Non-Canonical Functions of the Gamma-Tubulin Meshwork in the Regulation of the Nuclear Architecture
Simple Summary
Abstract
1. Introduction
2. Nuclear Morphology
2.1. Nuclear Envelope
2.2. Force Balance between Cytoskeleton and Nucleoskeleton
2.3. Nuclear Bodies
3. Show Me Your Nuclear Architecture and I Will Tell You Who You Are
3.1. The Organization of the Nucleus Influences Gene Expression
3.2. During Differentiation
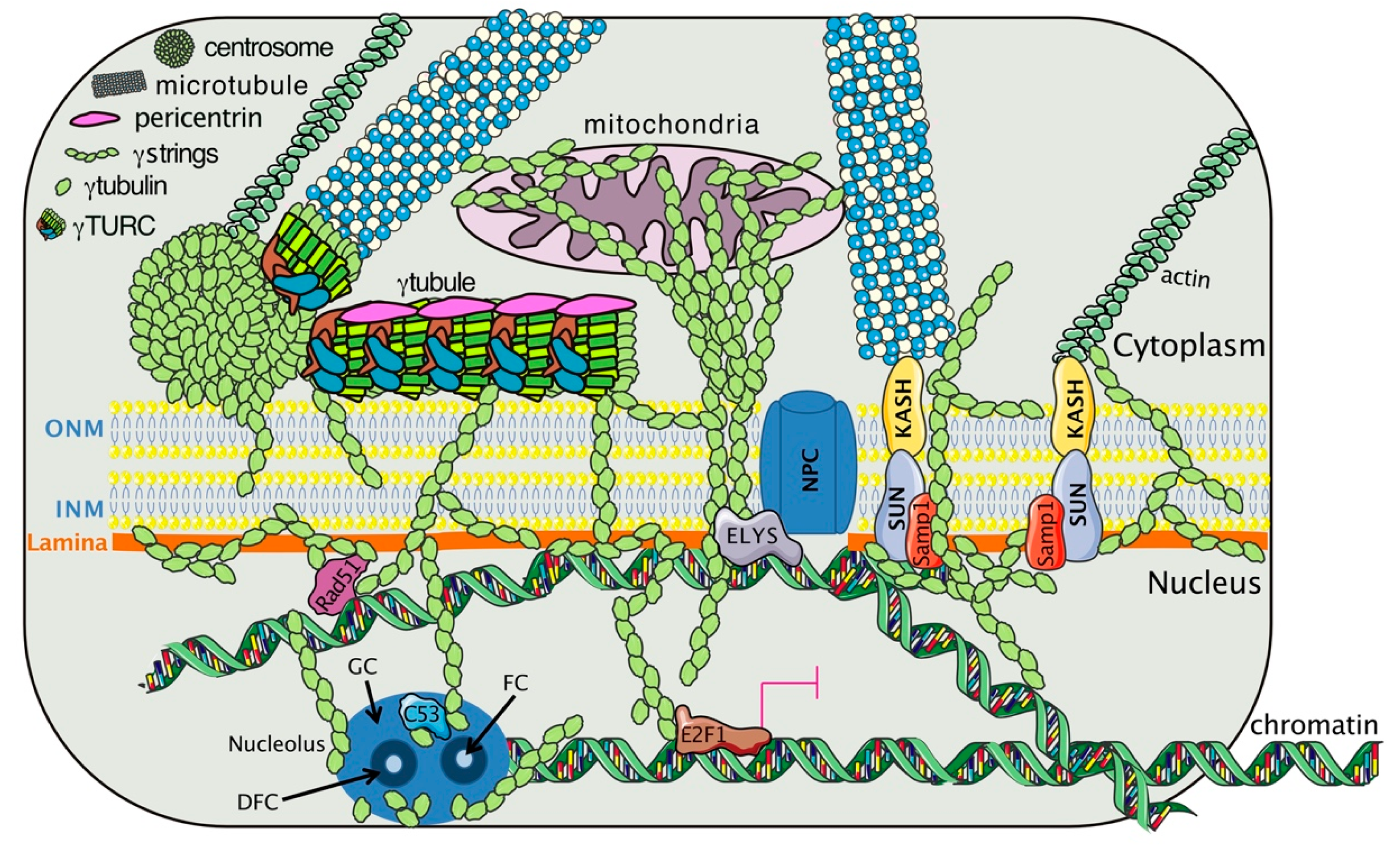
4. The Gamma-Tubulin Meshwork
4.1. Tubulins
| Protein/Complex | Reference | Comments |
|---|---|---|
| Centrosome | [82,83] | Cytoskeletal organizing centers |
| Microtubules | [92,93,94] | Polymers of tubulins |
| γTuRC | [87,89,90,91] | MT-nucleating unit |
| γTubule | [16] | γTubulin-rich filaments |
| γStrings | [17,18,96] | Thin cytosolic/nuclear γtubulin threads |
| Actin | [97] | Cytoskeletal/nuclearskeletal element |
| Intermediate filaments | [98] | Cytoskeleton |
| Lamin B | [17] | Nuclearskeleton |
| Ran | [99] | Regulates transport across NPC |
| Mel-28/ELYS | [99] | Required for NPC assembly |
| Nucleolin | [100,101] | RNA-binding protein at the nucleolus |
| E2F | [88,102,103] | Regulates gene expression |
| SUN | [33] | Links the nuclear lamina with NPCs |
| Samp1 | [104,105] | Inner nuclear membrane protein |
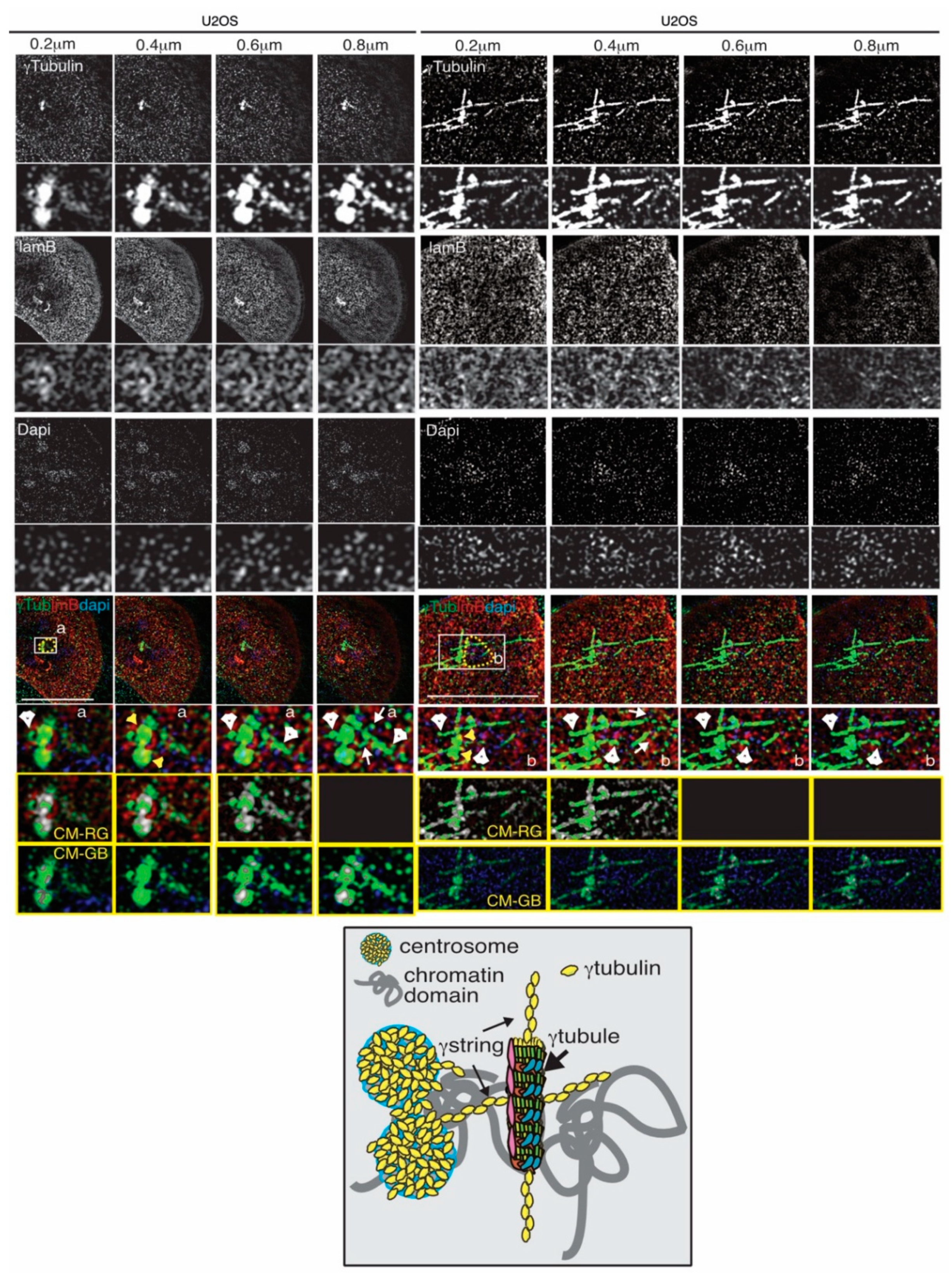
4.2. The Dynamics of the Gamma-Tubulin Meshwork
4.3. The Gamma-Tubulin Meshwork and Gene Transcription
4.4. The Gamma-Tubulin Meshwork and Nuclear Architecture
4.5. The Gamma-Tubulin Meshwork and Cell Differentiation
4.6. Gamma-Tubulin in Cancer
5. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ATR | Ataxia Telangiectasia and Rad3-related protein |
| BRCA | BReast CAncer type 1 susceptibility protein |
| CDA | Citral Dimethyl Acetyl |
| Chk | Checkpoint Kinase |
| CRM1 | ChRomosomal maintenance 1 |
| DFC | Dense Fibrillar Centre |
| DMF | DiMethyl Fumarate |
| DNA | DeoxyriboNucleic Acid |
| E2F | E2 promoter binding Factor |
| ELYS | Embryonic Large molecule derived from Yolk Sac |
| FC | Fibrillar Centre |
| FDA | Optune treatment Food and Drug Administration |
| GC | Granular Centre |
| GCP | Gamma tubulin Complex Proteins |
| GDP | Guanosine Diphosphate |
| GTP | Guanosine TriPhosphate |
| γTuRC | γTubulin Ring Complex |
| HGPS | Hutchinson–Gilford Progeria Syndrome |
| INM | Inner Nuclear Membrane |
| LAD | Lamin-associated DNA domains |
| LINC | LInker of Nucleo and Cytoskeleton |
| MT | MicroTubule |
| mtDNA | mitochondrial DNA |
| MYC | Myelocytomatosis viral oncogene homolog |
| MTOC | Microtubule Organizing Centre |
| NB | Nuclear Bodies |
| NE | Nuclear Envelope |
| NES | Nuclear Exclusion Signal |
| NLS | Nuclear Localization Sequence |
| NOR | Nucleolus Organizer Region |
| NB | Nuclear Basket |
| NPC | Nuclear Pore Complex |
| NSCLC | Non-Small Cell Lung Cancer |
| NTR | Nuclear Transport Receptors |
| NUP | NUcleoPorin |
| ONM | Outer Nuclear Membrane |
| PAP | PAPanicolaou |
| PCNA | Proliferating Cell Nuclear Antigen |
| PML | ProMyelocytic Leukemia protein |
| PRC1 | Polycomb Repressive Complexes 1 |
| r | Ribosomal |
| Ran | Small GTPase RAs related Nuclear |
| RANAP1 | RANGTPase-Activating Protein 1 |
| RANBP1 | RAN Binding Protein 1 |
| RB | RetinoBlastoma |
| RCC1 | Regulator of Chromosome Condensation 1 |
| RNA | RiboNucleic Acid |
| SAHF | Senescence Associated Heterochromatin Foci |
| SUN | Sad1 UNc-84 domain protein |
| TPR | Translocated Promoter Region |
References
- Lusk:, C.P.; King, M.C. The nucleus: Keeping it together by keeping it apart. Curr. Opin. Cell Biol. 2017, 44, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Schon, E.A.; DiMauro, S.; Hirano, M. Human mitochondrial DNA: Roles of inherited and somatic mutations. Nat. Rev. Genet. 2012, 13, 878–890. [Google Scholar] [CrossRef] [PubMed]
- Taanman, J.W. The mitochondrial genome: Structure, transcription, translation and replication. Biochim. Biophys. Acta 1999, 1410, 103–123. [Google Scholar] [CrossRef]
- Filipski, A.; Kumar, S. Comparative Genomics in Eukaryotes. In The Evolution of the Genome; Academic Press: Cambridge, MA, USA, 2005. [Google Scholar]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Simon, D.N.; Wilson, K.L. The nucleoskeleton as a genome-associated dynamic ‘network of networks’. Nat. Rev. Mol. Cell Biol. 2011, 12, 695–708. [Google Scholar] [CrossRef]
- Zhen, Y.Y.; Libotte, T.; Munck, M.; Noegel, A.A.; Korenbaum, E. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J. Cell Sci. 2002, 115, 3207–3222. [Google Scholar]
