C.; Bhatt, M.L.; Gupta, R.; Srivastava, K.; Gupta, S.; Dhawan, A.; Mishra, D.; et al. Circadian variation in radiation-induced intestinal mucositis in patients with cervical carcinoma. Cancer 2010, 116, 2031–2035. [Google Scholar] [CrossRef]
- Levi, F.A.; Zidani, R.; Vannetzel, J.M.; Perpoint, B.; Focan, C.; Faggiuolo, R.; Chollet, P.; Garufi, C.; Itzhaki, M.; Dogliotti, L.; et al. Chronomodulated versus fixed-infusion-rate delivery of ambulatory chemotherapy with oxaliplatin, fluorouracil, and folinic acid (leucovorin) in patients with colorectal cancer metastases: A randomized multi-institutional trial. J. Natl. Cancer Inst. 1994, 86, 1608–1617. [Google Scholar] [CrossRef]
- Qvortrup, C.; Jensen, B.V.; Fokstuen, T.; Nielsen, S.E.; Keldsen, N.; Glimelius, B.; Bjerregaard, B.; Mejer, J.; Larsen, F.O.; Pfeiffer, P. A randomized study comparing short-time infusion of oxaliplatin in combination with capecitabine XELOX(30) and chronomodulated XELOX(30) as first-line therapy in patients with advanced colorectal cancer. Ann. Oncol. 2010, 21, 87–91. [Google Scholar] [CrossRef]
- Innominato, P.F.; Ballesta, A.; Huang, Q.; Focan, C.; Chollet, P.; Karaboue, A.; Giacchetti, S.; Bouchahda, M.; Adam, R.; Garufi, C.; et al. Sex-dependent least toxic timing of irinotecan combined with chronomodulated chemotherapy for metastatic colorectal cancer: Randomized multicenter EORTC 05011 trial. Cancer Med. 2020, 9, 4148–4159. [Google Scholar] [CrossRef] [Green Version]
- Goyal, M.; Shukla, P.; Gupta, D.; Bisht, S.S.; Dhawan, A.; Gupta, S.; Pant, M.C.; Verma, N.S. Oral mucositis in morning vs. evening irradiated patients: A randomised prospective study. Int. J. Radiat. Biol. 2009, 85, 504–509. [Google Scholar] [CrossRef]
- Bjarnason, G.A.; Mackenzie, R.G.; Nabid, A.; Hodson, I.D.; El-Sayed, S.; Grimard, L.; Brundage, M.; Wright, J.; Hay, J.; Ganguly, P.; et al. Comparison of toxicity associated with early morning versus late afternoon radiotherapy in patients with head-and-neck cancer: A prospective randomized trial of the National Cancer Institute of Canada Clinical Trials Group (HN3). Int. J. Radiat. Oncol. Biol. Phys. 2009, 73, 166–172. [Google Scholar] [CrossRef]
- Tsuchiya, Y.; Ushijima, K.; Noguchi, T.; Okada, N.; Hayasaka, J.I.; Jinbu, Y.; Ando, H.; Mori, Y.; Kusama, M.; Fujimura, A. Influence of a dosing-time on toxicities induced by docetaxel, cisplatin and 5-fluorouracil in patients with oral squamous cell carcinoma; a cross-over pilot study. Chronobiol. Int. 2018, 35, 289–294. [Google Scholar] [CrossRef]
- Hsu, F.M.; Hou, W.H.; Huang, C.Y.; Wang, C.C.; Tsai, C.L.; Tsai, Y.C.; Yu, H.J.; Pu, Y.S.; Cheng, J.C. Differences in toxicity and outcome associated with circadian variations between patients undergoing daytime and evening radiotherapy for prostate adenocarcinoma. Chronobiol. Int. 2016, 33, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Dincol, D.; Samur, M.; Pamir, A.; Sencan, O.; Akbulut, H.; Yalcin, B.; Onur, H.; Demirkazik, A.; Senler, F.C.; Icli, F. Prospective randomized comparison of morning versus night daily single subcutaneous administration of granulocyte-macrophage-colony stimulating factor in patients with soft tissue or bone sarcoma. Cancer 2000, 88, 2033–2036. [Google Scholar] [CrossRef]
