Coordinated Expression of Ras Suppressor 1 (RSU-1) and Growth Differentiation Factor 15 (GDF15) Affects Glioma Cell Invasion
Abstract
1. Introduction
2. Results
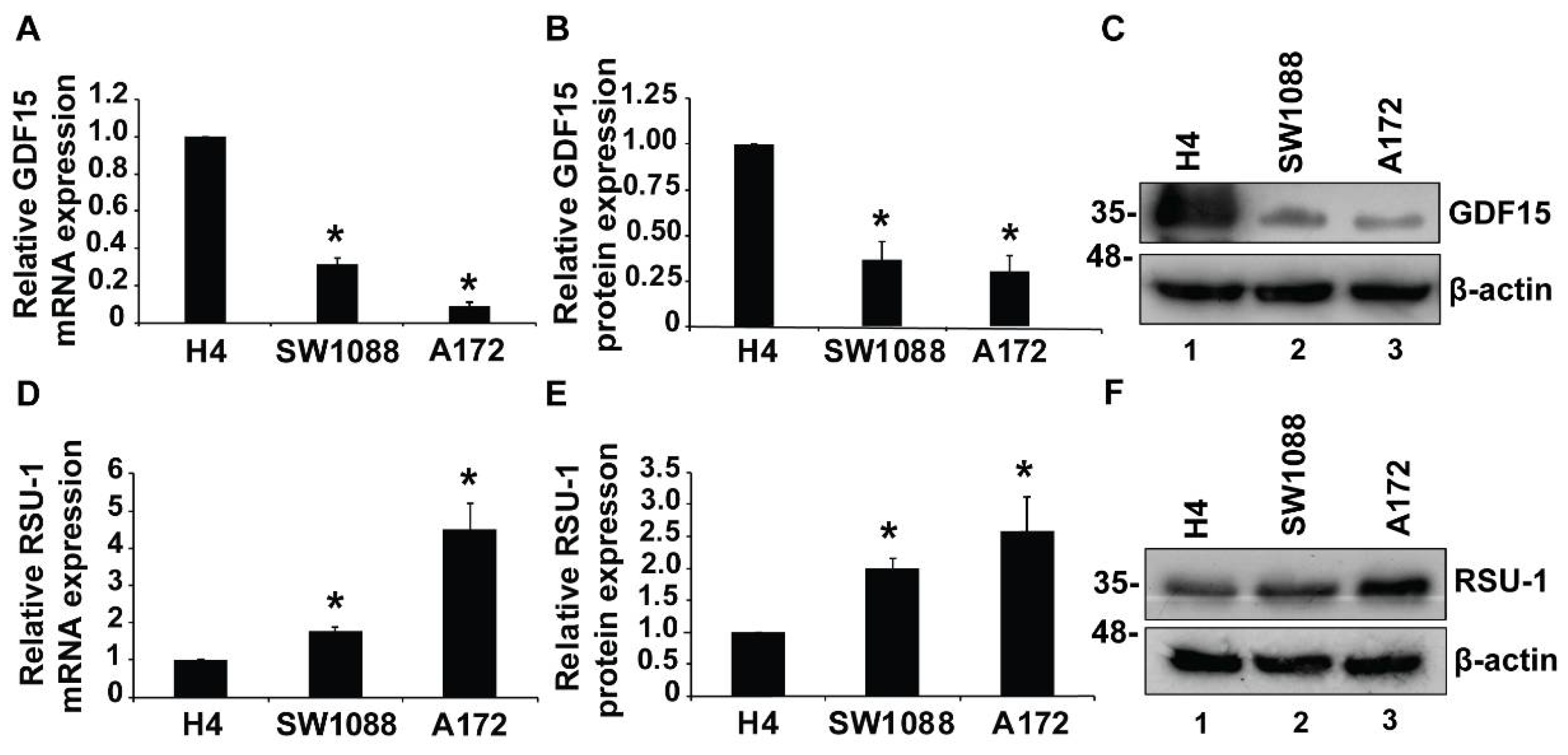
2.1. Growth Differentiation Factor 15 (GDF15) mRNA Expression is Reduced in More Aggressive Glioma Cells
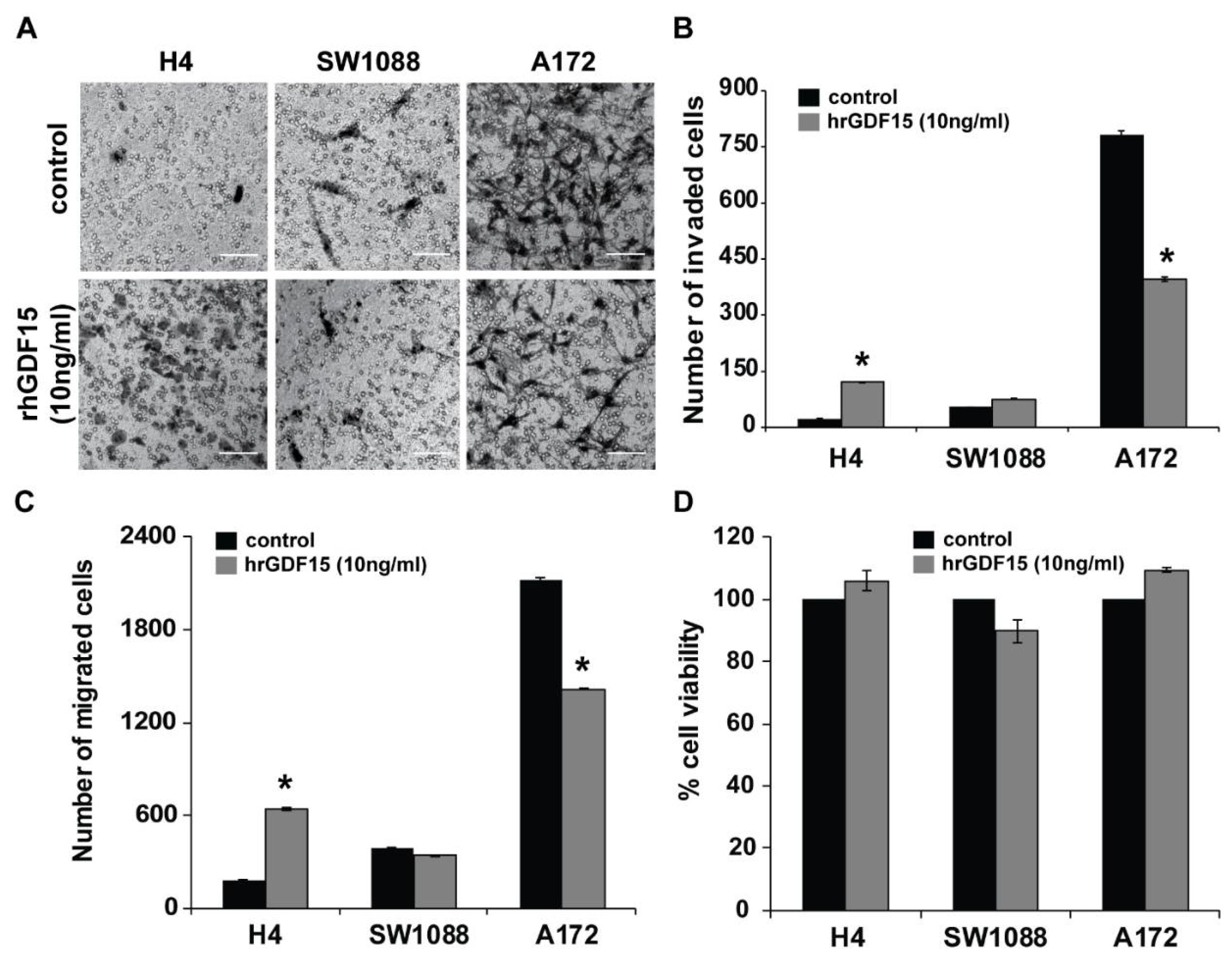
2.2. Human Recombinant GDF15 (hrGDF15) Treatment Protein Differentially Affects Motility and Invasive Capacity of Cells Depending on Cell Aggressiveness
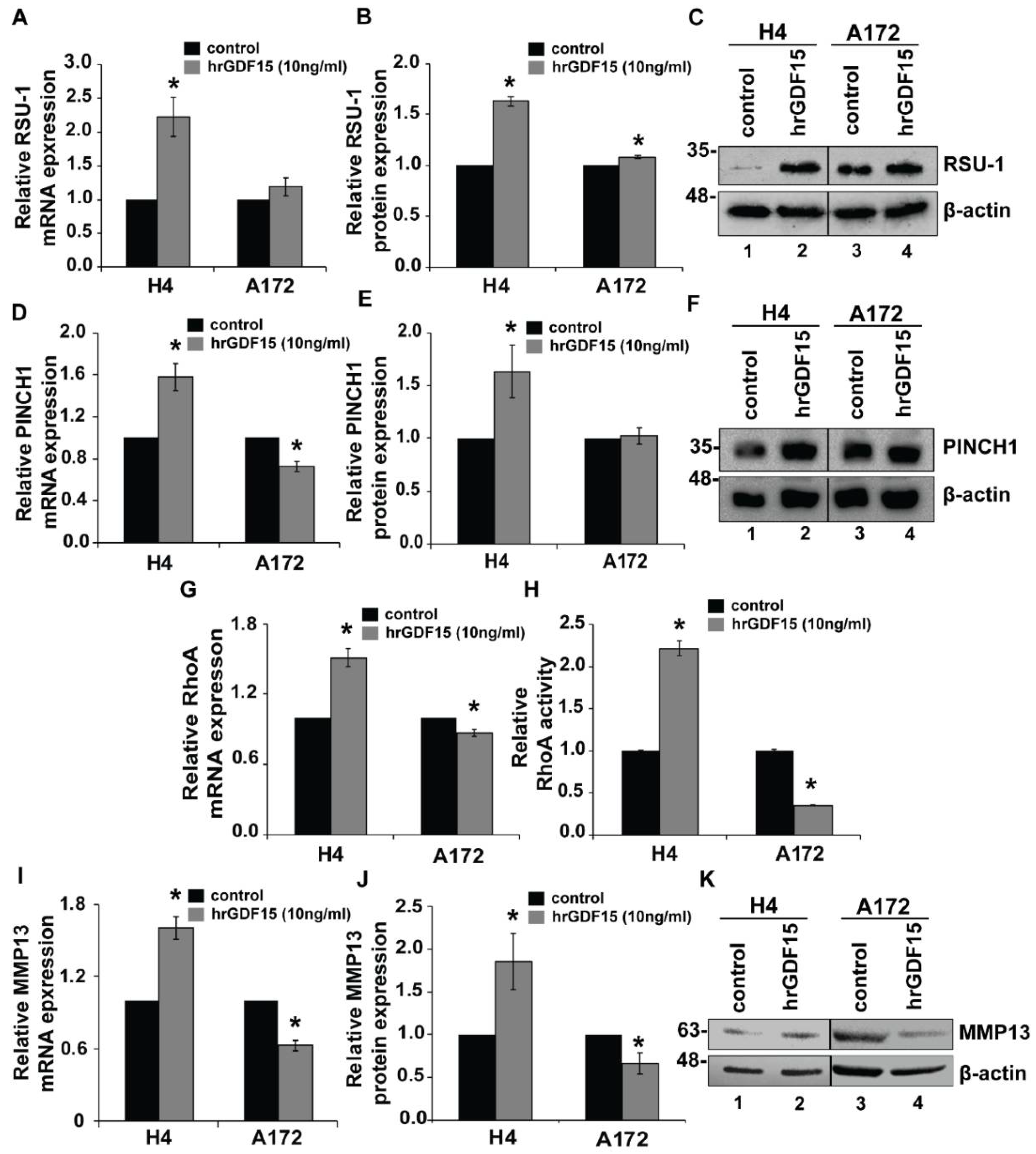
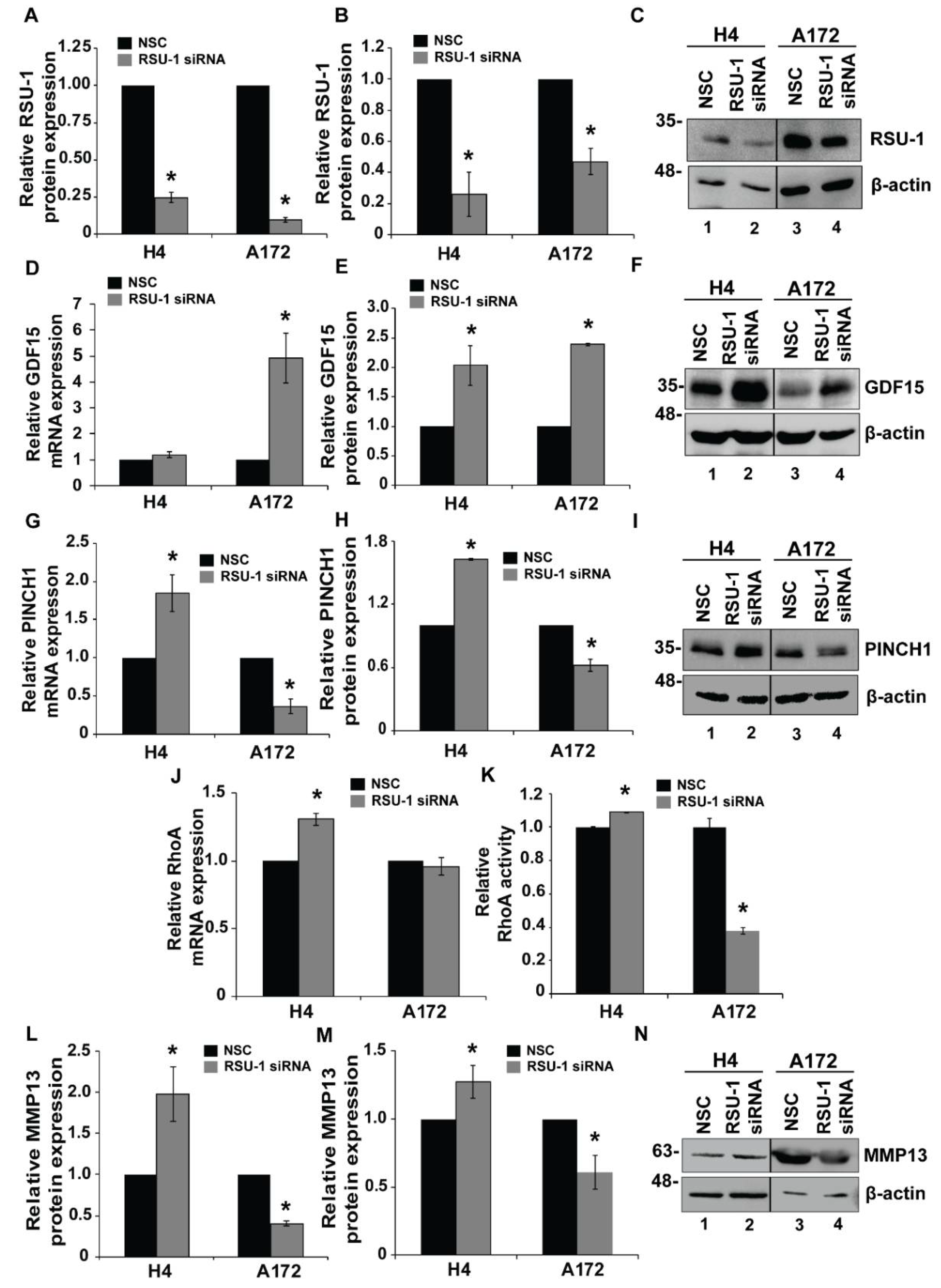
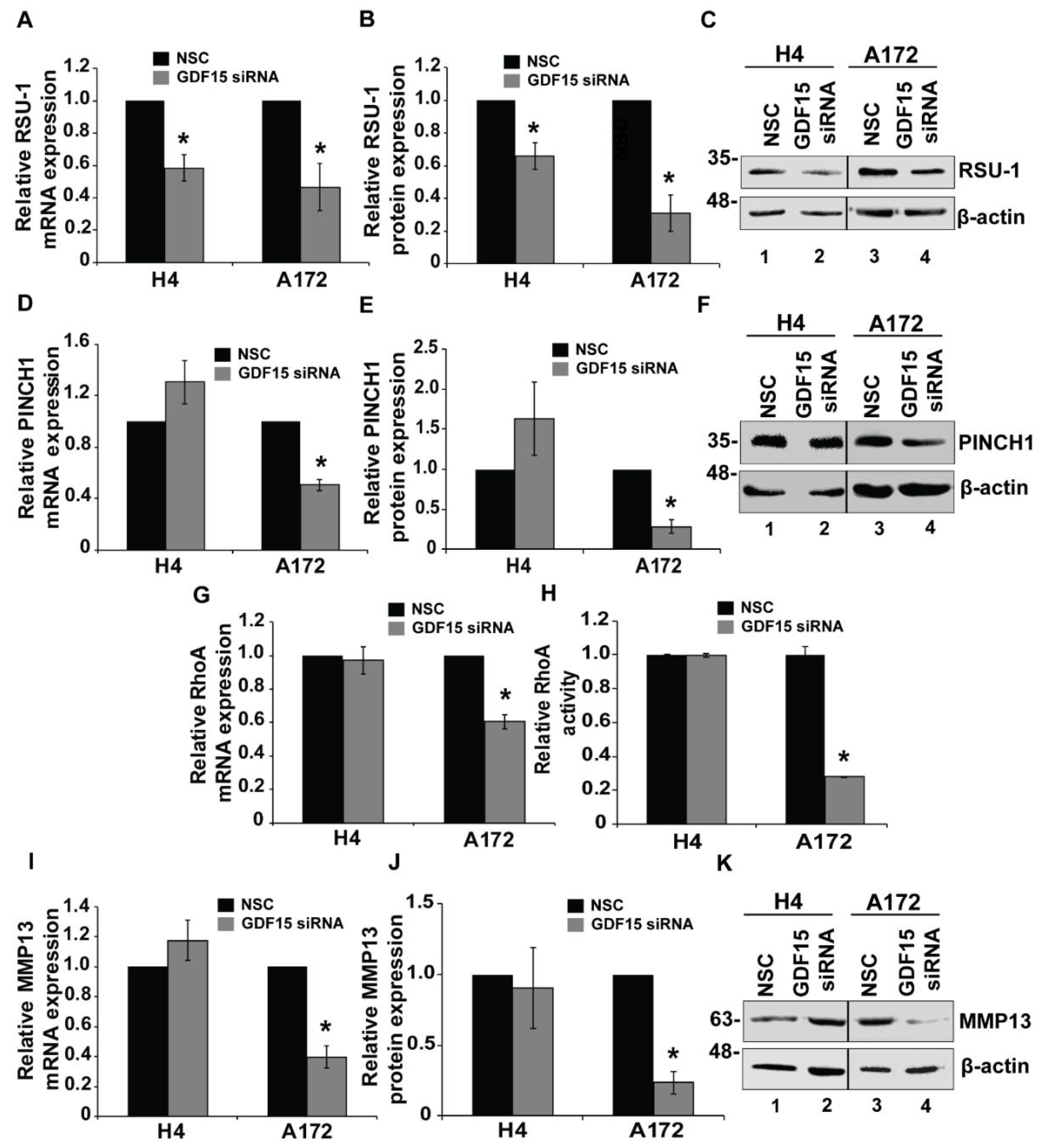
2.3. RSU-1 Silencing Regulates GDF15 Expression and Differentially Affectscell Invasiveness and the Expression of PINCH1, RhoA and MMP13
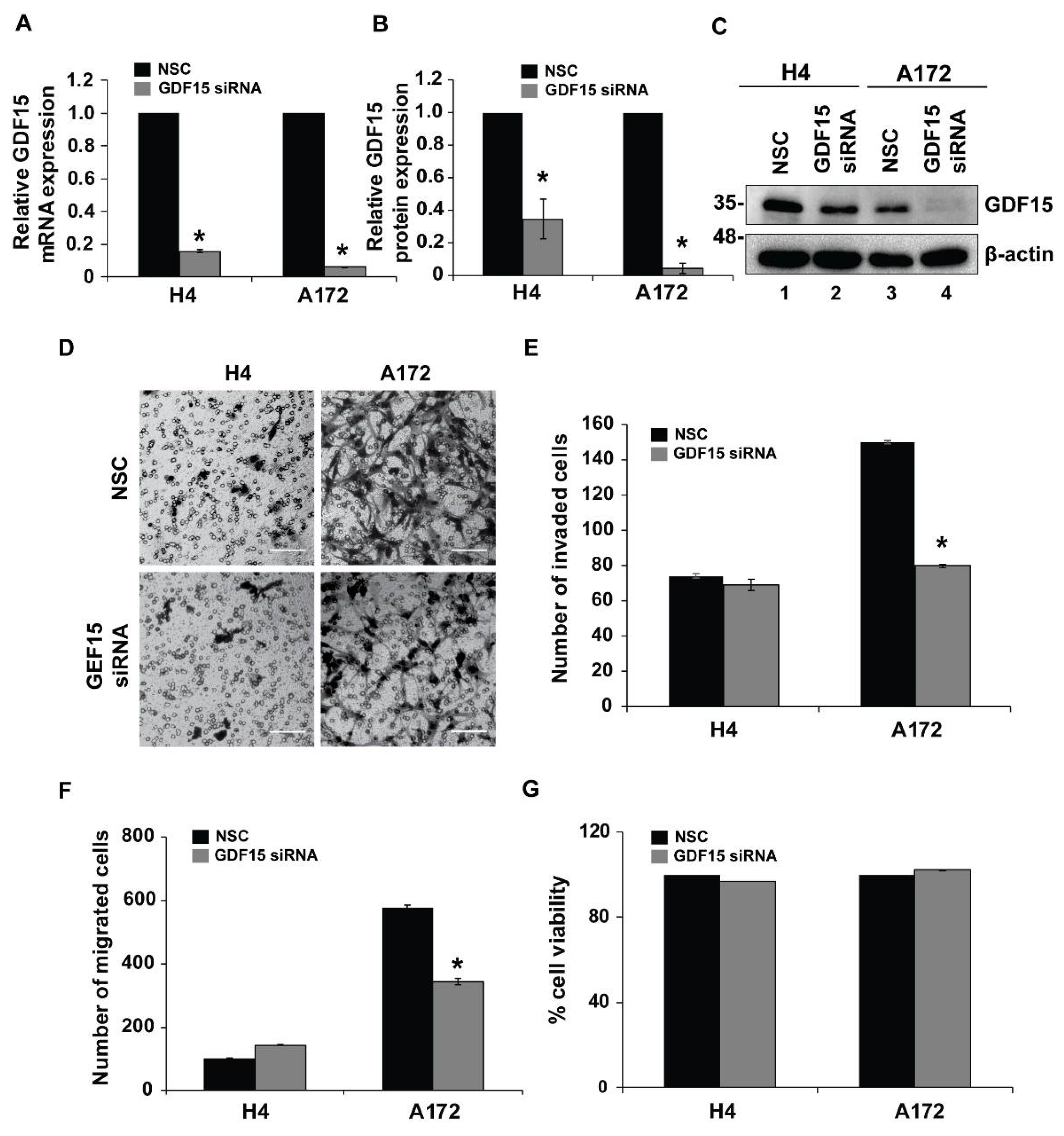
2.4. GDF15 Silencing Leads to Reduced Invasion in More Invasive Cells but Does Not Affect Less Invasive Cells
3. Discussion
4. Materials and Methods
4.1. Glioma Cell Lines
4.2. Antibodies and Reagents
4.3. Transwell Migration and Invasion Assays
4.4. Tumor Spheroids Formation in Collagen Gels
4.5. Transfection with siRNA
4.6. Treatment with hrGDF15
4.7. Cell Viability Assay
4.8. RNA Isolation and Real-Time Polymerase Chain Reaction (PCR)
4.9. Protein Extraction and Western Blotting
4.10. RhoA Activation Assay
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- De Groot, J.F.; Fuller, G.; Kumar, A.J.; Piao, Y.; Eterovic, K.; Ji, Y.; Conrad, C.A. Tumor invasion after treatment of glioblastoma with bevacizumab: Radiographic and pathologic correlation in humans and mice. Neuro Oncol. 2010, 12, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.R.; O’Neill, B.P. Glioblastoma survival in the United States before and during the temozolomide era. J. Neuro-Oncol. 2012, 107, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Buckner, J.C.; Brown, P.D.; O’Neill, B.P.; Meyer, F.B.; Wetmore, C.J.; Uhm, J.H. Central nervous system tumors. Mayo Clin. Proc. 2007, 82, 1271–1286. [Google Scholar] [CrossRef] [PubMed]
- Kleihues, P.; Sobin, L.H. World Health Organization classification of tumors. Cancer 2000, 88, 2887. [Google Scholar] [CrossRef]
- Ma, Y.S.; Wu, Z.J.; Bai, R.Z.; Dong, H.; Xie, B.X.; Wu, X.H.; Hang, X.S.; Liu, A.N.; Jiang, X.H.; Wang, G.R.; et al. DRR1 promotes glioblastoma cell invasion and epithelial-mesenchymal transition via regulating AKT activation. Cancer Lett. 2018, 423, 86–94. [Google Scholar] [CrossRef]