- Postel, R.; Ketema, M.; Kuikman, I.; de Pereda, J.M.; Sonnenberg, A. Nesprin-3 augments peripheral nuclear localization of intermediate filaments in zebrafish. J. Cell Sci. 2011, 124, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Gimpel, P.; Lee, Y.L.; Sobota, R.M.; Calvi, A.; Koullourou, V.; Patel, R.; Mamchaoui, K.; Nédélec, F.; Shackleton, S.; Schmoranzer, J.; et al. Nesprin-1α-Dependent Microtubule Nucleation from the Nuclear Envelope via Akap450 Is Necessary for Nuclear Positioning in Muscle Cells. Curr. Biol. 2017, 27, 2999–3009.e9. [Google Scholar] [CrossRef]
- Skinner, B.M.; Johnson, E.E. Nuclear morphologies: Their diversity and functional relevance. Chromosoma 2017, 126, 195–212. [Google Scholar] [CrossRef]
- Zink, D.; Fischer, A.H.; Nickerson, J.A. Nuclear structure in cancer cells. Nat. Rev. Cancer 2004, 4, 677–687. [Google Scholar] [CrossRef]
- Kim, H.J.; Taylor, J.P. Lost in Transportation: Nucleocytoplasmic Transport Defects in ALS and Other Neurodegenerative Diseases. Neuron 2017, 96, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Freibaum, B.D.; Lu, Y.; Lopez-Gonzalez, R.; Kim, N.C.; Almeida, S.; Lee, K.H.; Badders, N.; Valentine, M.; Miller, B.L.; Wong, P.C.; et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 2015, 525, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Rosselló, C.A.; Lindström, L.; Eklund, G.; Corvaisier, M.; Kristensson, M.A. γ-Tubulin–γ-Tubulin Interactions as the Basis for the Formation of a Meshwork. Int. J. Mol. Sci. 2018, 19, 3245. [Google Scholar] [CrossRef] [PubMed]
- Chumová, J.; Trögelová, L.; Kourová, H.; Volc, J.; Sulimenko, V.; Halada, P.; Kučera, O.; Benada, O.; Kuchařová, A.; Klebanovych, A.; et al. γ-Tubulin has a conserved intrinsic property of self-polymerization into double stranded filaments and fibrillar networks. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 734–748. [Google Scholar] [CrossRef]
- Lindstrom, L.; Alvarado-Kristensson, M. Characterization of gamma-tubulin filaments in mammalian cells. Biochim. Biophys. Acta 2018, 1865, 158–171. [Google Scholar] [CrossRef]
- Rossello, C.A.; Lindstrom, L.; Glindre, J.; Eklund, G.; Alvarado-Kristensson, M. Gamma-tubulin coordinates nuclear envelope assembly around chromatin. Heliyon 2016, 2, e00166. [Google Scholar] [CrossRef]
- Lindström, L.; Li, T.; Malycheva, D.; Kancharla, A.; Nilsson, H.; Vishnu, N.; Mulder, H.; Johansson, M.; Rosselló, C.A.; Alvarado-Kristensson, M. The GTPase domain of gamma-tubulin is required for normal mitochondrial function and spatial organization. Commun. Biol. 2018, 1, 37. [Google Scholar] [CrossRef]
- English, A.R.; Voeltz, G.K. Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb. Perspect. Biol. 2013, 5, a013227. [Google Scholar] [CrossRef]
- Hampoelz, B.; Andres-Pons, A.; Kastritis, P.; Beck, M. Structure and Assembly of the Nuclear Pore Complex. Annu. Rev. Biophys. 2019, 48, 515–536. [Google Scholar] [CrossRef]
- Mohr, D.; Frey, S.; Fischer, T.; Güttler, T.; Görlich, D. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 2009, 28, 2541–2553. [Google Scholar] [CrossRef]
- Cautain, B.; Hill, R.; de Pedro, N.; Link, W. Components and regulation of nuclear transport processes. FEBS J. 2015, 282, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Cook, A.; Bono, F.; Jinek, M.; Conti, E. Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 2007, 76, 647–671. [Google Scholar] [CrossRef] [PubMed]
- De Leeuw, R.; Gruenbaum, Y.; Medalia, O. Nuclear Lamins: Thin Filaments with Major Functions. Trends Cell Biol. 2018, 28, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, T.; Escalante-Alcalde, D.; Bhatt, H.; Anver, M.; Bhat, N.; Nagashima, K.; Stewart, C.L.; Burke, B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 1999, 147, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Nikolova, V.; Leimena, C.; McMahon, A.C.; Tan, J.C.; Chandar, S.; Jogia, D.; Kesteven, S.H.; Michalicek, J.; Otway, R.; Verheyen, F.; et al. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice. J. Clin. Investig. 2004, 113, 357–369. [Google Scholar] [CrossRef]
- Shimi, T.; Pfleghaar, K.; Kojima, S.; Pack, C.G.; Solovei, I.; Goldman, A.E.; Adam, S.A.; Shumaker, D.K.; Kinjo, M.; Cremer, T.; et al. The A- and B-type nuclear lamin networks: Microdomains involved in chromatin organization and transcription. Genes Dev. 2008, 22, 3409–3421. [Google Scholar] [CrossRef] [PubMed]
- Broers, J.L.; Peeters, E.A.; Kuijpers, H.J.; Endert, J.; Bouten, C.V.; Oomens, C.W.; Baaijens, F.P.; Ramaekers, F.C. Decreased mechanical stiffness in LMNA-/- cells is caused by defective nucleo-cytoskeletal integrity: Implications for the development of laminopathies. Hum. Mol. Genet. 2004, 13, 2567–2580. [Google Scholar] [CrossRef]
