Recent Advances in Monitoring Cell Behavior Using Cell-Based Impedance Spectroscopy
Abstract
1. Introduction
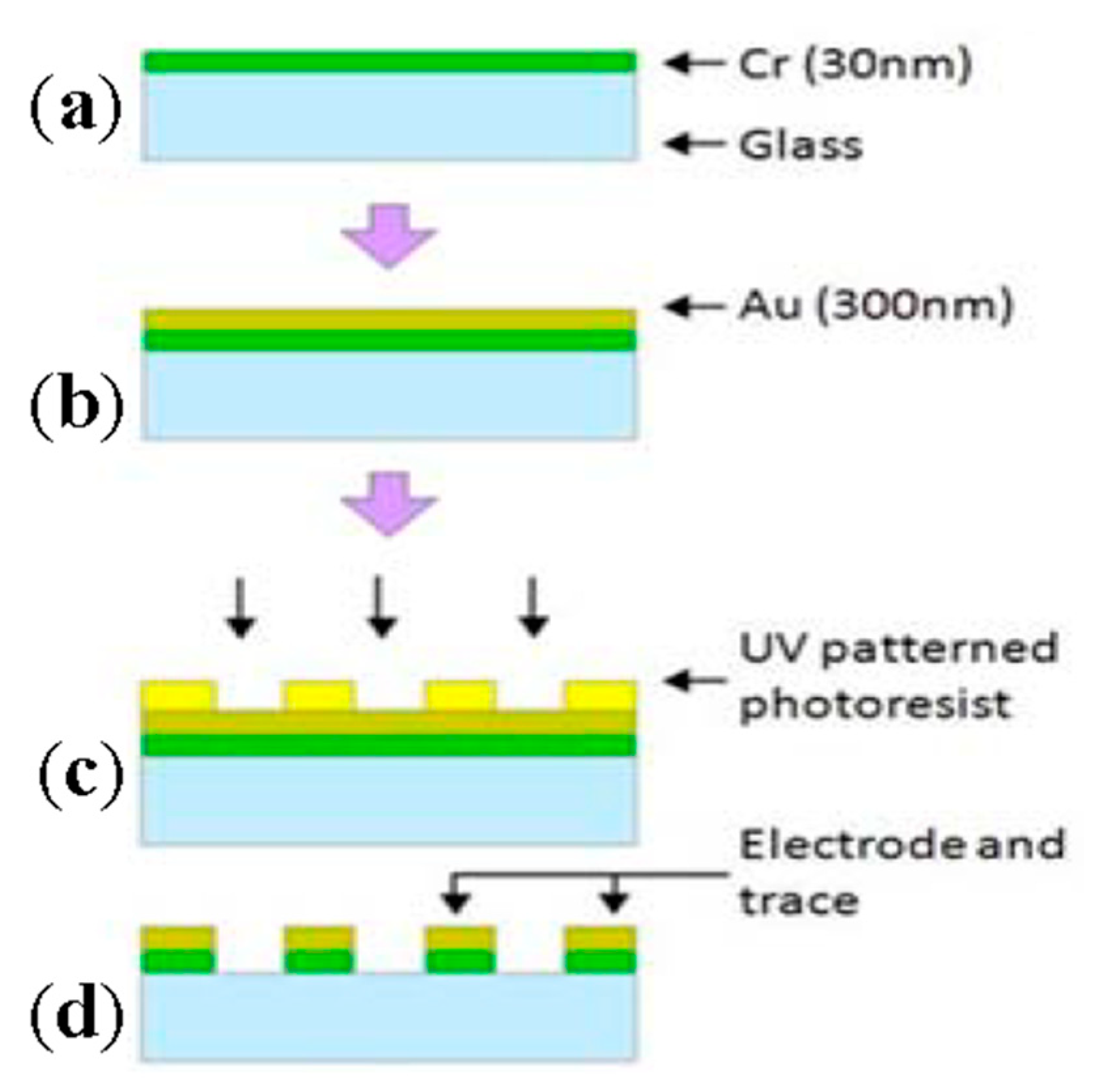
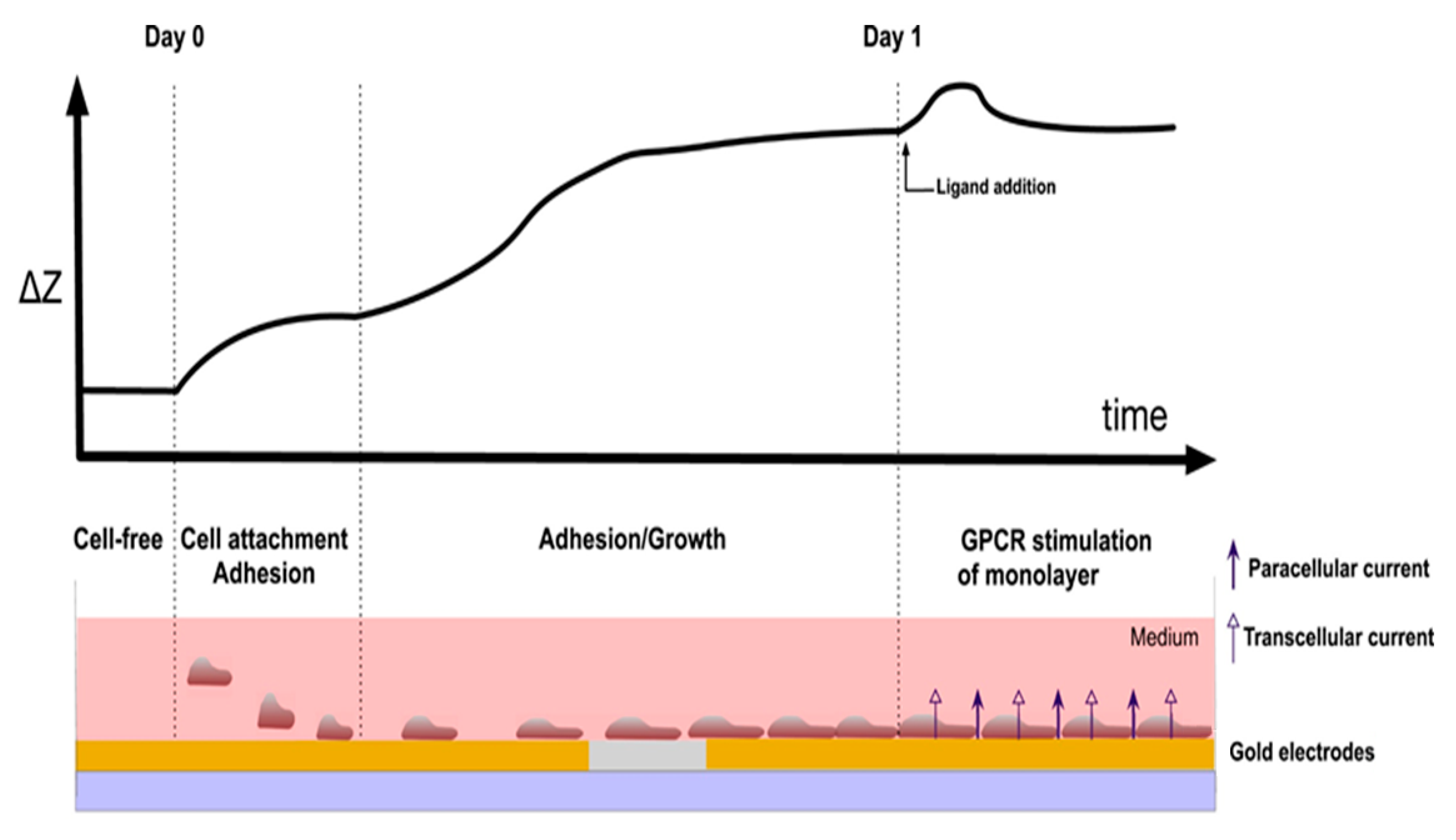