- Belot, N.; Rorive, S.; Doyen, I.; Lefranc, F.; Bruyneel, E.; Dedecker, R.; Micik, S.; Brotchi, J.; Decaestecker, C.; Salmon, I.; et al. Molecular characterization of cell substratum attachments in human glial tumors relates to prognostic features. Glia 2001, 36, 375–390. [Google Scholar] [CrossRef]
- Sun, L.H.; Yang, F.Q.; Zhang, C.B.; Wu, Y.P.; Liang, J.S.; Jin, S.; Wang, Z.; Wang, H.J.; Bao, Z.S.; Yang, Z.X.; et al. Overexpression of Paxillin Correlates with Tumor Progression and Predicts Poor Survival in Glioblastoma. CNS Neurosci. Ther. 2017, 23, 69–75. [Google Scholar] [CrossRef]
- Zaidel-Bar, R.; Itzkovitz, S.; Ma’ayan, A.; Iyengar, R.; Geiger, B. Functional atlas of the integrin adhesome. Nat. Cell Biol. 2007, 9, 858–867. [Google Scholar] [CrossRef]
- Bokel, C.; Brown, N.H. Integrins in development: Moving on, responding to, and sticking to the extracellular matrix. Dev. Cell 2002, 3, 311–321. [Google Scholar] [CrossRef]
- Hoffmann, M.; Schwarz, U.S. A kinetic model for RNA-interference of focal adhesions. BMC Syst. Biol. 2013, 7. [Google Scholar] [CrossRef]
- Gkretsi, V.; Stylianopoulos, T. Cell Adhesion and Matrix Stiffness: Coordinating Cancer Cell Invasion and Metastasis. Front. Oncol. 2018, 8, 145. [Google Scholar] [CrossRef] [PubMed]
- Izdebska, M.; Zielinska, W.; Grzanka, D.; Gagat, M. The Role of Actin Dynamics and Actin-Binding Proteins Expression in Epithelial-to-Mesenchymal Transition and Its Association with Cancer Progression and Evaluation of Possible Therapeutic Targets. BioMed Res. Int. 2018, 2018, 4578373. [Google Scholar] [CrossRef] [PubMed]
- Cutler, M.L.; Bassin, R.H.; Zanoni, L.; Talbot, N. Isolation of Rsp-1, a Novel Cdna Capable of Suppressing V-Ras Transformation. Mol. Cell. Biol. 1992, 12, 3750–3756. [Google Scholar] [CrossRef] [PubMed]
- Masuelli, L.; Cutler, M.L. Increased expression of the Ras suppressor Rsu-1 enhances Erk-2 activation and inhibits Jun kinase activation. Mol. Cell. Biol. 1996, 16, 5466–5476. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tsuda, T.; Cutler, M.L. Human RSU1 is highly homologous to mouse Rsu-1 and localizes to human chromosome 10. Genomics 1993, 18, 461–462. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, T.; Marinetti, M.R.; Masuelli, L.; Cutler, M.L. The Ras suppressor RSU-1 localizes to 10p13 and its expression in the U251 glioblastoma cell line correlates with a decrease in growth rate and tumorigenic potential. Oncogene 1995, 11, 397–403. [Google Scholar]