- Swift, J.; Ivanovska, I.L.; Buxboim, A.; Harada, T.; Dingal, P.C.; Pinter, J.; Pajerowski, J.D.; Spinler, K.R.; Shin, J.W.; Tewari, M.; et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 2013, 341, 1240104. [Google Scholar] [CrossRef] [PubMed]
- Isermann, P.; Lammerding, J. Nuclear mechanics and mechanotransduction in health and disease. Curr. Biol. 2013, 23, R1113–R1121. [Google Scholar] [CrossRef]
- Crisp, M.; Liu, Q.; Roux, K.; Rattner, J.B.; Shanahan, C.; Burke, B.; Stahl, P.D.; Hodzic, D. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J. Cell Biol. 2006, 172, 41–53. [Google Scholar] [CrossRef]
- Liu, Q.; Pante, N.; Misteli, T.; Elsagga, M.; Crisp, M.; Hodzic, D.; Burke, B.; Roux, K.J. Functional association of Sun1 with nuclear pore complexes. J. Cell Biol. 2007, 178, 785–798. [Google Scholar] [CrossRef]
- Chumová, J.; Kourová, H.; Trögelová, L.; Halada, P.; Binarová, P. Microtubular and Nuclear Functions of γ-Tubulin: Are They LINCed? Cells 2019, 8, 259. [Google Scholar] [CrossRef]
- Uhler, C.; Shivashankar, G.V. Nuclear Mechanopathology and Cancer Diagnosis. Trends Cancer 2018, 4, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, M.L.; Jaalouk, D.E.; Shanahan, C.M.; Burke, B.; Roux, K.J.; Lammerding, J. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J. Biol. Chem. 2011, 286, 26743–26753. [Google Scholar] [CrossRef]
- Tariq, Z.; Zhang, H.; Chia-Liu, A.; Shen, Y.; Gete, Y.; Xiong, Z.M.; Tocheny, C.; Campanello, L.; Wu, D.; Losert, W.; et al. Lamin A and microtubules collaborate to maintain nuclear morphology. Nucleus 2017, 8, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Hampoelz, B.; Azou-Gros, Y.; Fabre, R.; Markova, O.; Puech, P.H.; Lecuit, T. Microtubule-induced nuclear envelope fluctuations control chromatin dynamics in Drosophila embryos. Development 2011, 138, 3377–3386. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.S.; Zhang, B.; Spector, D.L. Biogenesis and function of nuclear bodies. Trends Genet. 2011, 27, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Shaw, P.J.; Jordan, E.G. The nucleolus. Annu. Rev. Cell Dev. Biol. 1995, 11, 93–121. [Google Scholar] [CrossRef]
- Boisvert, F.M.; van Koningsbruggen, S.; Navascués, J.; Lamond, A.I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007, 8, 574–585. [Google Scholar] [CrossRef]
- Németh, A.; Grummt, I. Dynamic regulation of nucleolar architecture. Curr. Opin. Cell Biol. 2018, 52, 105–111. [Google Scholar] [CrossRef]
- Matsumoto, A.; Sakamoto, C.; Matsumori, H.; Katahira, J.; Yasuda, Y.; Yoshidome, K.; Tsujimoto, M.; Goldberg, I.G.; Matsuura, N.; Nakao, M.; et al. Loss of the integral nuclear envelope protein SUN1 induces alteration of nucleoli. Nucleus 2016, 7, 68–83. [Google Scholar] [CrossRef] [PubMed]
- Satomi, E.; Ueda, M.; Katahira, J.; Hieda, M. The SUN1 splicing variants SUN1_888 and SUN1_916 differentially regulate nucleolar structure. Genes Cells 2020. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, E.; Hornáček, M.; Kováčik, L.; Mazel, T.; Schröfel, A.; Svidenská, S.; Skalníková, M.; Bartová, E.; Cmarko, D.; Raška, I. Reproduction of the FC/DFC units in nucleoli. Nucleus 2016, 7, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.M.; Lamond, A.I. The nucleolus under stress. Mol. Cell 2010, 40, 216–227. [Google Scholar] [CrossRef]
- Smith, E.R.; George, S.H.; Kobetz, E.; Xu, X.X. New biological research and understanding of Papanicolaou’s test. Diagn. Cytopathol. 2018, 46, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Cremer, T.; Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001, 2, 292–301. [Google Scholar] [CrossRef]
- Chen, H.; Chen, J.; Muir, L.A.; Ronquist, S.; Meixner, W.; Ljungman, M.; Ried, T.; Smale, S.; Rajapakse, I. Functional organization of the human 4D Nucleome. Proc. Natl. Acad. Sci. USA 2015, 112, 8002–8007. [Google Scholar] [CrossRef]
- Kozubek, S.; Lukásová, E.; Rýznar, L.; Kozubek, M.; Lisková, A.; Govorun, R.D.; Krasavin, E.A.; Horneck, G. Distribution of ABL and BCR genes in cell nuclei of normal and irradiated lymphocytes. Blood 1997, 89, 4537–4545. [Google Scholar] [CrossRef]
- Parreira, L.; Telhada, M.; Ramos, C.; Hernandez, R.; Neves, H.; Carmo-Fonseca, M. The spatial distribution of human immunoglobulin genes within the nucleus: Evidence for gene topography independent of cell type and transcriptional activity. Hum. Genet. 1997, 100, 588–594. [Google Scholar] [CrossRef]
- Weimer, R.; Haaf, T.; Krüger, J.; Poot, M.; Schmid, M. Characterization of centromere arrangements and test for random distribution in G0, G1, S, G2, G1, and early S’ phase in human lymphocytes. Hum. Genet. 1992, 88, 673–682. [Google Scholar] [CrossRef]