- Lin, H.X.; Hua, Y.J.; Chen, Q.Y.; Luo, D.H.; Sun, R.; Qiu, F.; Mo, H.Y.; Mai, H.Q.; Guo, X.; Xian, L.J.; et al. Randomized study of sinusoidal chronomodulated versus flat intermittent induction chemotherapy with cisplatin and 5-fluorouracil followed by traditional radiotherapy for locoregionally advanced nasopharyngeal carcinoma. Chin. J. Cancer 2013, 32, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Bi, T.; Jin, F.; Wu, W.; Long, J.; Li, Y.; Gong, X.; Luo, X.; Li, Z.; He, Q.; Qu, B. Phase II clinical trial of two different modes of administration of the induction chemotherapy for locally advanced nasopharyngeal carcinoma. Zhonghua Zhong Liu Za Zhi 2015, 37, 676–681. [Google Scholar]
- Zhang, P.X.; Jin, F.; Li, Z.L.; Wu, W.L.; Li, Y.Y.; Long, J.H.; Chen, G.Y.; Chen, X.X.; Gan, J.Y.; Gong, X.Y.; et al. A randomized phase II trial of induction chemotherapy followed by cisplatin chronotherapy versus constant rate delivery combined with radiotherapy. Chronobiol. Int. 2018, 35, 240–248. [Google Scholar] [CrossRef]
- Li, J.; Chen, R.; Ji, M.; Zou, S.L.; Zhu, L.N. Cisplatin-based chronotherapy for advanced non-small cell lung cancer patients: A randomized controlled study and its pharmacokinetics analysis. Cancer Chemother. Pharmacol. 2015, 76, 651–655. [Google Scholar] [CrossRef]
- Atluri, H.; Campian, J.L.; Talcott, G.; Meyer, M.; Slat, E.; Rubin, J.; Huang, J.; Chheda, M.G.; Johanns, T.M.; Tao, Y.; et al. Effect of temozolomide chronotherapy in patients with high-grade glioma. J. Clin. Oncol. 2020, 38, e14525. [Google Scholar] [CrossRef]
- Bishop, C.M. Neural Networks for Pattern Recognition; Clarendon Press: Oxford, UK, 1995; 482p. [Google Scholar]
- St Hilaire, M.A.; Klerman, E.B.; Khalsa, S.B.; Wright, K.P., Jr.; Czeisler, C.A.; Kronauer, R.E. Addition of a non-photic component to a light-based mathematical model of the human circadian pacemaker. J. Theor. Biol. 2007, 247, 583–599. [Google Scholar] [CrossRef] [Green Version]
- Gil, E.A.; Aubert, X.L.; Beersma, D.G.M. Ambulatory estimation of human circadian phase using models of varying complexity based on non-invasive signal modalities. In Proceedings of the 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014; pp. 2278–2281. [Google Scholar]
- Urwyler, P.; Rampa, L.; Stucki, R.; Buchler, M.; Muri, R.; Mosimann, U.P.; Nef, T. Recognition of activities of daily living in healthy subjects using two ad-hoc classifiers. Biomed. Eng. Online 2015, 14, 54. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhang, Y.; Jiang, F.; Zhao, H. A novel machine learning unsupervised algorithm for sleep/wake identification using actigraphy. Chronobiol. Int. 2020, 37, 1002–1015. [Google Scholar] [CrossRef]
- Stone, J.E.; Phillips, A.J.K.; Ftouni, S.; Magee, M.; Howard, M.; Lockley, S.W.; Sletten, T.L.; Anderson, C.; Rajaratnam, S.M.W.; Postnova, S. Generalizability of A Neural Network Model for Circadian Phase Prediction in Real-World Conditions. Sci. Rep. 2019, 9, 11001. [Google Scholar] [CrossRef]
- Gil, E.A.; Aubert, X.L.; Most, E.I.; Beersma, D.G. Human circadian phase estimation from signals collected in ambulatory conditions using an autoregressive model. J. Biol. Rhythm. 2013, 28, 152–163. [Google Scholar] [CrossRef] [Green Version]
- Kolodyazhniy, V.; Spati, J.; Frey, S.; Gotz, T.; Wirz-Justice, A.; Krauchi, K.; Cajochen, C.; Wilhelm, F.H. An improved method for estimating human circadian phase derived from multichannel ambulatory monitoring and artificial neural networks. Chronobiol. Int. 2012, 29, 1078–1097. [Google Scholar] [CrossRef]
- McDonald, M.J.; Rosbash, M. Microarray Analysis and Organization of Circadian Gene Expression in Drosophila. Cell 2001, 107, 567–578. [Google Scholar] [CrossRef] [Green Version]
- Akhtar, R.A.; Reddy, A.B.; Maywood, E.S.; Clayton, J.D.; King, V.M.; Smith, A.G.; Gant, T.W.; Hastings, M.H.; Kyriacou, C.P. Circadian Cycling of the Mouse Liver Transcriptome, as Revealed by cDNA Microarray, Is Driven by the Suprachiasmatic Nucleus. Curr. Biol. 2002, 12, 540–550. [Google Scholar] [CrossRef] [Green Version]
