Microdroplet Systems for Gene Transfer: From Fundamentals to Future Perspectives
Abstract
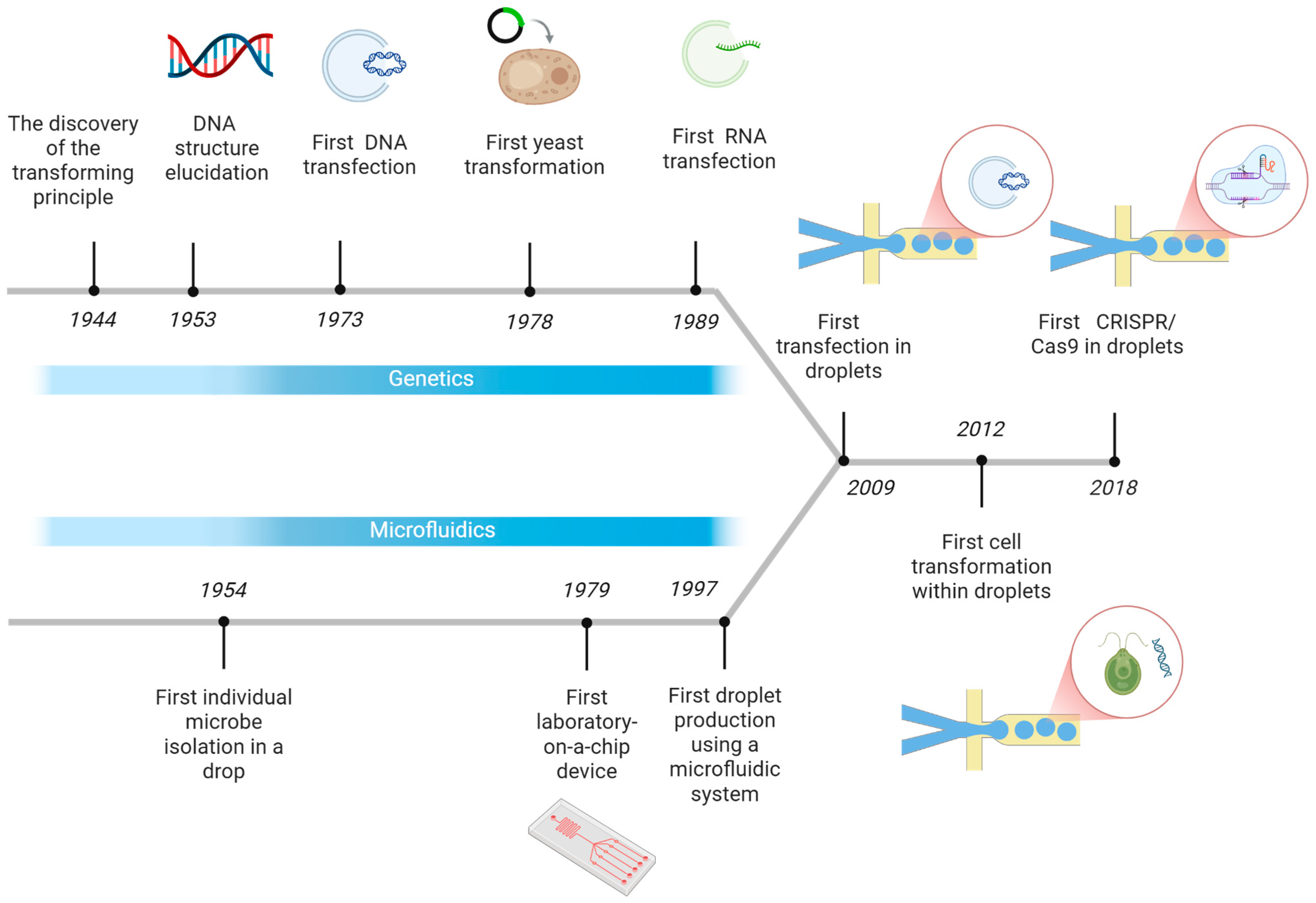
1. Introduction
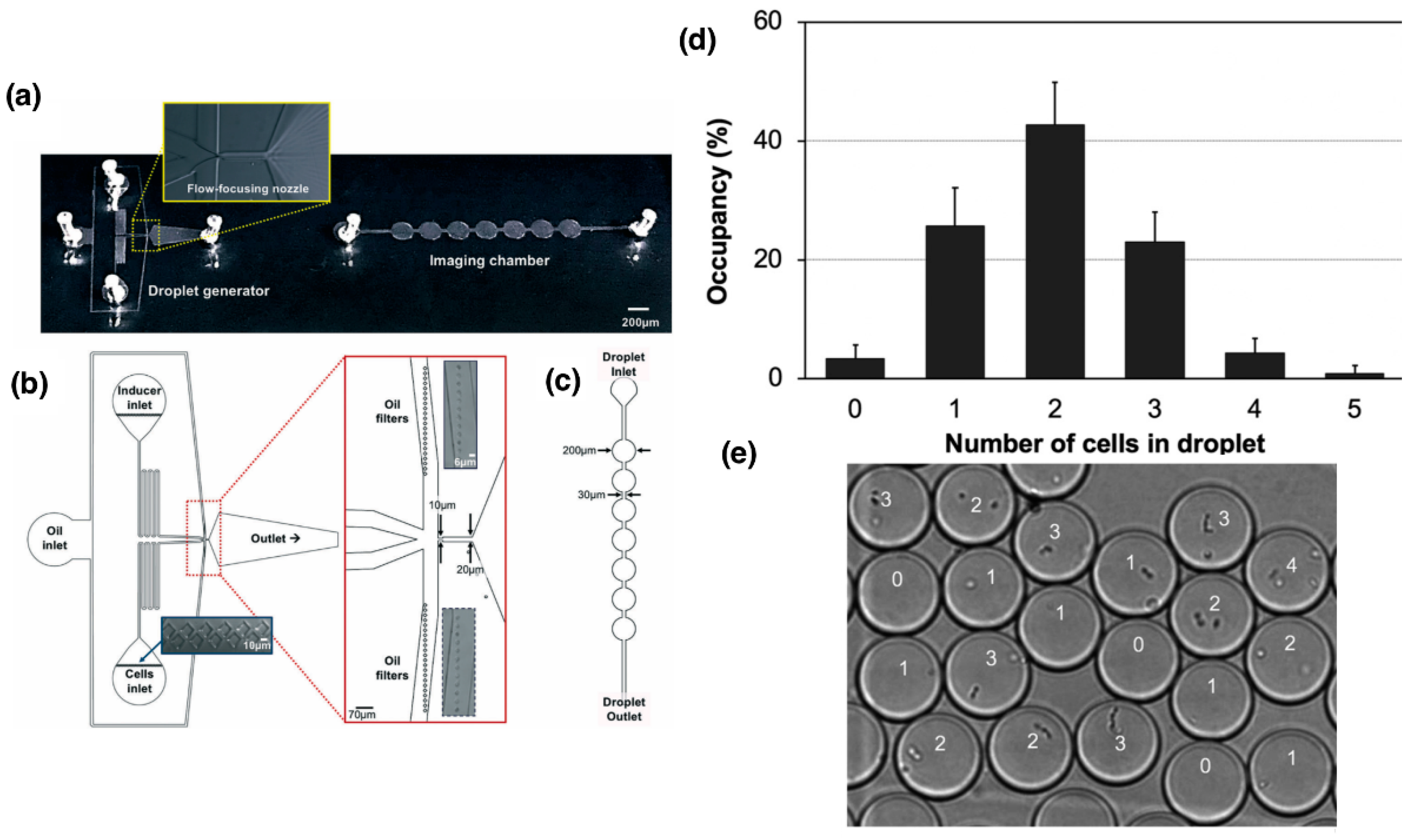
2. Transformation in Microdroplets
2.1. Streptococcus Pneumoniae
2.2. Algae
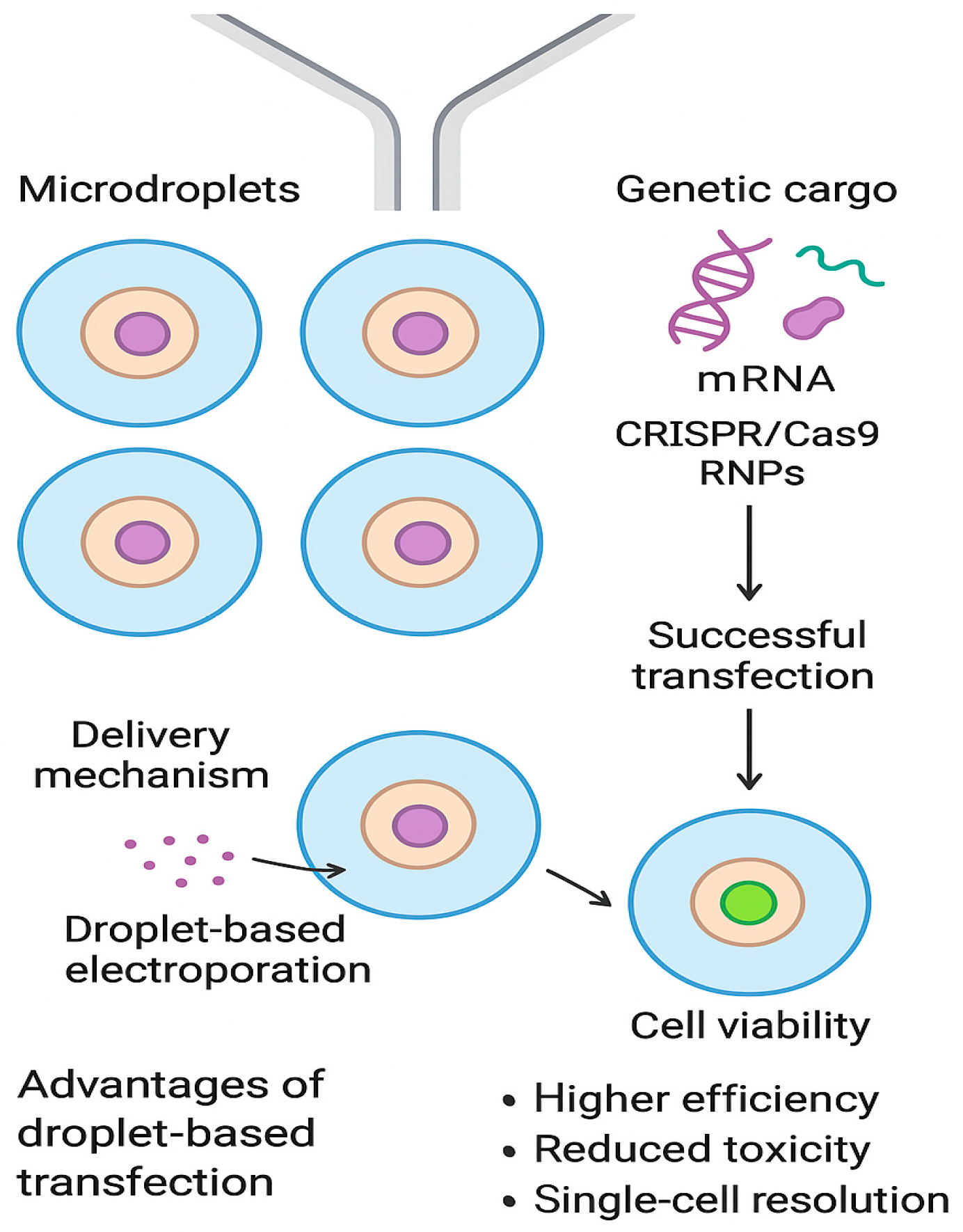
3. Transfection in Microdroplets
3.1. Non-Viral Transfection
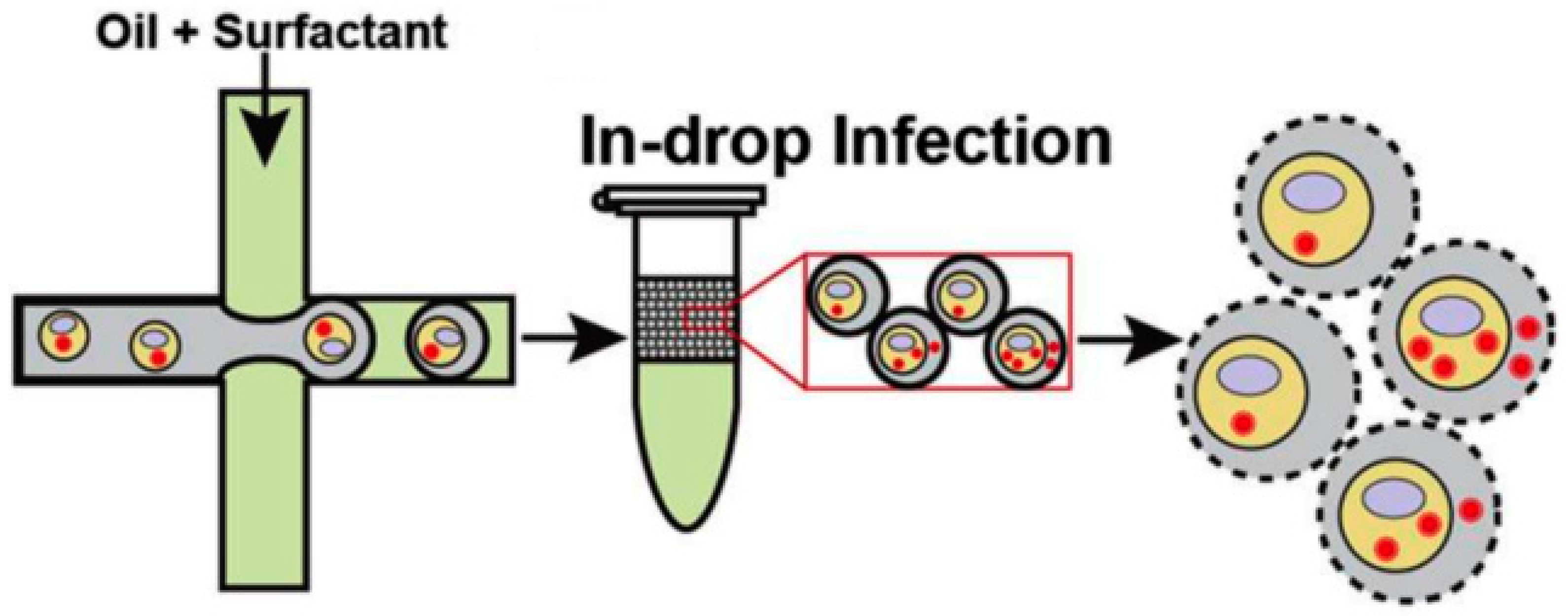
3.2. Viral Transfection
4. Hybrid and Integrative Approach
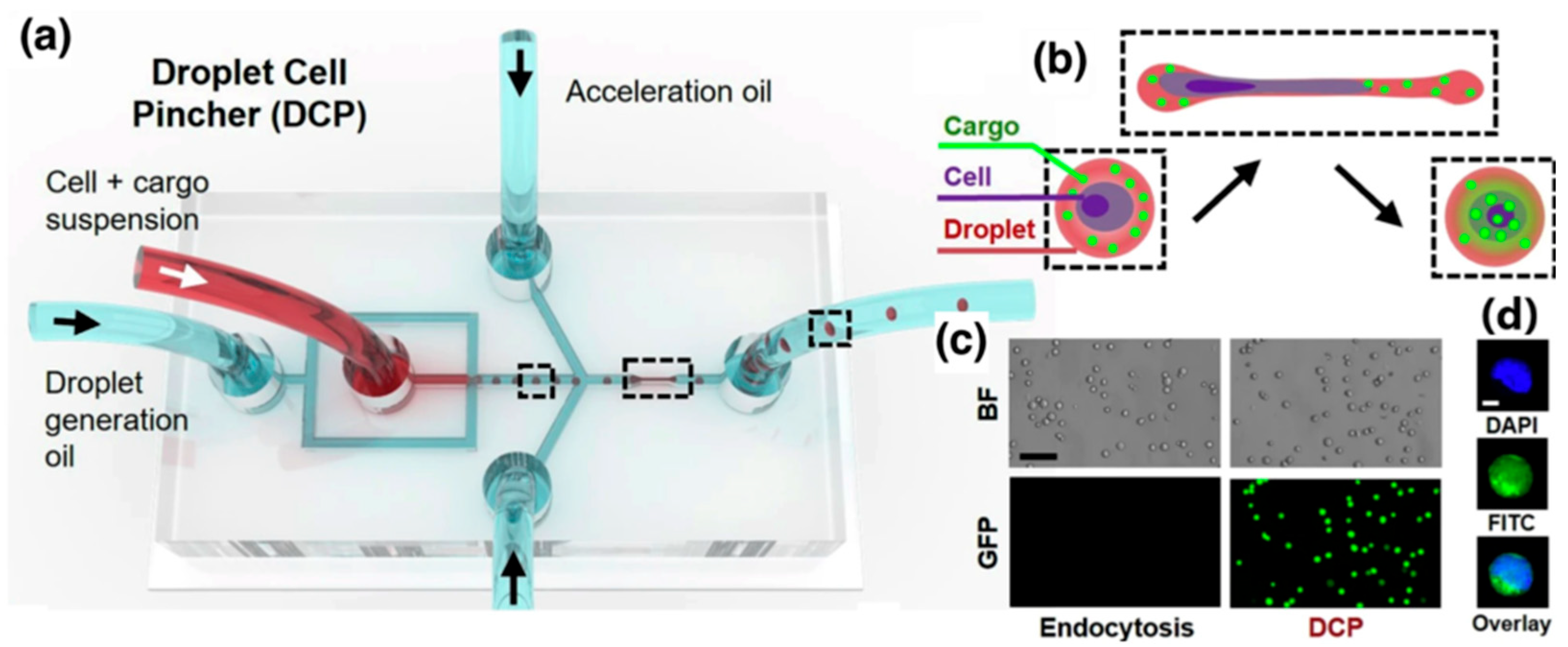
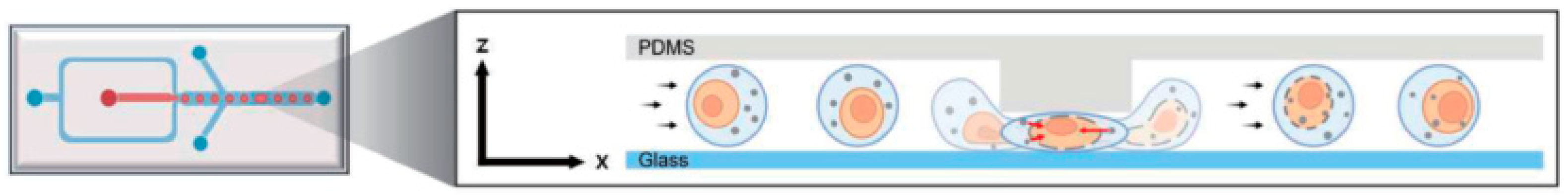
4.1. Cell Mechanoporation
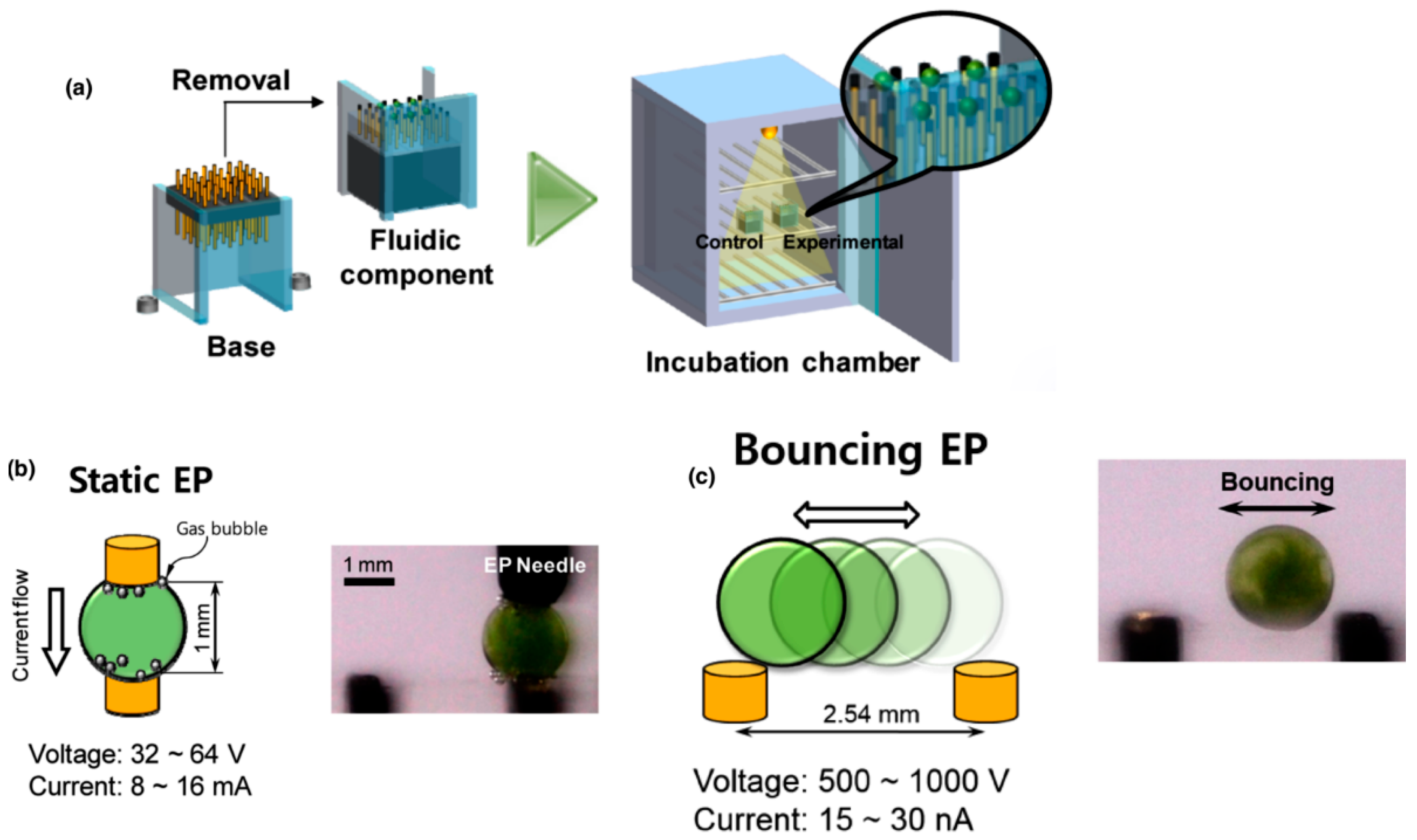
4.2. Electroporation
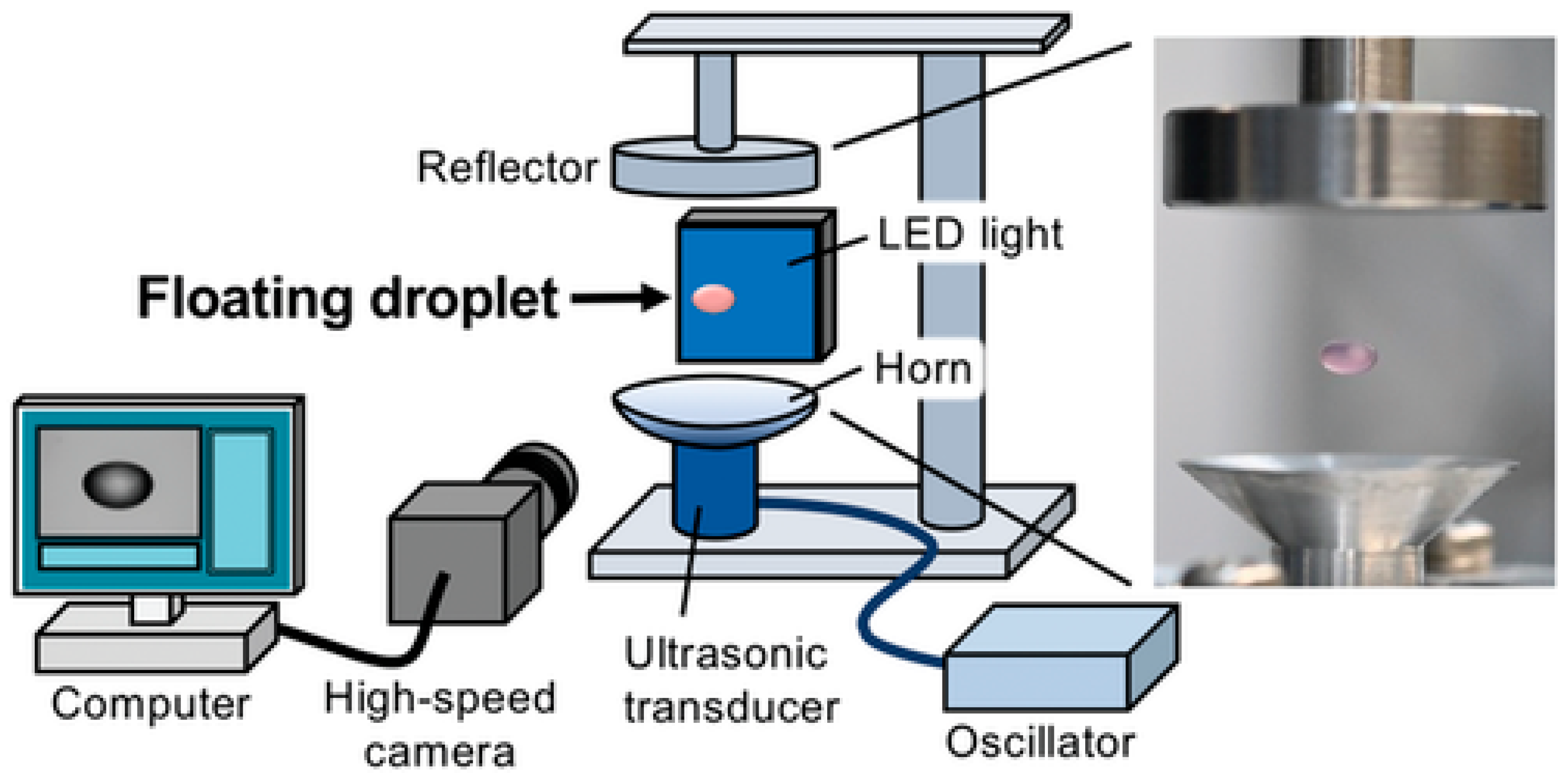
4.3. Ultrasonic Levitation
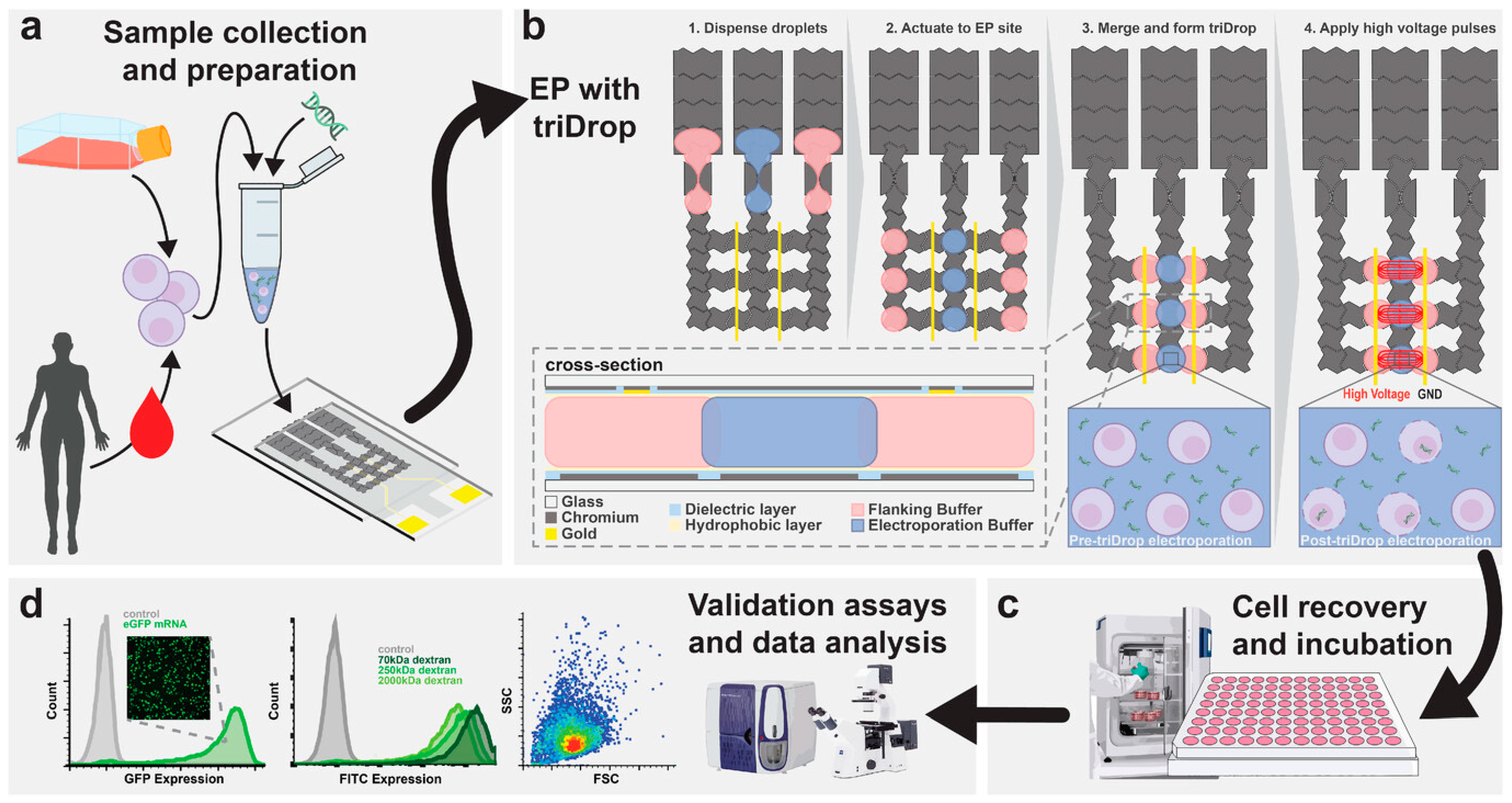
4.4. Voltage-Driven Digital Microfluidics
4.5. Acoustic Droplet Ejection Technology
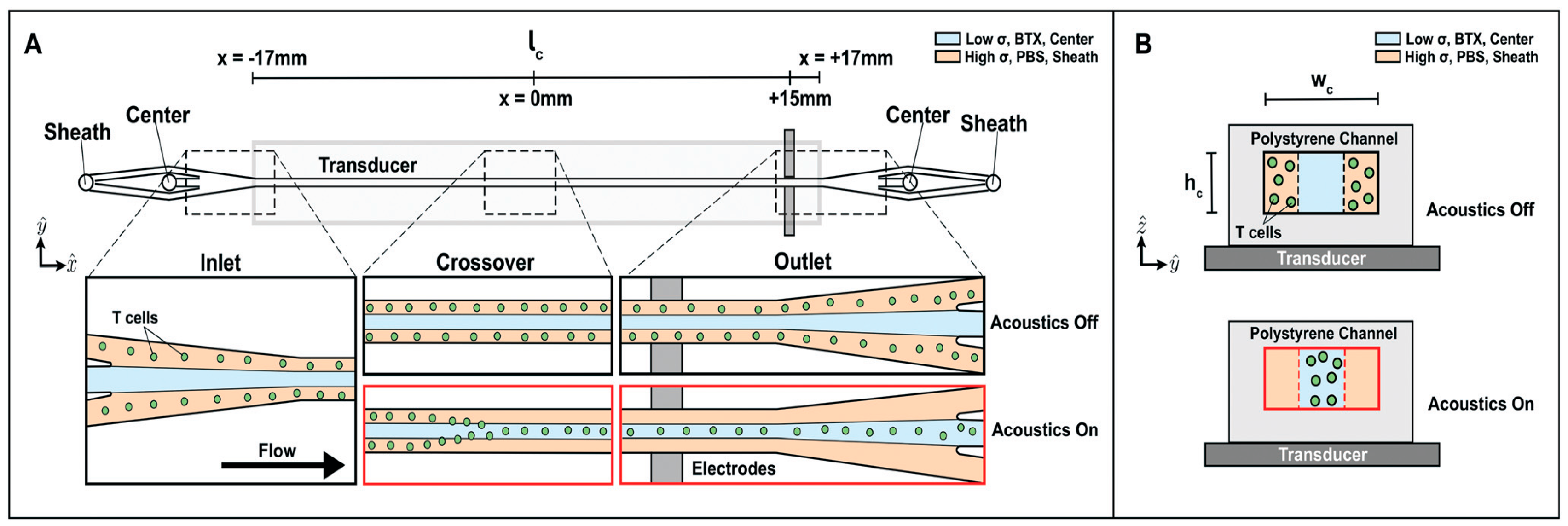
4.6. Acoustophoresis
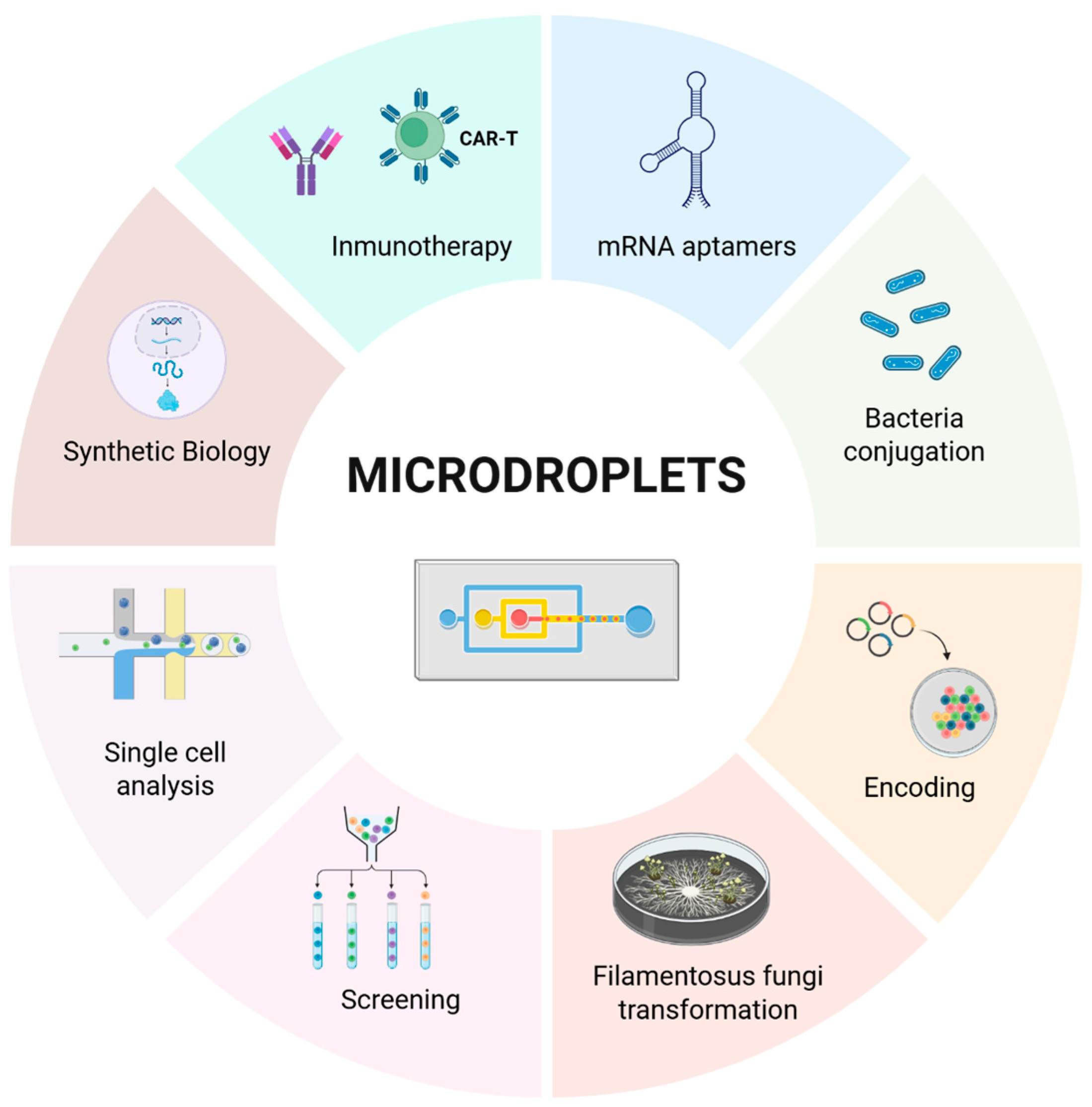
5. Applications Across Fields
5.1. Synthetic Biology
5.2. Immunology and Gene Therapy
5.3. Other Microdroplet Contributions
- -
- Microdroplet platforms for Bacterial Conjugation
- -
- Microdroplet-based workflow for filamentous fungi transformation
- -
- Microdroplets for screening of protoplast
- -
- Droplet-on-demand microfluidics for single-cell catalysis
- -
- Droplets for single cell isolation
- -
- Droplets for single cell—cell interaction
- -
- Droplet encoding
- -
- Microdroplets for RNA aptamers
6. Future Perspectives and Discussion
6.1. Comparative Performance of Microdroplet Gen Transfer Systems
6.2. Emerging Trends (Materials and AI Integration)
- -
- Materials to replace PDMS.
- -
- AI integration
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ASO | antisense oligonucleotides |
| CAR | chimeric antigen receptor |
| Cas9 | CRISPR-associated protein 9 |
| CFE | cell-free expression |
| CHO | Chinese hamster ovary |
| CHT | circular Hough transform |
| CNC | Computer Numerical Control |
| COC | Cyclic Olefin Copolymer |
| COP | Cyclic Olefin Polymer |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| DCP | Droplet Cell Pincher |
| DMF | Digital Microfluidics |
| EGFP | Enhanced Green Fluorescent Protein |
| EWOD | electrowetting-on-dielectric |
| HDR | homology-directed repair |
| hiPSCs | human induced pluripotent stem cells |
| miRNA | micro RNA |
| pDNA | plasmid DNA |
| PMMA | Poly(methyl methacrylate) |
| RNP | ribonucleoprotein |
| SCTTs | Single-Cell Transfection Technologies |
| sfGFP | superfolder green fluorescent protein |
| SG3 | Sustainable Goal 3 |
| shRNA | short hairpin RNA |
| siRNA | small interfering RNA |
| TR | transfection reagent |
References
- He, Y.; Yang, S.; Zhang, M.; Wang, Z.; Zhang, Y.; Gao, P. Advancements in the preparation and application of microdroplets: A review. Microchem. J. 2025, 214, 113901. [Google Scholar] [CrossRef]
- Jiang, Z.; Shi, H.; Tang, X.; Qin, J. Recent advances in droplet microfluidics for single-cell analysis. TrAC Trends Anal. Chem. 2023, 159, 116932. [Google Scholar] [CrossRef]
- Liu, L.; Duan, C.; Jiang, S.; Zhu, C.; Ma, Y.; Fu, T. Deformation and breakup of droplets in a double T-junction microdisperser with double input of the continuous phase. Chem. Eng. Process. Process Intensif. 2022, 180, 108674. [Google Scholar] [CrossRef]