- Guelen, L.; Pagie, L.; Brasset, E.; Meuleman, W.; Faza, M.B.; Talhout, W.; Eussen, B.H.; de Klein, A.; Wessels, L.; de Laat, W.; et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008, 453, 948–951. [Google Scholar] [CrossRef] [PubMed]
- Pickersgill, H.; Kalverda, B.; de Wit, E.; Talhout, W.; Fornerod, M.; van Steensel, B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 2006, 38, 1005–1014. [Google Scholar] [CrossRef]
- Kind, J.; Pagie, L.; de Vries, S.S.; Nahidiazar, L.; Dey, S.S.; Bienko, M.; Zhan, Y.; Lajoie, B.; de Graaf, C.A.; Amendola, M.; et al. Genome-wide maps of nuclear lamina interactions in single human cells. Cell 2015, 163, 134–147. [Google Scholar] [CrossRef]
- Maharana, S.; Iyer, K.V.; Jain, N.; Nagarajan, M.; Wang, Y.; Shivashankar, G.V. Chromosome intermingling-the physical basis of chromosome organization in differentiated cells. Nucleic Acids Res. 2016, 44, 5148–5160. [Google Scholar] [CrossRef]
- Meshorer, E.; Yellajoshula, D.; George, E.; Scambler, P.J.; Brown, D.T.; Misteli, T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell 2006, 10, 105–116. [Google Scholar] [CrossRef]
- Pajerowski, J.D.; Dahl, K.N.; Zhong, F.L.; Sammak, P.J.; Discher, D.E. Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. USA 2007, 104, 15619–15624. [Google Scholar] [CrossRef]
- Krull, S.; Dörries, J.; Boysen, B.; Reidenbach, S.; Magnius, L.; Norder, H.; Thyberg, J.; Cordes, V.C. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 2010, 29, 1659–1673. [Google Scholar] [CrossRef]
- Su, Y.; Pelz, C.; Huang, T.; Torkenczy, K.; Wang, X.; Cherry, A.; Daniel, C.J.; Liang, J.; Nan, X.; Dai, M.S.; et al. Post-translational modification localizes MYC to the nuclear pore basket to regulate a subset of target genes involved in cellular responses to environmental signals. Genes Dev. 2018, 32, 1398–1419. [Google Scholar] [CrossRef]
- Belyaeva, A.; Venkatachalapathy, S.; Nagarajan, M.; Shivashankar, G.V.; Uhler, C. Network analysis identifies chromosome intermingling regions as regulatory hotspots for transcription. Proc. Natl. Acad. Sci. USA 2017, 114, 13714–13719. [Google Scholar] [CrossRef] [PubMed]
- Boumendil, C.; Hari, P.; Olsen, K.C.F.; Acosta, J.C.; Bickmore, W.A. Nuclear pore density controls heterochromatin reorganization during senescence. Genes Dev. 2019, 33, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Underwood, J.C.E. Pathology of the Nucleus; Underwood, J.C.E., Ed.; Springer: Berlin/Heidelberg, Germany, 1990. [Google Scholar]
- Hnisz, D.; Weintraub, A.S.; Day, D.S.; Valton, A.L.; Bak, R.O.; Li, C.H.; Goldmann, J.; Lajoie, B.R.; Fan, Z.P.; Sigova, A.A.; et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 2016, 351, 1454–1458. [Google Scholar] [CrossRef]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef]
- McBeath, R.; Pirone, D.M.; Nelson, C.M.; Bhadriraju, K.; Chen, C.S. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell 2004, 6, 483–495. [Google Scholar] [CrossRef]
- Ruiz, S.A.; Chen, C.S. Emergence of patterned stem cell differentiation within multicellular structures. Stem Cells 2008, 26, 2921–2927. [Google Scholar] [CrossRef]
- Misu, S.; Takebayashi, M.; Miyamoto, K. Nuclear Actin in Development and Transcriptional Reprogramming. Front. Genet. 2017, 8, 27. [Google Scholar] [CrossRef]
- Goodson, H.V.; Jonasson, E.M. Microtubules and Microtubule-Associated Proteins. Cold Spring Harb. Perspect. Biol. 2018, 10. [Google Scholar] [CrossRef]
- Wiese, C.; Zheng, Y. A new function for the gamma-tubulin ring complex as a microtubule minus-end cap. Nat. Cell Biol. 2000, 2, 358–364. [Google Scholar] [CrossRef]
- Anders, A.; Sawin, K.E. Microtubule stabilization in vivo by nucleation-incompetent γ-tubulin complex. J. Cell Sci. 2011, 124, 1207–1213. [Google Scholar] [CrossRef]
- Bornens, M. The centrosome in cells and organisms. Science 2012, 335, 422–426. [Google Scholar] [CrossRef]
- Olins, A.L.; Olins, D.E. Cytoskeletal influences on nuclear shape in granulocytic HL-60 cells. BMC Cell Biol. 2004, 5, 30. [Google Scholar] [CrossRef]
- Peric-Hupkes, D.; Meuleman, W.; Pagie, L.; Bruggeman, S.W.; Solovei, I.; Brugman, W.; Gräf, S.; Flicek, P.; Kerkhoven, R.M.; van Lohuizen, M.; et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell 2010, 38, 603–613. [Google Scholar] [CrossRef]
- Brookes, E.; Riccio, A. Location, location, location: Nuclear structure regulates gene expression in neurons. Curr. Opin. Neurobiol. 2019, 59, 16–25. [Google Scholar] [CrossRef]
- Jacinto, F.V.; Benner, C.; Hetzer, M.W. The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev. 2015, 29, 1224–1238. [Google Scholar] [CrossRef]