- Vasaturo, F.; Dougherty, G.W.; Cutler, M.L. Ectopic expression of Rsu-1 results in elevation of p21CIP and inhibits anchorage-independent growth of MCF7 breast cancer cells. Breast Cancer Res. Treat. 2000, 61, 69–78. [Google Scholar] [CrossRef]
- Donthamsetty, S.; Bhave, V.S.; Mars, W.M.; Bowen, W.C.; Orr, A.; Haynes, M.M.; Wu, C.; Michalopoulos, G.K. Role of PINCH and its partner tumor suppressor Rsu-1 in regulating liver size and tumorigenesis. PLoS ONE 2013, 8, e74625. [Google Scholar] [CrossRef]
- Dougherty, G.W.; Chopp, T.; Qi, S.; Cutler, M.L. The Ras suppressor Rsu-1 binds to the LIM 5 domain of the adaptor protein PINCH1 and participates in adhesion-related functions. Exp. Cell Res. 2005, 306, 168–179. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, K.; Tu, Y.; Velyvis, A.; Yang, Y.; Qin, J.; Wu, C. Assembly of the PINCH-ILK-CH-ILKBP complex precedes and is essential for localization of each component to cell-matrix adhesion sites. J. Cell Sci. 2002, 115, 4777–4786. [Google Scholar] [CrossRef]
- Wu, C. The PINCH-ILK-parvin complexes: Assembly, functions and regulation. Biochim. Biophys. Acta 2004, 1692, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Wu, C. ILK: A pseudokinase in the center stage of cell-matrix adhesion and signaling. Curr. Opin. Cell Biol. 2012, 24, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Nobes, C.D.; Hall, A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 1995, 81, 53–62. [Google Scholar] [CrossRef]
- Bar-Sagi, D.; Hall, A. Ras and Rho GTPases: A family reunion. Cell 2000, 103, 227–238. [Google Scholar] [CrossRef]
- Fortin Ensign, S.P.; Mathews, I.T.; Symons, M.H.; Berens, M.E.; Tran, N.L. Implications of Rho GTPase Signaling in Glioma Cell Invasion and Tumor Progression. Front. Oncol. 2013, 3, 241. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.B.; Li, X.M.; Dixon, M.P.; Blagoi, E.L.; Nicolas, E.; Seeholzer, S.H.; Cheng, D.; He, Y.A.; Coudry, R.A.; Howard, S.D.; et al. APC +/− alters colonic fibroblast proteome in FAP. Oncotarget 2011, 2, 197–208. [Google Scholar] [CrossRef]