- Dubey, S.; Vishwakarma, P.; Ramarao, T.V.S.; Dubey, S.K.; Goel, S.; Javed, A. Artificial intelligence-based droplet size prediction for microfluidic system. Int. Jnl Num Meth HFF 2024, 34, 3045–3078. [Google Scholar] [CrossRef]
- Fan, R.; Wu, J.; Duan, S.; Jin, L.; Zhang, H.; Zhang, C.; Zheng, A. Droplet-based microfluidics for drug delivery applications. Int. J. Pharm. 2024, 663, 124551. [Google Scholar] [CrossRef] [PubMed]
- Moragues, T.; Arguijo, D.; Beneyton, T.; Modavi, C.; Simutis, K.; Abate, A.R.; Baret, J.-C.; deMello, A.J.; Densmore, D.; Griffiths, A.D. Droplet-based microfluidics. Nat. Rev. Methods Primers 2023, 3, 32. [Google Scholar] [CrossRef]
- van der Loh, M.; Schiffmann, M.; Polack, M.; Wink, K.; Belder, D. Coupling of droplet-on-demand microfluidcs with ESI/MS to study single-cell catalysis. RSC Adv. 2024, 14, 25337–25346. [Google Scholar] [CrossRef]
- Wippold, J.A.; Chu, M.; Renberg, R.; Li, Y.; Adams, B.; Han, A. XPORT ENTRAP: A droplet microfluidic platform for enhanced DNA transfer between microbial species. New. Biotechnol. 2024, 81, 10–19. [Google Scholar] [CrossRef]
- Pérez-Sosa, C.; Sanluis-Verdes, A.; Waisman, A.; Lombardi, A.; Rosero, G.; Greca, A.L.; Bhansali, S.; Bourguignon, N.; Luzzani, C.; Pérez, M.S.; et al. Single cell transfection of human-induced pluripotent stem cells using a droplet-based microfluidic system. R. Soc. Open Sci. 2022, 9, 211510. [Google Scholar] [CrossRef]
- Xiang, L.; Solarczek, J.; Krajka, V.; Liu, H.; Ahlborn, L.; Schallmey, A.; Constantinou, I. Evaluating the Potential of Microdroplet Flow in Two-Phase Biocatalysis: A Systematic Study. ACS Appl. Mater. Interfaces 2025, 17, 4776–4787. [Google Scholar] [CrossRef]
- Avery, O.T.; Macleod, C.M.; McCarty, M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Inductions of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 1944, 79, 137–158. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Lederberg, J. A simple method for isolating individual microbes. J. Bacteriol. 1954, 68, 258–259. [Google Scholar] [CrossRef] [PubMed]
- Terry, S.C.; Jerman, J.H.; Angell, J.B. A gas chromatographic air analyzer fabricated on a silicon wafer. IEEE Trans. Electron. Devices 1979, 26, 1880–1886. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Kikuchi, Y.; Nakajima, M. Regular-sized cell creation in microchannel emulsification by visual microprocessing method. J. Am. Oil Chem. Soc. 1997, 74, 317–321. [Google Scholar] [CrossRef]
- Graham, F.L.; van der Eb, A.J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 1973, 52, 456–467. [Google Scholar] [CrossRef]
- Hinnen, A.; Hicks, J.B.; Fink, G.R. Transformation of yeast. Proc. Natl. Acad. Sci. USA 1978, 75, 1929–1933. [Google Scholar] [CrossRef]
- Malone, R.W.; Felgner, P.L.; Verma, I.M. Cationic liposome-mediated RNA transfection. Proc. Natl. Acad. Sci. USA 1989, 86, 6077–6081. [Google Scholar] [CrossRef]
- Zhan, Y.; Wang, J.; Bao, N.; Lu, C. Electroporation of cells in microfluidic droplets. Anal. Chem. 2009, 81, 2027–2031. [Google Scholar] [CrossRef]
- Qu, B.; Eu, Y.-J.; Jeong, W.-J.; Kim, D.-P. Droplet electroporation in microfluidics for efficient cell transformation with or without cell wall removal. Lab Chip 2012, 12, 4483–4488. [Google Scholar] [CrossRef]