- Panda, S.; Antoch, M.P.; Miller, B.H.; Su, A.I.; Schook, A.B.; Straume, M.; Schultz, P.G.; Kay, S.A.; Takahashi, J.S.; Hogenesch, J.B. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 2002, 109, 307–320. [Google Scholar] [CrossRef] [Green Version]
- Hughes, M.E.; DiTacchio, L.; Hayes, K.R.; Vollmers, C.; Pulivarthy, S.; Baggs, J.E.; Panda, S.; Hogenesch, J.B. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009, 5, e1000442. [Google Scholar] [CrossRef] [Green Version]
- Duffield, G.E.; Best, J.D.; Meurers, B.H.; Bittner, A.; Loros, J.J.; Dunlap, J.C. Circadian Programs of Transcriptional Acitvation, Signaling and Protein Turnover Revealed by Microarray Analysis of Mammalian Cells. Curr. Biol. 2002, 12, 551–557. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, J.S.; Kumar, V.; Nakashe, P.; Koike, N.; Huang, H.C.; Green, C.B.; Kim, T.K. ChIP-seq and RNA-seq methods to study circadian control of transcription in mammals. Methods Enzymol. 2015, 551, 285–321. [Google Scholar] [CrossRef] [Green Version]
- Le Martelot, G.; Canella, D.; Symul, L.; Migliavacca, E.; Gilardi, F.; Liechti, R.; Martin, O.; Harshman, K.; Delorenzi, M.; Desvergne, B.; et al. Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 2012, 10, e1001442. [Google Scholar] [CrossRef] [Green Version]
- Hatanaka, F.; Matsubara, C.; Myung, J.; Yoritaka, T.; Kamimura, N.; Tsutsumi, S.; Kanai, A.; Suzuki, Y.; Sassone-Corsi, P.; Aburatani, H.; et al. Genome-wide profiling of the core clock protein BMAL1 targets reveals a strict relationship with metabolism. Mol. Cell Biol. 2010, 30, 5636–5648. [Google Scholar] [CrossRef] [Green Version]
- Deckard, A.; Anafi, R.C.; Hogenesch, J.B.; Haase, S.B.; Harer, J. Design and analysis of large-scale biological rhythm studies: A comparison of algorithms for detecting periodic signals in biological data. Bioinformatics 2013, 29, 3174–3180. [Google Scholar] [CrossRef] [Green Version]
- Hughes, M.E.; Abruzzi, K.C.; Allada, R.; Anafi, R.; Arpat, A.B.; Asher, G.; Baldi, P.; de Bekker, C.; Bell-Pedersen, D.; Blau, J.; et al. Guidelines for Genome-Scale Analysis of Biological Rhythms. J. Biol. Rhythm. 2017, 32, 380–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mei, W.; Jiang, Z.; Chen, Y.; Chen, L.; Sancar, A.; Jiang, Y. Genome-wide circadian rhythm detection methods: Systematic evaluations and practical guidelines. Brief. Bioinform. 2020, 1, bbaa135. [Google Scholar] [CrossRef]
- Laloum, D.; Robinson-Rechavi, M. Methods detecting rhythmic gene expression are biologically relevant only for strong signal. PLoS Comput. Biol. 2020, 16, e1007666. [Google Scholar] [CrossRef] [PubMed]
- Akashi, M.; Soma, H.; Yamamoto, T.; Tsugitomi, A.; Yamashita, S.; Yamamoto, T.; Nishida, E.; Yasuda, A.; Liao, J.K.; Node, K. Noninvasive method for assessing the human circadian clock using hair follicle cells. Proc. Natl. Acad. Sci. USA 2010, 107, 15643–15648. [Google Scholar] [CrossRef] [Green Version]
- Luck, S.; Thurley, K.; Thaben, P.F.; Westermark, P.O. Rhythmic degradation explains and unifies circadian transcriptome and proteome data. Cell Rep. 2014, 9, 741–751. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.; Su, Z. Analyzing circadian expression data by harmonic regression based on autoregressive spectral estimation. Bioinformatics 2010, 26, i168–i174. [Google Scholar] [CrossRef] [Green Version]
- Lomb, N.R. Least-squares frequency analysis of unequally spaced data. Astrophys. Space Sci. 1975, 39, 447–462. [Google Scholar] [CrossRef]