- Toda, T.; Hsu, J.Y.; Linker, S.B.; Hu, L.; Schafer, S.T.; Mertens, J.; Jacinto, F.V.; Hetzer, M.W.; Gage, F.H. Nup153 Interacts with Sox2 to Enable Bimodal Gene Regulation and Maintenance of Neural Progenitor Cells. Cell Stem Cell 2017, 21, 618–634.e7. [Google Scholar] [CrossRef]
- Gu, X.; Ebrahem, Q.; Mahfouz, R.Z.; Hasipek, M.; Enane, F.; Radivoyevitch, T.; Rapin, N.; Przychodzen, B.; Hu, Z.; Balusu, R.; et al. Leukemogenic nucleophosmin mutation disrupts the transcription factor hub that regulates granulomonocytic fates. J. Clin. Investig. 2018, 128, 4260–4279. [Google Scholar] [CrossRef]
- Enane, F.O.; Shuen, W.H.; Gu, X.; Quteba, E.; Przychodzen, B.; Makishima, H.; Bodo, J.; Ng, J.; Chee, C.L.; Ba, R.; et al. GATA4 loss of function in liver cancer impedes precursor to hepatocyte transition. J. Clin. Investig. 2017, 127, 3527–3542. [Google Scholar] [CrossRef]
- Jögi, A.; Vaapil, M.; Johansson, M.; Påhlman, S. Cancer cell differentiation heterogeneity and aggressive behavior in solid tumors. Ups. J. Med. Sci. 2012, 117, 217–224. [Google Scholar] [CrossRef]
- Fletcher, D.A.; Mullins, R.D. Cell mechanics and the cytoskeleton. Nature 2010, 463, 485–492. [Google Scholar] [CrossRef]
- Gönczy, P. Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 2012, 13, 425–435. [Google Scholar] [CrossRef]
- Alvarado-Kristensson, M. Choreography of the centrosome. Heliyon 2020, 6, e03238. [Google Scholar] [CrossRef]
- Dutcher, S.K. The tubulin fraternity: Alpha to eta. Curr. Opin. Cell. Biol. 2001, 13, 49–54. [Google Scholar] [CrossRef]
- Findeisen, P.; Muhlhausen, S.; Dempewolf, S.; Hertzog, J.; Zietlow, A.; Carlomagno, T.; Kollmar, M. Six subgroups and extensive recent duplications characterize the evolution of the eukaryotic tubulin protein family. Genome Biol. Evol. 2014, 6, 2274–2288. [Google Scholar] [CrossRef]
- Eklund, G.; Lang, S.; Glindre, J.; Ehlen, A.; Alvarado-Kristensson, M. The Nuclear Localization of gamma-Tubulin Is Regulated by SadB-mediated Phosphorylation. J. Biol. Chem. 2014, 289, 21360–21373. [Google Scholar] [CrossRef]
- Alvarado-Kristensson, M. γ-tubulin as a signal-transducing molecule and meshwork with therapeutic potential. Signal Transduct. Target Ther. 2018, 3, 24. [Google Scholar] [CrossRef]
- Yuba-Kubo, A.; Kubo, A.; Hata, M.; Tsukita, S. Gene knockout analysis of two gamma-tubulin isoforms in mice. Dev. Biol. 2005, 282, 361–373. [Google Scholar] [CrossRef]
- Höög, G.; Zarrizi, R.; von Stedingk, K.; Jonsson, K.; Alvarado-Kristensson, M. Nuclear localization of γ-tubulin affects E2F transcriptional activity and S-phase progression. FASEB J. 2011, 25, 3815–3827. [Google Scholar] [CrossRef]
- Hehnly, H.; Doxsey, S. Rab11 endosomes contribute to mitotic spindle organization and orientation. Dev. Cell 2014, 28, 497–507. [Google Scholar] [CrossRef]
- Draberova, E.; Sulimenko, V.; Vinopal, S.; Sulimenko, T.; Sladkova, V.; D’Agostino, L.; Sobol, M.; Hozak, P.; Kren, L.; Katsetos, C.D.; et al. Differential expression of human gamma-tubulin isotypes during neuronal development and oxidative stress points to a gamma-tubulin-2 prosurvival function. FASEB J. 2017, 31, 1828–1846. [Google Scholar] [CrossRef]
- Rios, R.M.; Sanchis, A.; Tassin, A.M.; Fedriani, C.; Bornens, M. GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell 2004, 118, 323–335. [Google Scholar] [CrossRef]
- Moritz, M.; Zheng, Y.; Alberts, B.M.; Oegema, K. Recruitment of the gamma-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J. Cell Biol. 1998, 142, 775–786. [Google Scholar] [CrossRef]
- Moritz, M.; Braunfeld, M.B.; Sedat, J.W.; Alberts, B.; Agard, D.A. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature 1995, 378, 638–640. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wong, M.L.; Alberts, B.; Mitchison, T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature 1995, 378, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Alvarado-Kristensson, M. A simple and fast method for fixation of cultured cell lines that preserves cellular structures containing gamma-tubulin. MethodsX 2018, 5, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Pouchucq, L.; Lobos-Ruiz, P.; Araya, G.; Valpuesta, J.M.; Monasterio, O. The chaperonin CCT promotes the formation of fibrillar aggregates of γ-tubulin. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 519–526. [Google Scholar] [CrossRef]
- Farina, F.; Gaillard, J.; Guérin, C.; Couté, Y.; Sillibourne, J.; Blanchoin, L.; Théry, M. The centrosome is an actin-organizing centre. Nat. Cell Biol. 2016, 18, 65–75. [Google Scholar] [CrossRef]
- Trevor, K.T.; McGuire, J.G.; Leonova, E.V. Association of vimentin intermediate filaments with the centrosome. J. Cell Sci. 1995, 108, 343–356. [Google Scholar]