- Gonzalez-Nieves, R.; Desantis, A.I.; Cutler, M.L. Rsu1 contributes to regulation of cell adhesion and spreading by PINCH1-dependent and—Independent mechanisms. J. Cell Commun. Signal. 2013, 7, 279–293. [Google Scholar] [CrossRef]
- Gkretsi, V.; Bogdanos, D.P. Elimination of Ras Suppressor-1 from hepatocellular carcinoma cells hinders their in vitro metastatic properties. Anticancer Res. 2015, 35, 1509–1512. [Google Scholar]
- Gkretsi, V.; Stylianou, A.; Louca, M.; Stylianopoulos, T. Identification of Ras suppressor-1 (RSU-1) as a potential breast cancer metastasis biomarker using a three-dimensional in vitro approach. Oncotarget 2017, 27364–27379. [Google Scholar] [CrossRef]
- Zacharia, L.C.; Stylianopoulos, T.; Gkretsi, V. Ras Suppressor-1 (RSU-1) in Cancer Cell Metastasis: Friend or Foe? Crit. Rev. Oncog. 2017, 22, 249–253. [Google Scholar] [CrossRef]
- Masuelli, L.; Ettenberg, S.; Vasaturo, F.; Vestergaard-Sykes, K.; Cutler, M.L. The ras suppressor, RSU-1, enhances nerve growth factor-induced differentiation of PC12 cells and induces p21CIP expression. Cell Growth Differ. Mol. Biol. J. Am. Assoc. Cancer Res. 1999, 10, 555–564. [Google Scholar]
- Porcheri, C.; Suter, U.; Jessberger, S. Dissecting integrin-dependent regulation of neural stem cell proliferation in the adult brain. J. Neurosci. Off. J. Soc. Neurosci. 2014, 34, 5222–5232. [Google Scholar] [CrossRef]
- Pierron, M.; Pinan-Lucarre, B.; Bessereau, J.L. Preventing Illegitimate Extrasynaptic Acetylcholine Receptor Clustering Requires the RSU-1 Protein. J. Neurosci. Off. J. Soc. Neurosci. 2016, 36, 6525–6537. [Google Scholar] [CrossRef]
- Louca, M.; Stylianou, A.; Minia, A.; Pliaka, V.; Alexopoulos, L.G.; Gkretsi, V.; Stylianopoulos, T. Ras suppressor-1 (RSU-1) promotes cell invasion in aggressive glioma cells and inhibits it in non-aggressive cells through STAT6 phospho-regulation. Sci. Rep. 2019, 9, 7782. [Google Scholar] [CrossRef]
- Roth, P.; Junker, M.; Tritschler, I.; Mittelbronn, M.; Dombrowski, Y.; Breit, S.N.; Tabatabai, G.; Wick, W.; Weller, M.; Wischhusen, J. GDF-15 contributes to proliferation and immune escape of malignant gliomas. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 3851–3859. [Google Scholar] [CrossRef]