- Thaben, P.F.; Westermark, P.O. Detecting rhythms in time series with RAIN. J. Biol. Rhythm. 2014, 29, 391–400. [Google Scholar] [CrossRef] [Green Version]
- Hutchison, A.L.; Maienschein-Cline, M.; Chiang, A.H.; Tabei, S.M.; Gudjonson, H.; Bahroos, N.; Allada, R.; Dinner, A.R. Improved statistical methods enable greater sensitivity in rhythm detection for genome-wide data. PLoS Comput. Biol. 2015, 11, e1004094. [Google Scholar] [CrossRef] [Green Version]
- Hughes, M.E.; Hogenesch, J.B.; Kornacker, K. JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythm. 2010, 25, 372–380. [Google Scholar] [CrossRef]
- Agostinelli, F.; Ceglia, N.; Shahbaba, B.; Sassone-Corsi, P.; Baldi, P. What time is it? Deep learning approaches for circadian rhythms. Bioinformatics 2016, 32, i8–i17. [Google Scholar] [CrossRef]
- Chudova, D.; Ihler, A.; Lin, K.K.; Andersen, B.; Smyth, P. Bayesian detection of non-sinusoidal periodic patterns in circadian expression data. Bioinformatics 2009, 25, 3114–3120. [Google Scholar] [CrossRef]
- Wichert, S.; Fokianos, K.; Strimmer, K. Identifying periodically expressed transcripts in microarray time series data. Bioinformatics 2004, 20, 5–20. [Google Scholar] [CrossRef] [Green Version]
- Leise, T.L.; Harrington, M.E. Wavelet-based time series analysis of circadian rhythms. J. Biol. Rhythm. 2011, 26, 454–463. [Google Scholar] [CrossRef]
- Leise, T.L. Wavelet-based analysis of circadian behavioral rhythms. Methods Enzymol. 2015, 551, 95–119. [Google Scholar] [CrossRef]
- Wu, G.; Anafi, R.C.; Hughes, M.E.; Kornacker, K.; Hogenesch, J.B. MetaCycle: An integrated R package to evaluate periodicity in large scale data. Bioinformatics 2016, 32, 3351–3353. [Google Scholar] [CrossRef] [Green Version]
- Scargle, J. Studies in astronomical time series analysis. II- Statistical aspects of spectral analysis of unevenly spaced data. Astrophys. J. 1982, 263, 835–853. [Google Scholar] [CrossRef]
- Cui, P.; Zhong, T.; Wang, Z.; Wang, T.; Zhao, H.; Liu, C.; Lu, H. Identification of human circadian genes based on time course gene expression profiles by using a deep learning method. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 2274–2283. [Google Scholar] [CrossRef] [PubMed]
- Chen, C. Ascent of machine learning in medicine. Nat. Mater. 2019, 18, 407. [Google Scholar] [CrossRef]
- Ambale-Venkatesh, B.; Yang, X.; Wu, C.O.; Liu, K.; Hundley, W.G.; McClelland, R.; Gomes, A.S.; Folsom, A.R.; Shea, S.; Guallar, E.; et al. Cardiovascular Event Prediction by Machine Learning: The Multi-Ethnic Study of Atherosclerosis. Circ. Res. 2017, 121, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Cuperlovic-Culf, M. Machine Learning Methods for Analysis of Metabolic Data and Metabolic Pathway Modeling. Metabolites 2018, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Erickson, B.J.; Korfiatis, P.; Akkus, Z.; Kline, T.L. Machine Learning for Medical Imaging. Radiographics 2017, 37, 505–515. [Google Scholar] [CrossRef]
- Martorell-Marugan, J.; Tabik, S.; Benhammou, Y.; Del Val, C.; Zwir, I.; Herrera, F.; Carmona-Saez, P. Deep Learning in Omics Data Analysis and Precision Medicine. In Computational Biology; Husi, H., Ed.; Codon Publications: Brisbane, Australia, 2019. [Google Scholar]
- Ekins, S.; Puhl, A.C.; Zorn, K.M.; Lane, T.R.; Russo, D.P.; Klein, J.J.; Hickey, A.J.; Clark, A.M. Exploiting machine learning for end-to-end drug discovery and development. Nat. Mater. 2019, 18, 435–441. [Google Scholar] [CrossRef]
- Talamanca, L.; Naef, F. How to tell time: Advances in decoding circadian phase from omics snapshots. F1000Research 2020, 9, 1150. [Google Scholar] [CrossRef]