- Yokoyama, H.; Koch, B.; Walczak, R.; Ciray-Duygu, F.; Gonzalez-Sanchez, J.C.; Devos, D.P.; Mattaj, I.W.; Gruss, O.J. The nucleoporin MEL-28 promotes RanGTP-dependent gamma-tubulin recruitment and microtubule nucleation in mitotic spindle formation. Nat. Commun. 2014, 5, 3270. [Google Scholar] [CrossRef]
- Hořejší, B.; Vinopal, S.; Sládková, V.; Dráberová, E.; Sulimenko, V.; Sulimenko, T.; Vosecká, V.; Philimonenko, A.; Hozák, P.; Katsetos, C.D.; et al. Nuclear γ-tubulin associates with nucleoli and interacts with tumor suppressor protein C53. J. Cell. Physiol. 2012, 227, 367–382. [Google Scholar] [CrossRef]
- Andersen, J.S.; Lyon, C.E.; Fox, A.H.; Leung, A.K.; Lam, Y.W.; Steen, H.; Mann, M.; Lamond, A.I. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002, 12, 1–11. [Google Scholar] [CrossRef]
- Kállai, B.M.; Kourová, H.; Chumová, J.; Papdi, C.; Trögelová, L.; Kofroňová, O.; Hozák, P.; Filimonenko, V.; Mészáros, T.; Magyar, Z.; et al. γ-Tubulin interacts with E2F transcription factors to regulate proliferation and endocycling in Arabidopsis. J. Exp. Bot. 2020, 71, 1265–1277. [Google Scholar] [CrossRef]
- Lindström, L.; Villoutreix, B.O.; Lehn, S.; Hellsten, R.; Nilsson, E.; Crneta, E.; Olsson, R.; Alvarado-Kristensson, M. Therapeutic Targeting of Nuclear γ-Tubulin in RB1-Negative Tumors. Mol. Cancer Res. 2015, 13, 1073–1082. [Google Scholar] [CrossRef]
- Larsson, V.J.; Jafferali, M.H.; Vijayaraghavan, B.; Figueroa, R.A.; Hallberg, E. Mitotic spindle assembly and gamma-tubulin localisation depend on the integral nuclear membrane protein Samp1. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef] [PubMed]
- Gudise, S.; Figueroa, R.A.; Lindberg, R.; Larsson, V.; Hallberg, E. Samp1 is functionally associated with the LINC complex and A-type lamina networks. J. Cell Sci. 2011, 124, 2077–2085. [Google Scholar] [CrossRef] [PubMed]
- Lesca, C.; Germanier, M.; Raynaud-Messina, B.; Pichereaux, C.; Etievant, C.; Emond, S.; Burlet-Schiltz, O.; Monsarrat, B.; Wright, M.; Defais, M. DNA damage induce gamma-tubulin-RAD51 nuclear complexes in mammalian cells. Oncogene 2005, 24, 5165–5172. [Google Scholar] [CrossRef] [PubMed]
- Korver, W.; Guevara, C.; Chen, Y.; Neuteboom, S.; Bookstein, R.; Tavtigian, S.; Lees, E. The product of the candidate prostate cancer susceptibility gene ELAC2 interacts with the gamma-tubulin complex. Int. J. Cancer 2003, 104, 283–288. [Google Scholar] [CrossRef]
- Ehlen, A.; Rossello, C.A.; von Stedingk, K.; Hoog, G.; Nilsson, E.; Pettersson, H.M.; Jirstrom, K.; Alvarado-Kristensson, M. Tumors with nonfunctional retinoblastoma protein are killed by reduced gamma-tubulin levels. J. Biol. Chem. 2012, 287, 17241–17247. [Google Scholar] [CrossRef]
- Draberova, E.; D’Agostino, L.; Caracciolo, V.; Sladkova, V.; Sulimenko, T.; Sulimenko, V.; Sobol, M.; Maounis, N.F.; Tzelepis, E.; Mahera, E.; et al. Overexpression and Nucleolar Localization of gamma-Tubulin Small Complex Proteins GCP2 and GCP3 in Glioblastoma. J. Neuropathol. Exp. Neurol. 2015, 74, 723–742. [Google Scholar] [CrossRef]
- Alvarado-Kristensson, M.; Rodriguez, M.J.; Silio, V.; Valpuesta, J.M.; Carrera, A.C. SADB phosphorylation of gamma-tubulin regulates centrosome duplication. Nat. Cell Biol. 2009, 11, 1081–1092. [Google Scholar] [CrossRef]
- Carrera, A.C.; Alvarado-Kristensson, M. SADB kinases license centrosome replication. Cell Cycle 2009, 8, 4005–4006. [Google Scholar] [CrossRef] [PubMed]
- Lowe, J.; Li, H.; Downing, K.H.; Nogales, E. Refined structure of alpha beta-tubulin at 3.5 a resolution. J. Mol. Biol. 2001, 313, 1045–1057. [Google Scholar] [CrossRef]
- Aldaz, H.; Rice, L.M.; Stearns, T.; Agard, D.A. Insights into microtubule nucleation from the crystal structure of human gamma-tubulin. Nature 2005, 435, 523–527. [Google Scholar] [CrossRef]
- Vázquez, M.; Cooper, M.T.; Zurita, M.; Kennison, J.A. gammaTub23C interacts genetically with brahma chromatin-remodeling complexes in Drosophila melanogaster. Genetics 2008, 180, 835–843. [Google Scholar] [CrossRef]
- Meraldi, P.; Lukas, J.; Fry, A.M.; Bartek, J.; Nigg, E.A. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1999, 1, 88–93. [Google Scholar] [CrossRef]
- Brehm, A.; Miska, E.A.; McCance, D.J.; Reid, J.L.; Bannister, A.J.; Kouzarides, T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 1998, 391, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Dunaief, J.L.; Strober, B.E.; Guha, S.; Khavari, P.A.; Alin, K.; Luban, J.; Begemann, M.; Crabtree, G.R.; Goff, S.P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 1994, 79, 119–130. [Google Scholar] [CrossRef]