- Hromas, R.; Hufford, M.; Sutton, J.; Xu, D.; Li, Y.; Lu, L. PLAB, a novel placental bone morphogenetic protein. Biochim. Biophys. Acta 1997, 1354, 40–44. [Google Scholar] [CrossRef]
- Li, P.X.; Wong, J.; Ayed, A.; Ngo, D.; Brade, A.M.; Arrowsmith, C.; Austin, R.C.; Klamut, H.J. Placental transforming growth factor-beta is a downstream mediator of the growth arrest and apoptotic response of tumor cells to DNA damage and p53 overexpression. J. Biol. Chem. 2000, 275, 20127–20135. [Google Scholar] [CrossRef]
- Paralkar, V.M.; Vail, A.L.; Grasser, W.A.; Brown, T.A.; Xu, H.; Vukicevic, S.; Ke, H.Z.; Qi, H.; Owen, T.A.; Thompson, D.D. Cloning and characterization of a novel member of the transforming growth factor-beta/bone morphogenetic protein family. J. Biol. Chem. 1998, 273, 13760–13767. [Google Scholar] [CrossRef]
- Baek, S.J.; Kim, K.S.; Nixon, J.B.; Wilson, L.C.; Eling, T.E. Cyclooxygenase inhibitors regulate the expression of a TGF-beta superfamily member that has proapoptotic and antitumorigenic activities. Mol. Pharmacol. 2001, 59, 901–908. [Google Scholar] [CrossRef]
- Segerer, S.E.; Rieger, L.; Kapp, M.; Dombrowski, Y.; Muller, N.; Dietl, J.; Kammerer, U. MIC-1 (a multifunctional modulator of dendritic cell phenotype and function) is produced by decidual stromal cells and trophoblasts. Hum. Reprod. 2012, 27, 200–209. [Google Scholar] [CrossRef]
- Aw Yong, K.M.; Zeng, Y.; Vindivich, D.; Phillip, J.M.; Wu, P.H.; Wirtz, D.; Getzenberg, R.H. Morphological effects on expression of growth differentiation factor 15 (GDF15), a marker of metastasis. J. Cell. Physiol. 2014, 229, 362–373. [Google Scholar] [CrossRef]
- Kalli, M.; Minia, A.; Pliaka, V.; Fotis, C.; Alexopoulos, L.G.; Stylianopoulos, T. Solid stress-induced migration is mediated by GDF15 through Akt pathway activation in pancreatic cancer cells. Sci. Rep. 2019, 9, 978. [Google Scholar] [CrossRef]
- Kalli, M.; Papageorgis, P.; Gkretsi, V.; Stylianopoulos, T. Solid Stress Facilitates Fibroblasts Activation to Promote Pancreatic Cancer Cell Migration. Ann. Biomed. Eng. 2018, 46, 657–669. [Google Scholar] [CrossRef]
- Xu, Q.; Xu, H.X.; Li, J.P.; Wang, S.; Fu, Z.; Jia, J.; Wang, L.; Zhu, Z.F.; Lu, R.; Yao, Z. Growth differentiation factor 15 induces growth and metastasis of human liver cancer stem-like cells via AKT/GSK-3beta/beta-catenin signaling. Oncotarget 2017, 8, 16972–16987. [Google Scholar] [CrossRef]