- Sinha, H.; Quach, A.B.V.; Vo, P.Q.N.; Shih, S.C.C. An automated microfluidic gene-editing platform for deciphering cancer genes. Lab Chip 2018, 18, 2300–2312. [Google Scholar] [CrossRef] [PubMed]
- Iwai, K.; Wehrs, M.; Garber, M.; Sustarich, J.; Washburn, L.; Costello, Z.; Kim, P.W.; Ando, D.; Gaillard, W.R.; Hillson, N.J.; et al. Scalable and automated CRISPR-based strain engineering using droplet microfluidics. Microsyst. Nanoeng. 2022, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Onan, D.; Özder, M.; Sipahi, M.İ.; Poyraz, N.; Apaydın, C.; Erel-Akbaba, G.; Akbaba, H. Microfluidics based particle and droplet generation for gene and drug delivery approaches. J. Biomed. Mater. Res. Part B Appl. Biomater. 2025, 113, e35530. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Veroniaina, H.; Su, N.; Sha, K.; Jiang, F.; Wu, Z.; Qi, X. Applications and developments of gene therapy drug delivery systems for genetic diseases. Asian J. Pharm. Sci. 2021, 16, 687–703. [Google Scholar] [CrossRef]
- Luu, X.C.; Shida, Y.; Suzuki, Y.; Sato, N.; Nakumura, A.; Ogasawara, W. A novel high-throughput approach for transforming filamentous fungi employing a droplet-based microfluidic platform. New. Biotechnol. 2022, 72, 149–158. [Google Scholar] [CrossRef]
- Orevi, T.; Sørensen, S.J.; Kashtan, N. Droplet size and surface hydrophobicity enhance bacterial plasmid transfer rates in microscopic surface wetness. ISME Commun. 2022, 2, 72. [Google Scholar] [CrossRef]
- Sun, J.; Jiao, Y.; Pan, F.; Cheng, S.H.; Sun, D. A High-Throughput Microdroplet-Based Single Cell Transfection Method for Gene Knockout Based on the CRISPR/Cas9 System. IEEE Trans. Nanobiosci. 2024, 23, 378–388. [Google Scholar] [CrossRef]
- Sun, M.; Duan, X. Recent advances in micro/nanoscale intracellular delivery. Nanotechnol. Precis. Eng. 2020, 3, 18–31. [Google Scholar] [CrossRef]
- Duckert, B.; Vinkx, S.; Braeken, D.; Fauvart, M. Single-cell transfection technologies for cell therapies and gene editing. J. Control. Release 2021, 330, 963–975. [Google Scholar] [CrossRef]
- Zhang, Y.; Yao, Y.; Liu, S.; Chen, Y.; Zhou, S.; Wang, K.; Yang, X.; Liu, J. Coacervate microdroplet protocell-mediated gene transfection for nitric oxide production and induction of cell apoptosis. J. Mater. Chem. B Mater. Biol. Med. 2021, 9, 9784–9793. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Yun, D.; Lee, J.K.; Jung, C.; Chung, A.J. Highly efficient CRISPR-mediated genome editing through microfluidic droplet cell mechanoporation. Nat. Commun. 2024, 15, 8099. [Google Scholar] [CrossRef] [PubMed]
- Croucher, N.J.; Harris, S.R.; Barquist, L.; Parkhill, J.; Bentley, S.D. A high-resolution view of genome-wide pneumococcal transformation. PLoS Pathog. 2012, 8, e1002745. [Google Scholar] [CrossRef] [PubMed]
- Griffith, F. The significance of pneumococcal types. J. Hyg. 1928, 27, 113–159. [Google Scholar] [CrossRef] [PubMed]
- Chaguza, C.; Cornick, J.E.; Everett, D.B. Mechanisms and impact of genetic recombination in the evolution of Streptococcus pneumoniae. Comput. Struct. Biotechnol. J. 2015, 13, 241–247. [Google Scholar] [CrossRef]
- Lam, T.; Brennan, M.D.; Morrison, D.A.; Eddington, D.T. Femtoliter droplet confinement of Streptococcus pneumoniae: Bacterial genetic transformation by cell-cell interaction in droplets. Lab Chip 2019, 19, 682–692. [Google Scholar] [CrossRef]