- Robertson, K.D.; Ait-Si-Ali, S.; Yokochi, T.; Wade, P.A.; Jones, P.L.; Wolffe, A.P. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 2000, 25, 338–342. [Google Scholar] [CrossRef]
- Hsu, L.C.; White, R.L. BRCA1 is associated with the centrosome during mitosis. Proc. Natl. Acad. Sci. USA 1998, 95, 12983–12988. [Google Scholar] [CrossRef] [PubMed]
- Hubert, T.; Vandekerckhove, J.; Gettemans, J. Cdk1 and BRCA1 target gamma-tubulin to microtubule domains. Biochem. Biophys. Res. Commun. 2011, 414, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Starita, L.M.; Machida, Y.; Sankaran, S.; Elias, J.E.; Griffin, K.; Schlegel, B.P.; Gygi, S.P.; Parvin, J.D. BRCA1-dependent ubiquitination of gamma-tubulin regulates centrosome number. Mol. Cell. Biol. 2004, 24, 8457–8466. [Google Scholar] [CrossRef]
- Zhang, S.; Hemmerich, P.; Grosse, F. Centrosomal localization of DNA damage checkpoint proteins. J. Cell. Biochem. 2007, 101, 451–465. [Google Scholar] [CrossRef]
- Morris, V.B.; Brammall, J.; Noble, J.; Reddel, R. p53 localizes to the centrosomes and spindles of mitotic cells in the embryonic chick epiblast, human cell lines, and a human primary culture: An immunofluorescence study. Exp. Cell Res. 2000, 256, 122–130. [Google Scholar] [CrossRef]
- Kanai, M.; Tong, W.M.; Sugihara, E.; Wang, Z.Q.; Fukasawa, K.; Miwa, M. Involvement of poly(ADP-Ribose) polymerase 1 and poly(ADP-Ribosyl)ation in regulation of centrosome function. Mol. Cell. Biol. 2003, 23, 2451–2462. [Google Scholar] [CrossRef]
- Chouinard, G.; Clement, I.; Lafontaine, J.; Rodier, F.; Schmitt, E. Cell cycle-dependent localization of CHK2 at centrosomes during mitosis. Cell Div. 2013, 8, 7. [Google Scholar] [CrossRef]
- Rasala, B.A.; Orjalo, A.V.; Shen, Z.; Briggs, S.; Forbes, D.J. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. USA 2006, 103, 17801–17806. [Google Scholar] [CrossRef]
- Batzenschlager, M.; Masoud, K.; Janski, N.; Houlne, G.; Herzog, E.; Evrard, J.L.; Baumberger, N.; Erhardt, M.; Nomine, Y.; Kieffer, B.; et al. The GIP gamma-tubulin complex-associated proteins are involved in nuclear architecture in Arabidopsis thaliana. Front. Plant Sci. 2013, 4, 480. [Google Scholar] [CrossRef]
- Xue, J.Z.; Woo, E.M.; Postow, L.; Chait, B.T.; Funabiki, H. Chromatin-bound Xenopus Dppa2 shapes the nucleus by locally inhibiting microtubule assembly. Dev. Cell 2013, 27, 47–59. [Google Scholar] [CrossRef]
- Zheng, L.; Cardaci, S.; Jerby, L.; MacKenzie, E.D.; Sciacovelli, M.; Johnson, T.I.; Gaude, E.; King, A.; Leach, J.D.; Edrada-Ebel, R.; et al. Fumarate induces redox-dependent senescence by modifying glutathione metabolism. Nat. Commun. 2015, 6, 6001. [Google Scholar] [CrossRef]
- Ivanova, E.L.; Gilet, J.G.; Sulimenko, V.; Duchon, A.; Rudolf, G.; Runge, K.; Collins, S.C.; Asselin, L.; Broix, L.; Drouot, N.; et al. TUBG1 missense variants underlying cortical malformations disrupt neuronal locomotion and microtubule dynamics but not neurogenesis. Nat. Commun. 2019, 10, 2129. [Google Scholar] [CrossRef]
- Poirier, K.; Lebrun, N.; Broix, L.; Tian, G.; Saillour, Y.; Boscheron, C.; Parrini, E.; Valence, S.; Pierre, B.S.; Oger, M.; et al. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat. Genet. 2013, 45, 639–647. [Google Scholar] [CrossRef]
- Brock, S.; Stouffs, K.; Scalais, E.; D’Hooghe, M.; Keymolen, K.; Guerrini, R.; Dobyns, W.B.; Di Donato, N.; Jansen, A.C. Tubulinopathies continued: Refining the phenotypic spectrum associated with variants in TUBG1. Eur. J. Hum. Genet. 2018, 26, 1132–1142. [Google Scholar] [CrossRef]
- Stiess, M.; Maghelli, N.; Kapitein, L.C.; Gomis-Rüth, S.; Wilsch-Bräuninger, M.; Hoogenraad, C.C.; Tolić-Nørrelykke, I.M.; Bradke, F. Axon extension occurs independently of centrosomal microtubule nucleation. Science 2010, 327, 704–707. [Google Scholar] [CrossRef]
- Sánchez-Huertas, C.; Freixo, F.; Viais, R.; Lacasa, C.; Soriano, E.; Lüders, J. Non-centrosomal nucleation mediated by augmin organizes microtubules in post-mitotic neurons and controls axonal microtubule polarity. Nat. Commun. 2016, 7, 12187. [Google Scholar] [CrossRef]
- Müller, H.; Fogeron, M.L.; Lehmann, V.; Lehrach, H.; Lange, B.M. A centrosome-independent role for gamma-TuRC proteins in the spindle assembly checkpoint. Science 2006, 314, 654–657. [Google Scholar] [CrossRef]
- Yang, R.; Feldman, J.L. SPD-2/CEP192 and CDK Are Limiting for Microtubule-Organizing Center Function at the Centrosome. Curr. Biol. 2015, 25, 1924–1931. [Google Scholar] [CrossRef]
- Muroyama, A.; Seldin, L.; Lechler, T. Divergent regulation of functionally distinct γ-tubulin complexes during differentiation. J. Cell Biol. 2016, 213, 679–692. [Google Scholar] [CrossRef]