- Bastanlar, Y.; Ozuysal, M. Introduction to machine learning. Methods Mol. Biol. 2014, 1107, 105–128. [Google Scholar] [CrossRef] [Green Version]
- Madani, A.; Ong, J.R.; Tibrewal, A.; Mofrad, M.R.K. Deep echocardiography: Data-efficient supervised and semi-supervised deep learning towards automated diagnosis of cardiac disease. NPJ Digit. Med. 2018, 1, 59. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, Y.; Therneau, T.M.; Atkinson, E.J.; Tafti, A.P.; Zhang, N.; Amin, S.; Limper, A.H.; Khosla, S.; Liu, H. Unsupervised machine learning for the discovery of latent disease clusters and patient subgroups using electronic health records. J. Biomed. Inform. 2020, 102, 103364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueda, H.R.; Chen, W.; Minami, Y.; Honma, S.; Honma, K.; Iino, M.; Hashimoto, S. Molecular-timetable methods for detection of body time and rhythm disorders from single-time-point genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2004, 101, 11227–11232. [Google Scholar] [CrossRef] [Green Version]
- Anafi, R.C.; Lee, Y.; Sato, T.K.; Venkataraman, A.; Ramanathan, C.; Kavakli, I.H.; Hughes, M.E.; Baggs, J.E.; Growe, J.; Liu, A.C.; et al. Machine learning helps identify CHRONO as a circadian clock component. PLoS Biol. 2014, 12, e1001840. [Google Scholar] [CrossRef] [PubMed]
- Hughey, J.J.; Hastie, T.; Butte, A.J. ZeitZeiger: Supervised learning for high-dimensional data from an oscillatory system. Nucleic Acids Res. 2016, 44, e80. [Google Scholar] [CrossRef] [PubMed]
- Anafi, R.C.; Francey, L.J.; Hogenesch, J.B.; Kim, J. CYCLOPS reveals human transcriptional rhythms in health and disease. Proc. Natl. Acad. Sci. USA 2017, 114, 5312–5317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlachou, D.; Bjarnason, G.A.; Giacchetti, S.; Lévi, F.; Rand, D.A. TimeTeller: A New Tool for Precision Circadian Medicine and Cancer Prognosis. bioRxiv 2020, 622050. [Google Scholar] [CrossRef]
- Minami, Y.; Kasukawa, T.; Kakazu, Y.; Iigo, M.; Sugimoto, M.; Ikeda, S.; Yasui, A.; van der Horst, G.T.; Soga, T.; Ueda, H.R. Measurement of internal body time by blood metabolomics. Proc. Natl. Acad. Sci. USA 2009, 106, 9890–9895. [Google Scholar] [CrossRef] [Green Version]
- Kasukawa, T.; Sugimoto, M.; Hida, A.; Minami, Y.; Mori, M.; Honma, S.; Honma, K.; Mishima, K.; Soga, T.; Ueda, H.R. Human blood metabolite timetable indicates internal body time. Proc. Natl. Acad. Sci. USA 2012, 109, 15036–15041. [Google Scholar] [CrossRef] [Green Version]
- Sutskever, I.; Martens, J.; Dahl, G.; Hinton, G. On the importance of initialization and momentum in deep learning. In Proceedings of the 30th International Conference on Machine Learning, Atlanta, GA, USA, 17–19 June 2013; pp. 1139–1147. [Google Scholar]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Martinez-Nicolas, A.; Martinez-Madrid, M.J.; Almaida-Pagan, P.F.; Bonmati-Carrion, M.A.; Madrid, J.A.; Rol, M.A. Assessing Chronotypes by Ambulatory Circadian Monitoring. Front. Physiol. 2019, 10, 1396. [Google Scholar] [CrossRef] [Green Version]