- Ning, R.; Fan, J.; Kong, L.; Jiang, X.; Qian, Y.; Du, T.; Zhang, G.; Wu, W. Recent developments of droplets-based microfluidics for bacterial analysis. Chin. Chem. Lett. 2021, 33, 2243–2252. [Google Scholar] [CrossRef]
- Lam, T.; Maienschein-Cline, M.; Eddington, D.T.; Morrison, D.A. Multiplex gene transfer by genetic transformation between isolated S. pneumoniae cells confined in microfluidic droplets. Integr. Biol. 2019, 11, 415–424. [Google Scholar] [CrossRef]
- Pérez-Sosa, C.; Pérez, M.S.; Vallejo-Janeta, A.P.; Bhansali, S.; Miriuka, S.; Lerner, B. Droplets for Gene Editing Using CRISPR-Cas9 and Clonal Selection Improvement Using Hydrogels. Micromachines 2024, 15, 413. [Google Scholar] [CrossRef]
- Carvalho, B.G.; Vit, F.F.; Carvalho, H.F.; Han, S.W.; de la Torre, L.G. Layer-by-Layer Biomimetic Microgels for 3D Cell Culture and Nonviral Gene Delivery. Biomacromolecules 2022, 23, 1545–1556. [Google Scholar] [CrossRef]
- Chong, Z.X.; Yeap, S.K.; Ho, W.Y. Transfection types, methods and strategies: A technical review. PeerJ 2021, 9, e11165. [Google Scholar] [CrossRef]
- Dan, L.; Kang-Zheng, L. Optimizing viral transduction in immune cell therapy manufacturing: Key process design considerations. J. Transl. Med. 2025, 23, 501. [Google Scholar] [CrossRef]
- Quach, A.B.V.; Little, S.R.; Shih, S.C.C. Viral generation, packaging, and transduction on a digital microfluidic platform. Anal. Chem. 2022, 94, 4039–4047. [Google Scholar] [CrossRef]
- Li, X.; Le, Y.; Zhang, Z.; Nian, X.; Liu, B.; Yang, X. Viral Vector-Based Gene Therapy. Int. J. Mol. Sci. 2023, 24, 7736. [Google Scholar] [CrossRef] [PubMed]
- Loveday, E.K.; Sanchez, H.S.; Thomas, M.M.; Chang, C.B. Single-Cell Infection of Influenza A Virus Using Drop-Based Microfluidics. Microbiol. Spectr. 2022, 10, e0099322. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, P.; Gupta, P.; Illath, K.; Kar, S.; Nagai, M.; Tseng, F.-G.; Santra, T.S. Microfluidic mechanoporation for cellular delivery and analysis. Mater. Today Bio 2022, 13, 100193. [Google Scholar] [CrossRef] [PubMed]
- De Vry, J.; Martínez-Martínez, P.; Losen, M.; Temel, Y.; Steckler, T.; Steinbusch, H.W.M.; De Baets, M.H.; Prickaerts, J. In vivo electroporation of the central nervous system: A non-viral approach for targeted gene delivery. Prog. Neurobiol. 2010, 92, 227–244. [Google Scholar] [CrossRef]
- Im, D.J.; Jeong, S.-N.; Yoo, B.S.; Kim, B.; Kim, D.-P.; Jeong, W.-J.; Kang, I.S. Digital Microfluidic Approach for Efficient Electroporation with High Productivity: Transgene Expression of Microalgae without Cell Wall Removal. Anal. Chem. 2015, 87, 6592–6599. [Google Scholar] [CrossRef]
- Kim, Y.H.; Kwon, S.G.; Bae, S.J.; Park, S.J.; Im, D.J. Optimization of the droplet electroporation method for wild type Chlamydomonas reinhardtii transformation. Bioelectrochemistry 2019, 126, 29–37. [Google Scholar] [CrossRef]
- Arai, T.; Sato, T.; Matsubara, T. Effective cell transfection in an ultrasonically levitated droplet for sustainable technology. Adv. Sci. 2022, 9, e2203576. [Google Scholar] [CrossRef]
- Little, S.R.; Leung, Z.; Quach, A.B.V.; Hirukawa, A.; Gholizadeh, F.; Hajiaghayi, M.; Darlington, P.J.; Shih, S.C.C. A tri-droplet liquid structure for highly efficient intracellular delivery in primary mammalian cells using digital microfluidics. Adv. Mater. Technol. 2023, 8, 2300719. [Google Scholar] [CrossRef]