- Pimenta-Marques, A.; Bento, I.; Lopes, C.A.; Duarte, P.; Jana, S.C.; Bettencourt-Dias, M. A mechanism for the elimination of the female gamete centrosome in Drosophila melanogaster. Science 2016, 353, aaf4866. [Google Scholar] [CrossRef]
- Sen, G.L.; Reuter, J.A.; Webster, D.E.; Zhu, L.; Khavari, P.A. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 2010, 463, 563–567. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, M.H.; Wu, X.; Kodani, A.; Fan, J.; Doan, R.; Ozawa, M.; Ma, J.; Yoshida, N.; Reiter, J.F.; et al. Cell-Type-Specific Alternative Splicing Governs Cell Fate in the Developing Cerebral Cortex. Cell 2016, 166, 1147–1162.e15. [Google Scholar] [CrossRef]
- Zhao, T.; Graham, O.S.; Raposo, A.; St Johnston, D. Growing microtubules push the oocyte nucleus to polarize the Drosophila dorsal-ventral axis. Science 2012, 336, 999–1003. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, E.S.; Nambiar, R.; Rosario, S.R.; Smiraglia, D.J.; Goodrich, D.W.; Witkiewicz, A.K. Pan-cancer molecular analysis of the RB tumor suppressor pathway. Commun. Biol. 2020, 3, 158. [Google Scholar] [CrossRef]
- Knudsen, E.S.; Wang, J.Y. Targeting the RB-pathway in cancer therapy. Clin. Cancer Res. 2010, 16, 1094–1099. [Google Scholar] [CrossRef] [PubMed]
- Katsetos, C.D.; Reddy, G.; Dráberová, E.; Smejkalová, B.; Del Valle, L.; Ashraf, Q.; Tadevosyan, A.; Yelin, K.; Maraziotis, T.; Mishra, O.P.; et al. Altered cellular distribution and subcellular sorting of gamma-tubulin in diffuse astrocytic gliomas and human glioblastoma cell lines. J. Neuropathol. Exp. Neurol. 2006, 65, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Caracciolo, V.; D’Agostino, L.; Dráberová, E.; Sládková, V.; Crozier-Fitzgerald, C.; Agamanolis, D.P.; de Chadarévian, J.P.; Legido, A.; Giordano, A.; Dráber, P.; et al. Differential expression and cellular distribution of gamma-tubulin and betaIII-tubulin in medulloblastomas and human medulloblastoma cell lines. J. Cell Physiol. 2010, 223, 519–529. [Google Scholar] [CrossRef]
- Johnson, I.S.; Armstrong, J.G.; Gorman, M.; Burnett, J.P. The vinca alkaloids: A new class of oncolytic agents. Cancer Res. 1963, 23, 1390–1427. [Google Scholar]
- Zhou, J.; Giannakakou, P. Targeting microtubules for cancer chemotherapy. Curr. Med. Chem. Anticancer Agents 2005, 5, 65–71. [Google Scholar] [CrossRef]
- Hotchkiss, K.A.; Ashton, A.W.; Mahmood, R.; Russell, R.G.; Sparano, J.A.; Schwartz, E.L. Inhibition of endothelial cell function in vitro and angiogenesis in vivo by docetaxel (Taxotere): Association with impaired repositioning of the microtubule organizing center. Mol. Cancer Ther. 2002, 1, 1191–1200. [Google Scholar]
- Chinen, T.; Liu, P.; Shioda, S.; Pagel, J.; Cerikan, B.; Lin, T.C.; Gruss, O.; Hayashi, Y.; Takeno, H.; Shima, T.; et al. The γ-tubulin-specific inhibitor gatastatin reveals temporal requirements of microtubule nucleation during the cell cycle. Nat. Commun. 2015, 6, 8722. [Google Scholar] [CrossRef]
- Chaimovitsh, D.; Abu-Abied, M.; Belausov, E.; Rubin, B.; Dudai, N.; Sadot, E. Microtubules are an intracellular target of the plant terpene citral. Plant J. Cell Mol. Biol. 2010, 61, 399–408. [Google Scholar] [CrossRef]
- Linker, R.A.; Gold, R. Dimethyl fumarate for treatment of multiple sclerosis: Mechanism of action, effectiveness, and side effects. Curr. Neurol. Neurosci. Rep. 2013, 13, 394. [Google Scholar] [CrossRef] [PubMed]
- Mrowietz, U.; Asadullah, K. Dimethylfumarate for psoriasis: More than a dietary curiosity. Trends Mol. Med. 2005, 11, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Loewe, R.; Valero, T.; Kremling, S.; Pratscher, B.; Kunstfeld, R.; Pehamberger, H.; Petzelbauer, P. Dimethylfumarate impairs melanoma growth and metastasis. Cancer Res. 2006, 66, 11888–11896. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corvaisier, M.; Alvarado-Kristensson, M. Non-Canonical Functions of the Gamma-Tubulin Meshwork in the Regulation of the Nuclear Architecture. Cancers 2020, 12, 3102. https://doi.org/10.3390/cancers12113102
Corvaisier M, Alvarado-Kristensson M. Non-Canonical Functions of the Gamma-Tubulin Meshwork in the Regulation of the Nuclear Architecture. Cancers. 2020; 12(11):3102. https://doi.org/10.3390/cancers12113102
Chicago/Turabian StyleCorvaisier, Matthieu, and Maria Alvarado-Kristensson. 2020. "Non-Canonical Functions of the Gamma-Tubulin Meshwork in the Regulation of the Nuclear Architecture" Cancers 12, no. 11: 3102. https://doi.org/10.3390/cancers12113102
APA StyleCorvaisier, M., & Alvarado-Kristensson, M. (2020). Non-Canonical Functions of the Gamma-Tubulin Meshwork in the Regulation of the Nuclear Architecture. Cancers, 12(11), 3102. https://doi.org/10.3390/cancers12113102