- Lewy, A.J.; Sack, R.L. The dim light melatonin onset as a marker for circadian phase position. Chronobiol. Int. 1989, 6, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Xie, P.; Dong, Y.; Liu, Z.; Zhou, F.; Pan, D.; Huang, Z.; Zhai, Q.; Gu, Y.; Wu, Q.; et al. High-throughput discovery of genetic determinants of circadian misalignment. PLoS Genet. 2020, 16, e1008577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dean, D.A., 2nd; Goldberger, A.L.; Mueller, R.; Kim, M.; Rueschman, M.; Mobley, D.; Sahoo, S.S.; Jayapandian, C.P.; Cui, L.; Morrical, M.G.; et al. Scaling Up Scientific Discovery in Sleep Medicine: The National Sleep Research Resource. Sleep 2016, 39, 1151–1164. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.Q.; Cui, L.; Mueller, R.; Tao, S.; Kim, M.; Rueschman, M.; Mariani, S.; Mobley, D.; Redline, S. The National Sleep Research Resource: Towards a sleep data commons. J. Am. Med. Inform. Assoc. 2018, 25, 1351–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woller, A.; Duez, H.; Staels, B.; Lefranc, M. A Mathematical Model of the Liver Circadian Clock Linking Feeding and Fasting Cycles to Clock Function. Cell Rep. 2016, 17, 1087–1097. [Google Scholar] [CrossRef] [Green Version]
- Schmelling, N.M.; Axmann, I.M. Computational modelling unravels the precise clockwork of cyanobacteria. Interface Focus 2018, 8, 20180038. [Google Scholar] [CrossRef] [PubMed]
- Bujdoso, N.; Davis, S.J. Mathematical modeling of an oscillating gene circuit to unravel the circadian clock network of Arabidopsis thaliana. Front. Plant Sci. 2013, 4, 3. [Google Scholar] [CrossRef] [Green Version]
- Asgari-Targhi, A.; Klerman, E.B. Mathematical modeling of circadian rhythms. Wiley Interdiscip. Rev. Syst. Biol. Med. 2019, 11, e1439. [Google Scholar] [CrossRef]
- Jewett, M.E.; Forger, D.B.; Kronauer, R.E. Revised limit cycle oscillator model of human circadian pacemaker. J. Biol. Rhythm. 1999, 14, 493–499. [Google Scholar] [CrossRef]
- Roenneberg, T.; Merrow, M. The Circadian Clock and Human Health. Curr. Biol. 2016, 26, R432–R443. [Google Scholar] [CrossRef] [PubMed]
- Reppert, S.M.; Weaver, D.R. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 2001, 63, 647–676. [Google Scholar] [CrossRef]
- Goodwin, B.C. Oscillatory behavior in enzymatic control processes. Adv. Enzym. Regul. 1965, 3, 425–438. [Google Scholar] [CrossRef]
- Leloup, J.C.; Goldbeter, A. Toward a detailed computational model for the mammalian circadian clock. Proc. Natl. Acad. Sci. USA 2003, 100, 7051–7056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forger, D.B.; Peskin, C.S. A detailed predictive model of the mammalian circadian clock. Proc. Natl. Acad. Sci. USA 2003, 100, 14806–14811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker-Weimann, S.; Wolf, J.; Herzel, H.; Kramer, A. Modeling feedback loops of the Mammalian circadian oscillator. Biophys. J. 2004, 87, 3023–3034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirsky, H.P.; Liu, A.C.; Welsh, D.K.; Kay, S.A.; Doyle, F.J., 3rd. A model of the cell-autonomous mammalian circadian clock. Proc. Natl. Acad. Sci. USA 2009, 106, 11107–11112. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.K.; Forger, D.B. A mechanism for robust circadian timekeeping via stoichiometric balance. Mol. Syst. Biol. 2012, 8, 630. [Google Scholar] [CrossRef]
- Gonze, D. Modeling circadian clocks: From equations to oscillations. Open Life Sci. 2011, 6, 699–711. [Google Scholar] [CrossRef]
- Ballesta, A.; Lopez, J.; Popgeorgiev, N.; Gonzalo, P.; Doumic, M.; Gillet, G. Data-driven modeling of SRC control on the mitochondrial pathway of apoptosis: Implication for anticancer therapy optimization. PLoS Comput. Biol. 2013, 9, e1003011. [Google Scholar] [CrossRef]
- Leloup, J.C.; Goldbeter, A. Modeling the mammalian circadian clock: Sensitivity analysis and multiplicity of oscillatory mechanisms. J. Theor. Biol. 2004, 230, 541–562. [Google Scholar] [CrossRef] [PubMed]
- Battogtokh, D.; Tyson, J.J. Deciphering the Dynamics of Interlocked Feedback Loops in a Model of the Mammalian Circadian Clock. Biophys. J. 2018, 115, 2055–2066. [Google Scholar] [CrossRef] [Green Version]