- Colin, B.; Deprez, B.; Couturier, C. High-Throughput DNA Plasmid Transfection Using Acoustic Droplet Ejection Technology. SLAS Discov. 2019, 24, 492–500. [Google Scholar] [CrossRef]
- Hsi, P.; Christianson, R.J.; Dubay, R.A.; Lissandrello, C.A.; Fiering, J.; Balestrini, J.L.; Tandon, V. Acoustophoretic rapid media exchange and continuous-flow electrotransfection of primary human T cells for applications in automated cellular therapy manufacturing. Lab Chip 2019, 19, 2978–2992. [Google Scholar] [CrossRef]
- Gach, P.C.; Iwai, K.; Kim, P.W.; Hillson, N.J.; Singh, A.K. Droplet microfluidics for synthetic biology. Lab Chip 2017, 17, 3388–3400. [Google Scholar] [CrossRef]
- Siltanen, C.A.; Cole, R.H.; Poust, S.; Chao, L.; Tyerman, J.; Kaufmann-Malaga, B.; Ubersax, J.; Gartner, Z.J.; Abate, A.R. An Oil-Free Picodrop Bioassay Platform for Synthetic Biology. Sci. Rep. 2018, 8, 7913. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wu, H.; Zhang, R.; Zhao, D.; Wei, W.; Yang, Q.; Wang, Z.; Shi, T.; Wang, Y. Ingenious integration of synthetic biology and droplet microfluidics. Chem. Eng. J. 2025, 520, 165813. [Google Scholar] [CrossRef]
- Ngocho, K.; Yang, X.; Wang, Z.; Hu, C.; Yang, X.; Shi, H.; Wang, K.; Liu, J. Synthetic Cells from Droplet-Based Microfluidics for Biosensing and Biomedical Applications. Small 2024, 20, e2400086. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, K. Development of Artificial Cell Models Using Microfluidic Technology and Synthetic Biology. Micromachines 2020, 11, 559. [Google Scholar] [CrossRef]
- Dimitriou, P. Microfluidic Construction and Operation of Artificial Cell Chassis Encapsulating Living Cells and Pharmaceutical Compounds Towards Their Controlled Interaction. Ph.D. Thesis, Cardiff University, Cardiff, UK, 2023. [Google Scholar]
- Seo, H.; Lee, H. Recent developments in microfluidic synthesis of artificial cell-like polymersomes and liposomes for functional bioreactors. Biomicrofluidics 2021, 15, 021301. [Google Scholar] [CrossRef]
- Yagui, T.R.T. High-Throughput Screening of Cell-Free Riboswitches for Chemical Communication Between Microdroplets. Ph.D. Thesis, Okinawa Institute of Science and Technology Graduate University, Onna, Japan, 2022. [Google Scholar]
- Salvail, H.; Breaker, R.R. Riboswitches. Curr. Biol. 2023, 33, R343–R348. [Google Scholar] [CrossRef]
- Joo, B.; Hur, J.; Kim, G.-B.; Yun, S.G.; Chung, A.J. Highly Efficient Transfection of Human Primary T Lymphocytes Using Droplet-Enabled Mechanoporation. ACS Nano 2021, 15, 12888–12898. [Google Scholar] [CrossRef]
- Loo, J.; Sicher, I.; Goff, A.; Kim, O.; Clary, N.; Alexeev, A.; Sulchek, T.; Zamarayeva, A.; Han, S.; Calero-Garcia, M. Microfluidic transfection of mRNA into human primary lymphocytes and hematopoietic stem and progenitor cells using ultra-fast physical deformations. Sci. Rep. 2021, 11, 21407. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Chung, A. Highly efficient mRNA transfection with droplet cell squeezing for cellular engineering. In Proceedings of the 2023 IEEE 16th International Conference on Nano/Molecular Medicine & Engineering (NANOMED), Okinawa, Japan, 5–8 December 2023; IEEE: New York, NY, USA, 2023; pp. 133–136. [Google Scholar]