- Sasahara, A.; Tominaga, K.; Nishimura, T.; Yano, M.; Kiyokawa, E.; Noguchi, M.; Noguchi, M.; Kanauchi, H.; Ogawa, T.; Minato, H.; et al. An autocrine/paracrine circuit of growth differentiation factor (GDF) 15 has a role for maintenance of breast cancer stem-like cells. Oncotarget 2017, 8, 24869–24881. [Google Scholar] [CrossRef]
- Ji, H.; Lu, H.W.; Li, Y.M.; Lu, L.; Wang, J.L.; Zhang, Y.F.; Shang, H. Twist promotes invasion and cisplatin resistance in pancreatic cancer cells through growth differentiation factor 15. Mol. Med. Rep. 2015, 12, 3841–3848. [Google Scholar] [CrossRef]
- Wallin, U.; Glimelius, B.; Jirstrom, K.; Darmanis, S.; Nong, R.Y.; Ponten, F.; Johansson, C.; Pahlman, L.; Birgisson, H. Growth differentiation factor 15: A prognostic marker for recurrence in colorectal cancer. Br. J. Cancer 2011, 104, 1619–1627. [Google Scholar] [CrossRef]
- Senapati, S.; Rachagani, S.; Chaudhary, K.; Johansson, S.L.; Singh, R.K.; Batra, S.K. Overexpression of macrophage inhibitory cytokine-1 induces metastasis of human prostate cancer cells through the FAK-RhoA signaling pathway. Oncogene 2010, 29, 1293–1302. [Google Scholar] [CrossRef]
- Codo, P.; Weller, M.; Kaulich, K.; Schraivogel, D.; Silginer, M.; Reifenberger, G.; Meister, G.; Roth, P. Control of glioma cell migration and invasiveness by GDF-15. Oncotarget 2016, 7, 7732–7746. [Google Scholar] [CrossRef]
- Yoshioka, H.; Kamitani, H.; Watanabe, T.; Eling, T.E. Nonsteroidal anti-inflammatory drug-activated gene (NAG-1/GDF15) expression is increased by the histone deacetylase inhibitor trichostatin A. J. Biol. Chem. 2008, 283, 33129–33137. [Google Scholar] [CrossRef]
- Kadowaki, M.; Yoshioka, H.; Kamitani, H.; Watanabe, T.; Wade, P.A.; Eling, T.E. DNA methylation-mediated silencing of nonsteroidal anti-inflammatory drug-activated gene (NAG-1/GDF15) in glioma cell lines. Int. J. Cancer 2012, 130, 267–277. [Google Scholar] [CrossRef]
- Strelau, J.; Schmeer, C.; Peterziel, H.; Sackmann, T.; Herold-Mende, C.; Steiner, H.; Weller, M.; Unsicker, K. Expression and putative functions of GDF-15, a member of the TGF-beta superfamily, in human glioma and glioblastoma cell lines. Cancer Lett. 2008, 270, 30–39. [Google Scholar] [CrossRef]