- Jolley, C.C.; Ukai-Tadenuma, M.; Perrin, D.; Ueda, H.R. A mammalian circadian clock model incorporating daytime expression elements. Biophys. J. 2014, 107, 1462–1473. [Google Scholar] [CrossRef] [Green Version]
- Ballesta, A.; Dulong, S.; Abbara, C.; Cohen, B.; Okyar, A.; Clairambault, J.; Levi, F. A combined experimental and mathematical approach for molecular-based optimization of irinotecan circadian delivery. PLoS Comput. Biol. 2011, 7, e1002143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dulong, S.; Ballesta, A.; Okyar, A.; Levi, F. Identification of Circadian Determinants of Cancer Chronotherapy through In Vitro Chronopharmacology and Mathematical Modeling. Mol. Cancer Ther. 2015, 14, 2154–2164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjornsti, M.A.; Kaufmann, S.H. Topoisomerases and cancer chemotherapy: Recent advances and unanswered questions. F1000Research 2019, 8, 1704. [Google Scholar] [CrossRef]
- Thomas, A.; Pommier, Y. Targeting Topoisomerase I in the Era of Precision Medicine. Clin. Cancer Res. 2019, 25, 6581–6589. [Google Scholar] [CrossRef]
- Selfridge, J.M.; Gotoh, T.; Schiffhauer, S.; Liu, J.; Stauffer, P.E.; Li, A.; Capelluto, D.G.; Finkielstein, C.V. Chronotherapy: Intuitive, Sound, Founded. But Not Broadly Applied. Drugs 2016, 76, 1507–1521. [Google Scholar] [CrossRef] [Green Version]
- Ruben, M.D.; Francey, L.J.; Guo, Y.; Wu, G.; Cooper, E.B.; Shah, A.S.; Hogenesch, J.B.; Smith, D.F. A large-scale study reveals 24-h operational rhythms in hospital treatment. Proc. Natl. Acad. Sci. USA 2019, 116, 20953–20958. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Kettner, N.M. The circadian clock in cancer development and therapy. Prog. Mol. Biol. Transl. Sci. 2013, 119, 221–282. [Google Scholar] [CrossRef] [Green Version]
- Masri, S.; Sassone-Corsi, P. The emerging link between cancer, metabolism, and circadian rhythms. Nat. Med. 2018, 24, 1795–1803. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.N.; Dvuchbabny, S.; Martinez, C.-A.; Kerr, B.; Cistulli, P.A.; Cook, K.M. The Cancer Clock Is (Not) Ticking: Links between Circadian Rhythms and Cancer. Clocks Sleep 2019, 1, 435–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahar, S.; Sassone-Corsi, P. Metabolism and cancer: The circadian clock connection. Nat. Rev. Cancer 2009, 9, 886–896. [Google Scholar] [CrossRef] [PubMed]
- Sulli, G.; Lam, M.T.Y.; Panda, S. Interplay between Circadian Clock and Cancer: New Frontiers for Cancer Treatment. Trends Cancer 2019, 5, 475–494. [Google Scholar] [CrossRef]
- Tsuchiya, Y.; Umemura, Y.; Yagita, K. Circadian clock and cancer: From a viewpoint of cellular differentiation. Int. J. Urol. 2020, 27, 518–524. [Google Scholar] [CrossRef]
- Furtado, A.; Astaburuaga, R.; Costa, A.; Duarte, A.C.; Goncalves, I.; Cipolla-Neto, J.; Lemos, M.C.; Carro, E.; Relogio, A.; Santos, C.R.A.; et al. The Rhythmicity of Clock Genes is Disrupted in the Choroid Plexus of the APP/PS1 Mouse Model of Alzheimer’s Disease. J. Alzheimers Dis. 2020, 77, 795–806. [Google Scholar] [CrossRef]
- Castellana, S.; Mazza, T.; Capocefalo, D.; Genov, N.; Biagini, T.; Fusilli, C.; Scholkmann, F.; Relogio, A.; Hogenesch, J.B.; Mazzoccoli, G. Systematic Analysis of Mouse Genome Reveals Distinct Evolutionary and Functional Properties among Circadian and Ultradian Genes. Front. Physiol. 2018, 9, 1178. [Google Scholar] [CrossRef] [Green Version]