- Li, X.; Aghaamoo, M.; Liu, S.; Lee, D.-H.; Lee, A.P. Lipoplex-Mediated Single-Cell Transfection via Droplet Microfluidics. Small 2018, 14, e1802055. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jin, R.; Shen, B.; Li, N.; Zhou, H.; Wang, W.; Zhao, Y.; Huang, M.; Fang, P.; Wang, S.; et al. High-throughput functional screening for next-generation cancer immunotherapy using droplet-based microfluidics. Sci. Adv. 2021, 7, eabe3839. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.J.; Wippold, J.A.; Renberg, R.; Hurley, M.; Adams, B.L.; Han, A. A High-Throughput Droplet-based Method to Facilitate Microbial Conjugation. Bio Protoc. 2024, 14, e5120. [Google Scholar] [CrossRef]
- Alberti, F.; Foster, G.D.; Bailey, A.M. Natural products from filamentous fungi and production by heterologous expression. Appl. Microbiol. Biotechnol. 2017, 101, 493–500. [Google Scholar] [CrossRef]
- Yu, Z.; Boehm, C.R.; Hibberd, J.M.; Abell, C.; Haseloff, J.; Burgess, S.J.; Reyna-Llorens, I. Droplet-based microfluidic analysis and screening of single plant cells. PLoS ONE 2018, 13, e0196810. [Google Scholar] [CrossRef]
- Samlali, K.; Ahmadi, F.; Quach, A.B.V.; Soffer, G.; Shih, S.C.C. One cell, one drop, one click: Hybrid microfluidics for mammalian single cell isolation. Small 2020, 16, e2002400. [Google Scholar] [CrossRef]
- Shang, Y.; Xing, G.; Lin, J.; Li, Y.; Lin, Y.; Chen, S.; Lin, J.-M. Multiplex bacteria detection using one-pot CRISPR/Cas13a-based droplet microfluidics. Biosens. Bioelectron. 2024, 243, 115771. [Google Scholar] [CrossRef]
- Kinghorn, A.B.; Guo, W.; Wang, L.; Tang, M.Y.H.; Wang, F.; Shiu, S.C.-C.; Lau, K.K.; Jinata, C.; Poonam, A.D.; Shum, H.C.; et al. Evolution driven microscale combinatorial chemistry in intracellular mimicking droplets to engineer thermostable RNA for cellular imaging. Small 2025, 21, e2409911. [Google Scholar] [CrossRef]
- Zhu, J.; Meng, Y.; Gao, W.; Yang, S.; Zhu, W.; Ji, X.; Zhai, X.; Liu, W.-Q.; Luo, Y.; Ling, S.; et al. AI-driven high-throughput droplet screening of cell-free gene expression. Nat. Commun. 2025, 16, 2720. [Google Scholar] [CrossRef]
- Kitte, R.; Rabel, M.; Geczy, R.; Park, S.; Fricke, S.; Koehl, U.; Tretbar, U.S. Lipid nanoparticles outperform electroporation in mRNA-based CAR T cell engineering. Mol. Ther. Methods Clin. Dev. 2023, 31, 101139. [Google Scholar] [CrossRef] [PubMed]
- Prommersberger, S.; Reiser, M.; Beckmann, J.; Danhof, S.; Amberger, M.; Quade-Lyssy, P.; Einsele, H.; Hudecek, M.; Bonig, H.; Ivics, Z. CARAMBA: A first-in-human clinical trial with SLAMF7 CAR-T cells prepared by virus-free Sleeping Beauty gene transfer to treat multiple myeloma. Gene Ther. 2021, 28, 560–571. [Google Scholar] [CrossRef] [PubMed]
- Jayasooriya, V.; Ringwelski, B.; Dorsam, G.; Nawarathna, D. mRNA-based CAR T-cells manufactured by miniaturized two-step electroporation produce selective cytotoxicity toward target cancer cells. Lab Chip 2021, 21, 3748–3761. [Google Scholar] [CrossRef] [PubMed]
- Gomes-Silva, D.; Mukherjee, M.; Srinivasan, M.; Krenciute, G.; Dakhova, O.; Zheng, Y.; Cabral, J.M.S.; Rooney, C.M.; Orange, J.S.; Brenner, M.K.; et al. Tonic 4-1BB Costimulation in Chimeric Antigen Receptors Impedes T Cell Survival and Is Vector-Dependent. Cell Rep. 2017, 21, 17–26. [Google Scholar] [CrossRef]