- Zimmers, T.A.; Jin, X.; Gutierrez, J.C.; Acosta, C.; McKillop, I.H.; Pierce, R.H.; Koniaris, L.G. Effect of in vivo loss of GDF-15 on hepatocellular carcinogenesis. J. Cancer Res. Clin. Oncol. 2008, 134, 753–759. [Google Scholar] [CrossRef]
- Gkretsi, V.; Louca, M.; Stylianou, A.; Minadakis, G.; Spyrou, G.M.; Stylianopoulos, T. Inhibition of Breast Cancer Cell Invasion by Ras Suppressor-1 (RSU-1) Silencing Is Reversed by Growth Differentiation Factor-15 (GDF-15). Int. J. Mol. Sci. 2019, 20, 163. [Google Scholar] [CrossRef]
- Ulu, A.; Frost, J.A. Regulation of RhoA activation and cell motility by c-Jun N-terminal kinases and Net1. Small GTPases 2018, 1–7. [Google Scholar] [CrossRef]
- Law, M.E.; Templeton, K.L.; Kitange, G.; Smith, J.; Misra, A.; Feuerstein, B.G.; Jenkins, R.B. Molecular cytogenetic analysis of chromosomes 1 and 19 in glioma cell lines. Cancer Genet. Cytogenet. 2005, 160, 1–14. [Google Scholar] [CrossRef]
- Kim, T.A.; Ota, S.; Jiang, S.; Pasztor, L.M.; White, R.A.; Avraham, S. Genomic organization, chromosomal localization and regulation of expression of the neuronal nuclear matrix protein NRP/B in human brain tumors. Gene 2000, 255, 105–116. [Google Scholar] [CrossRef]
- Ridley, A.J. Rho GTPase signalling in cell migration. Curr. Opin. Cell Biol. 2015, 36, 103–112. [Google Scholar] [CrossRef]
- Gong, L.; Xia, Y.; Qian, Z.; Shi, J.; Luo, J.; Song, G.; Xu, J.; Ye, Z. Overexpression of MYC binding protein promotes invasion and migration in gastric cancer. Oncol. Lett. 2018, 15, 5243–5249. [Google Scholar] [CrossRef]
- Wang, K.; Jin, W.; Song, Y.; Fei, X. LncRNA RP11-436H11.5, functioning as a competitive endogenous RNA, upregulates BCL-W expression by sponging miR-335-5p and promotes proliferation and invasion in renal cell carcinoma. Mol. Cancer 2017, 16, 166. [Google Scholar] [CrossRef]
- Chen, L.; Li, H.; Han, L.; Zhang, K.; Wang, G.; Wang, Y.; Liu, Y.; Zheng, Y.; Jiang, T.; Pu, P.; et al. Expression and function of miR-27b in human glioma. Oncol. Rep. 2011, 26, 1617–1621. [Google Scholar] [CrossRef]
- Wen, M.; Wang, H.; Zhang, X.; Long, J.; Lv, Z.; Kong, Q.; An, Y. Cytokine-like 1 is involved in the growth and metastasis of neuroblastoma cells. Int. J. Oncol. 2012, 41, 1419–1424. [Google Scholar] [CrossRef]