- Genov, N.; Castellana, S.; Scholkmann, F.; Capocefalo, D.; Truglio, M.; Rosati, J.; Turco, E.M.; Biagini, T.; Carbone, A.; Mazza, T.; et al. A Multi-Layered Study on Harmonic Oscillations in Mammalian Genomics and Proteomics. Int. J. Mol. Sci. 2019, 20, 4585. [Google Scholar] [CrossRef] [Green Version]
| Questionnaire | Abbreviation | Number of Questions | Results |
|---|---|---|---|
| Morningness-Eveningness Questionnaire | MEQ | 19 | five categories: definite evening type, moderate evening type, neither type, moderate morning type and definite morning type |
| Composite Scale of Morningness | CSM | 13 | three categories: evening type, intermediate type and morning type. |
| Munich ChronoType Questionnaire | MCTQ | 32 | population-specific continuous distributions featuring different categories |
| Drug | Indications | Drug Target Genes | Organs in Which Targets Oscillate |
|---|---|---|---|
| Filgrastim | Acute myeloid leukemia | Csf3r | lung |
| Rituximab | Non-Hodgkin’s lymphoma | Fcgr2b, Ms4a1, Fcgr3 | liver, kidney, skeletal muscle |
| Bevacizumab | Colorectal cancer, Non-small cell lung cancer | Fcgr2b, Vegfa, Fcgr3 | heart, brown fat, liver, kidney, skeletal muscle, aorta |
| Trastuzumab | Breast cancer | Fcgr2b, Erbb2, Egfr, Fcgr3 | heart, liver, kidney |
| Imatinib | Chronic myeloid leukemia | Ptgs1, Kit, Slc22a2, Abcg2, Pdgfra, Pdgfrb, Ddr1, Abca3, Abl1, Ret, Abcb1a | lung, heart, brainstem, white fat, adrenal gland, brown fat, liver, kidney |
| Pemetrexed | Non-small cell lung cancer | Tyms, Atic, Gart, Slc29a1 | lung, heart, brainstem, brown fat, liver, kidney, aorta |
| Filgrastim | Acute myeloid leukemia | Csf3r | lung |
| Capecitabine | Breast cancer, colorectal cancer | Cda, Tymp, Tyms, Ces1g, Dpyd | lung, adrenal gland, liver, brown fat, kidney, aorta |
| Type of Cancer | Type of Intervention: | Reference |
|---|---|---|
| Brain metastasis in non-small-cell lung carcinoma | Radiosurgery | [101,102] |
| Multiple brain metastases | Radiotherapy | [103] |
| Breast cancer | Radiotherapy | [104] |
| Breast carcinoma, hepatocellular carcinoma, Cholangiocarcinoma | Chemotherapy | [105] |
| Painful bone metastases | Radiotherapy | [106] |
| Cervical carcinoma | Radiotherapy | [107] |
| Colorectal | Chemotherapy | [5,6,98,108,109,110] |
| Head and neck | Radiotherapy | [111,112] |
| Oral squamous cell carcinoma | Chemotherapy | [113] |
| Prostate adenocarcinoma | Radiotherapy | [114] |
| Soft tissue, bone sarcoma | Chemotherapy | [115] |
| Nasopharyngeal carcinoma | Chemotherapy | [116,117,118] |
| Non-small-cell lung carcinoma | Chemotherapy | [119] |
| High-grade glioma | Chemotherapy | [120] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hesse, J.; Malhan, D.; Yalҫin, M.; Aboumanify, O.; Basti, A.; Relógio, A. An Optimal Time for Treatment—Predicting Circadian Time by Machine Learning and Mathematical Modelling. Cancers 2020, 12, 3103. https://doi.org/10.3390/cancers12113103
Hesse J, Malhan D, Yalҫin M, Aboumanify O, Basti A, Relógio A. An Optimal Time for Treatment—Predicting Circadian Time by Machine Learning and Mathematical Modelling. Cancers. 2020; 12(11):3103. https://doi.org/10.3390/cancers12113103
Chicago/Turabian StyleHesse, Janina, Deeksha Malhan, Müge Yalҫin, Ouda Aboumanify, Alireza Basti, and Angela Relógio. 2020. "An Optimal Time for Treatment—Predicting Circadian Time by Machine Learning and Mathematical Modelling" Cancers 12, no. 11: 3103. https://doi.org/10.3390/cancers12113103
APA StyleHesse, J., Malhan, D., Yalҫin, M., Aboumanify, O., Basti, A., & Relógio, A. (2020). An Optimal Time for Treatment—Predicting Circadian Time by Machine Learning and Mathematical Modelling. Cancers, 12(11), 3103. https://doi.org/10.3390/cancers12113103