- O’Sullivan, A.; Case, S.; McCrudden, A.; Hackett, E.; Gallagher, L.; Martin, D.; Johnson, G.P.; Mahadik, K.; Kienzle, T.; Lim, J.K.; et al. Increased Functional Potency of Multi-Edited CAR-T Cells Manufactured by a Non-Viral Transfection System. Mol. Ther. Methods Clin. Dev 2025, 33, 101389. [Google Scholar] [CrossRef]
- Zhang, Z.; Qiu, S.; Zhang, X.; Chen, W. Optimized DNA electroporation for primary human T cell engineering. BMC Biotechnol. 2018, 18, 4. [Google Scholar] [CrossRef]
- de Los Milagros Bassani Molinas, M.; Beer, C.; Hesse, F.; Wirth, M.; Wagner, R. Optimizing the transient transfection process of HEK-293 suspension cells for protein production by nucleotide ratio monitoring. Cytotechnology 2014, 66, 493–514. [Google Scholar] [CrossRef]
- Harris, E.; Zimmerman, D.; Warga, E.; Bamezai, A.; Elmer, J. Nonviral gene delivery to T cells with Lipofectamine LTX. Biotechnol. Bioeng. 2021, 118, 1693–1706. [Google Scholar] [CrossRef]
- Cao, Y.; Ma, E.; Cestellos-Blanco, S.; Zhang, B.; Qiu, R.; Su, Y.; Doudna, J.A.; Yang, P. Nontoxic nanopore electroporation for effective intracellular delivery of biological macromolecules. Proc. Natl. Acad. Sci. USA 2019, 116, 7899–7904. [Google Scholar] [CrossRef]
- Morbioli, G.G.; Speller, N.C.; Stockton, A.M. A practical guide to rapid-prototyping of PDMS-based microfluidic devices: A tutorial. Anal. Chim. Acta 2020, 1135, 150–174. [Google Scholar] [CrossRef]
- Peñaherrera-Pazmiño, A.B.; Rosero, G.I.; Pérez, M.; Lerner, B. Photopolymer flexographic printing plate mold for PDMS microfluidic manufacture. Polymers 2025, 17, 1723. [Google Scholar] [CrossRef]
- Shakeri, A.; Khan, S.; Jarad, N.A.; Didar, T.F. The fabrication and bonding of thermoplastic microfluidics: A review. Materials 2022, 15, 6478. [Google Scholar] [CrossRef] [PubMed]
- Lissandrello, C.A.; Santos, J.A.; Hsi, P.; Welch, M.; Mott, V.L.; Kim, E.S.; Chesin, J.; Haroutunian, N.J.; Stoddard, A.G.; Czarnecki, A.; et al. High-throughput continuous-flow microfluidic electroporation of mRNA into primary human T cells for applications in cellular therapy manufacturing. Sci. Rep. 2020, 10, 18045. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.G.; Condelipes, P.G.M.; Rosa, R.R.; Chu, V.; Conde, J.P. Scalable processing of cyclic olefin copolymer (COC) microfluidic biochips. Micromachines 2023, 14, 1837. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Sun, A.; Brodský, J.; Gablech, I.; Lednický, T.; Vopařilová, P.; Zítka, O.; Zeng, W.; Neužil, P. Microfluidics chips fabrication techniques comparison. Sci. Rep. 2024, 14, 28793. [Google Scholar] [CrossRef]
- Hwang, Y.-H.; Shepherd, S.J.; Kim, D.; Mukalel, A.J.; Mitchell, M.J.; Issadore, D.A.; Lee, D. Robust, Scalable Microfluidic Manufacturing of RNA-Lipid Nanoparticles Using Immobilized Antifouling Lubricant Coating. ACS Nano 2025, 19, 1090–1102. [Google Scholar] [CrossRef]
- Sun, H.; Xie, W.; Mo, J.; Huang, Y.; Dong, H. Deep learning with microfluidics for on-chip droplet generation, control, and analysis. Front. Bioeng. Biotechnol. 2023, 11, 1208648. [Google Scholar] [CrossRef]
- Song, Y.; Lim, S.; Kim, Y.T.; Park, Y.M.; Jo, D.A.; Bae, N.H.; Lee, S.J.; Choi, B.G.; Im, S.G.; Kim, H.U.; et al. Deep learning enables accurate analysis of images generated from droplet-based digital polymerase chain reaction (dPCR). Sens. Actuators B Chem. 2023, 379, 133241. [Google Scholar] [CrossRef]