- Gao, S.; Jin, L.; Liu, G.; Wang, P.; Sun, Z.; Cao, Y.; Shi, H.; Liu, X.; Shi, Q.; Zhou, X.; et al. Overexpression of RASD1 inhibits glioma cell migration/invasion and inactivates the AKT/mTOR signaling pathway. Sci. Rep. 2017, 7, 3202. [Google Scholar] [CrossRef]
- Foty, R. A simple hanging drop cell culture protocol for generation of 3D spheroids. J. Vis. Exp. 2011. [Google Scholar] [CrossRef]
- Bernstein, J.J.; Woodard, C.A. Glioblastoma cells do not intravasate into blood vessels. Neurosurgery 1995, 36, 124–132. [Google Scholar] [CrossRef]
- Dougherty, G.W.; Jose, C.; Gimona, M.; Cutler, M.L. The Rsu-1-PINCH1-ILK complex is regulated by Ras activation in tumor cells. Eur. J. Cell Biol. 2008, 87, 721–734. [Google Scholar] [CrossRef]

| Primer Name | Sequence |
|---|---|
| GDF15 | Forward: 5′- TCAAGGTCGTGGGACGTGACA-3′ Reverse: 5′-GCCGTGCGGACGAAGATTCT-3′ |
| MMP13 | Forward: 5′-TGGCATTGCTGACATCATGA-3′ Reverse: 5′-GCCAGAGGGCCCATCAA-3′ |
| PINCH1 | Forward: 5′-CCGCTGAGAAGATCGTGAAC-3′ Reverse: 5′-GGGCAAAGAGCATCTGAAAG-3′ |
| RhoA | Forward: 5′-CGGGAGCTAGCCAAGATGAAG-3′ Reverse: 5′-CCTTGCAGAGCAGCTCTCGTA-3′ |
| RSU-1 | Forward: 5′-AGGCCACAGAGCAAGGTCTA-3′ Reverse: 5′-CGTGCAATCTCAAAAGCTCA-3′ |
| β-actin | Forward:5′-CGAGCACAGAGCCTCGCCTTTGCC-3′ Reverse: 5′-TGTCGACGACGAGCGCGGCGATAT-3′ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Louca, M.; Gkretsi, V.; Stylianopoulos, T. Coordinated Expression of Ras Suppressor 1 (RSU-1) and Growth Differentiation Factor 15 (GDF15) Affects Glioma Cell Invasion. Cancers 2019, 11, 1159. https://doi.org/10.3390/cancers11081159
Louca M, Gkretsi V, Stylianopoulos T. Coordinated Expression of Ras Suppressor 1 (RSU-1) and Growth Differentiation Factor 15 (GDF15) Affects Glioma Cell Invasion. Cancers. 2019; 11(8):1159. https://doi.org/10.3390/cancers11081159
Chicago/Turabian StyleLouca, Maria, Vasiliki Gkretsi, and Triantafyllos Stylianopoulos. 2019. "Coordinated Expression of Ras Suppressor 1 (RSU-1) and Growth Differentiation Factor 15 (GDF15) Affects Glioma Cell Invasion" Cancers 11, no. 8: 1159. https://doi.org/10.3390/cancers11081159
APA StyleLouca, M., Gkretsi, V., & Stylianopoulos, T. (2019). Coordinated Expression of Ras Suppressor 1 (RSU-1) and Growth Differentiation Factor 15 (GDF15) Affects Glioma Cell Invasion. Cancers, 11(8), 1159. https://doi.org/10.3390/cancers11081159