- Park, I.; Choi, S.; Gwak, Y.; Kim, J.; Min, G.; Lim, D.; Lee, S.W. Microfluidic Electroporation Arrays for Investigating Electroporation-Induced Cellular Rupture Dynamics. Biosensors 2024, 14, 242. [Google Scholar] [CrossRef]

| Part A. Transformation | ||||||
| Biological Sample | Target Cargo | Concentration/Size | Viability (%) | Efficiency (%) or Ratio 1 | Throughput | Ref. |
| C. reinhardtii | DNA fragments | 40 µg/mL | 81 | 0.081, ratio: 8.14 ± 0.20 × 10−4 | - | [20] |
| C. reinhardtii | pDNA | 1 µg/mL | 80–90 | 20.7 ± 6.2 | High | [47] |
| C. reinhardtii | pDNA | 8104 bp | >80 | ≈10–20 (1–5% stable transformation) | High | [48] |
| E. coli | pDNA | 10 pmol/µL | - | 98 ± 3 | High | [22] |
| S. pneumoniae (CP2204 recipient + CP2215 donor) | Native chromosomal DNA | - | 40 (Donor) 60 (Recipient) | - | ≈50% | [35] |
| S. pneumoniae (CP2204 recipient + CP2215 donor) | Genomic DNA | 5 µg/mL | - | - | ≈50% | [37] |
| Part B. Transfection | ||||||
| Biological Sample | Target Cargo | Concentration/Size | Viability (%) | Efficiency (%) | Throughput | Ref. |
| CHO cells | pDNA | 100 µg/mL | ≈68 | ≈11 | - | [19] |
| K562 | mRNA | 20 µg/mL | >80 | ≈100 | High | [62] |
| Human primary T lymphocytes | mRNA | 20 µg/mL | >82 | ≈90 | High | |
| K562 | pDNA | 7.9 kbp | ≈80 | 45% | High | |
| K562 | mRNA | 20 µg/mL | >98 | 1 × 106 cells/mL | [64] | |
| H1299 | pDNA | 10.5 kbb | - | ≈70% | - | [21] |
| SMMC-7721 | pNOS | 25 ug/mL | 51 | 61.5 | - | [30] |
| Jurkart cells K562 HEK-293T NK-92 | >75 | >90 | 6 × 107 cells/mL | [31] | ||
| HEK293 | mRNA | 2 pg/cell | >90 | >90 | High | [50] |
| pDNA | ≈5 kb | 90 | 71 | |||
| HeLa | pDNA | ≈5 kb | >90 | ≈60 | ||
| HEK293T | pDNA | 5 ng/µL | >80 | 60 | High | [27] |
| MCF7 | pDNA | 20 µg/mL | >80 | 40–60% | High | |
| hiPSC | pDNA-CAG-mCerulean + PiggyBac | 2.5–10 µg/mL | >85 | 70 | High | [9] |
| hiPSC | CRISPR/Cas9 pDNA | 10 µg/mL | >80 | 94–96 | High | [38] |
| K562 | CRISPR-Cas9/mRNA y pDNA | 100 µg/mL | >75 | >62 | High | [38] |
| Huh-7 | pDNA | 60 µg/mL | - | Qualitative | Qualitative | [49] |
| Primary human T cells CART | mRNA | 50 µg/mL | - | >60 (2× to conventional technics) | - | [52] |
| HeLa cells | pDNA tdTomato and mVenus | 100 ng/μL | - | 90 Co-transfection ≈ 100 | High | [51] |
| HeLa cells | pDNA | 5 ng/μL and 30 ng/μL | - | ≈100 | High | [52] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Criollo, M.; Layedra, G.; Pérez-Sosa, C.; Rosero, G.; Peñaherrera-Pazmiño, A.B. Microdroplet Systems for Gene Transfer: From Fundamentals to Future Perspectives. Micromachines 2025, 16, 1245. https://doi.org/10.3390/mi16111245
Criollo M, Layedra G, Pérez-Sosa C, Rosero G, Peñaherrera-Pazmiño AB. Microdroplet Systems for Gene Transfer: From Fundamentals to Future Perspectives. Micromachines. 2025; 16(11):1245. https://doi.org/10.3390/mi16111245
Chicago/Turabian StyleCriollo, Mishell, Gina Layedra, Camilo Pérez-Sosa, Gustavo Rosero, and Ana Belén Peñaherrera-Pazmiño. 2025. "Microdroplet Systems for Gene Transfer: From Fundamentals to Future Perspectives" Micromachines 16, no. 11: 1245. https://doi.org/10.3390/mi16111245
APA StyleCriollo, M., Layedra, G., Pérez-Sosa, C., Rosero, G., & Peñaherrera-Pazmiño, A. B. (2025). Microdroplet Systems for Gene Transfer: From Fundamentals to Future Perspectives. Micromachines, 16(11), 1245. https://doi.org/10.3390/mi16111245