2. Instrumentation and Detection Principle of CBI
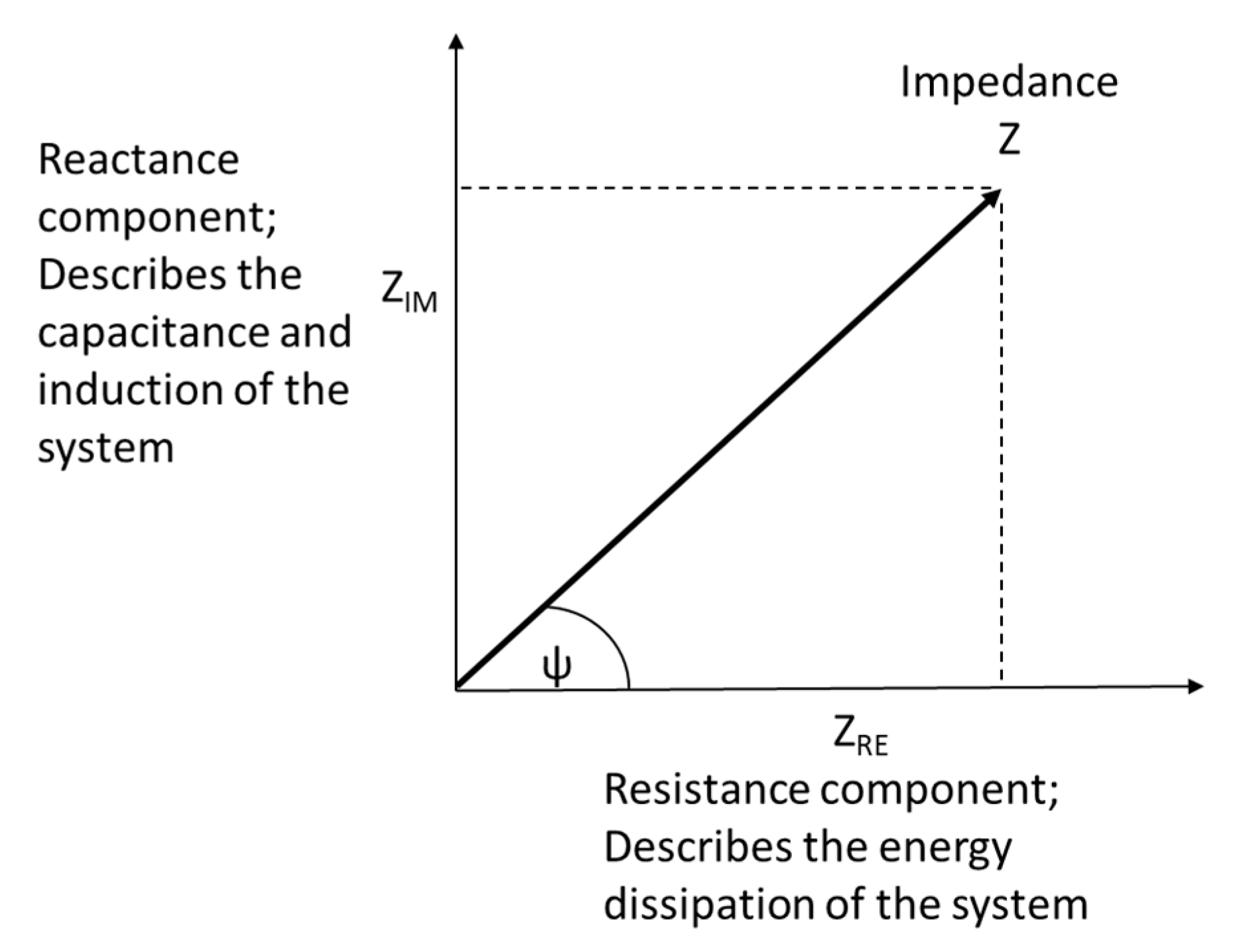
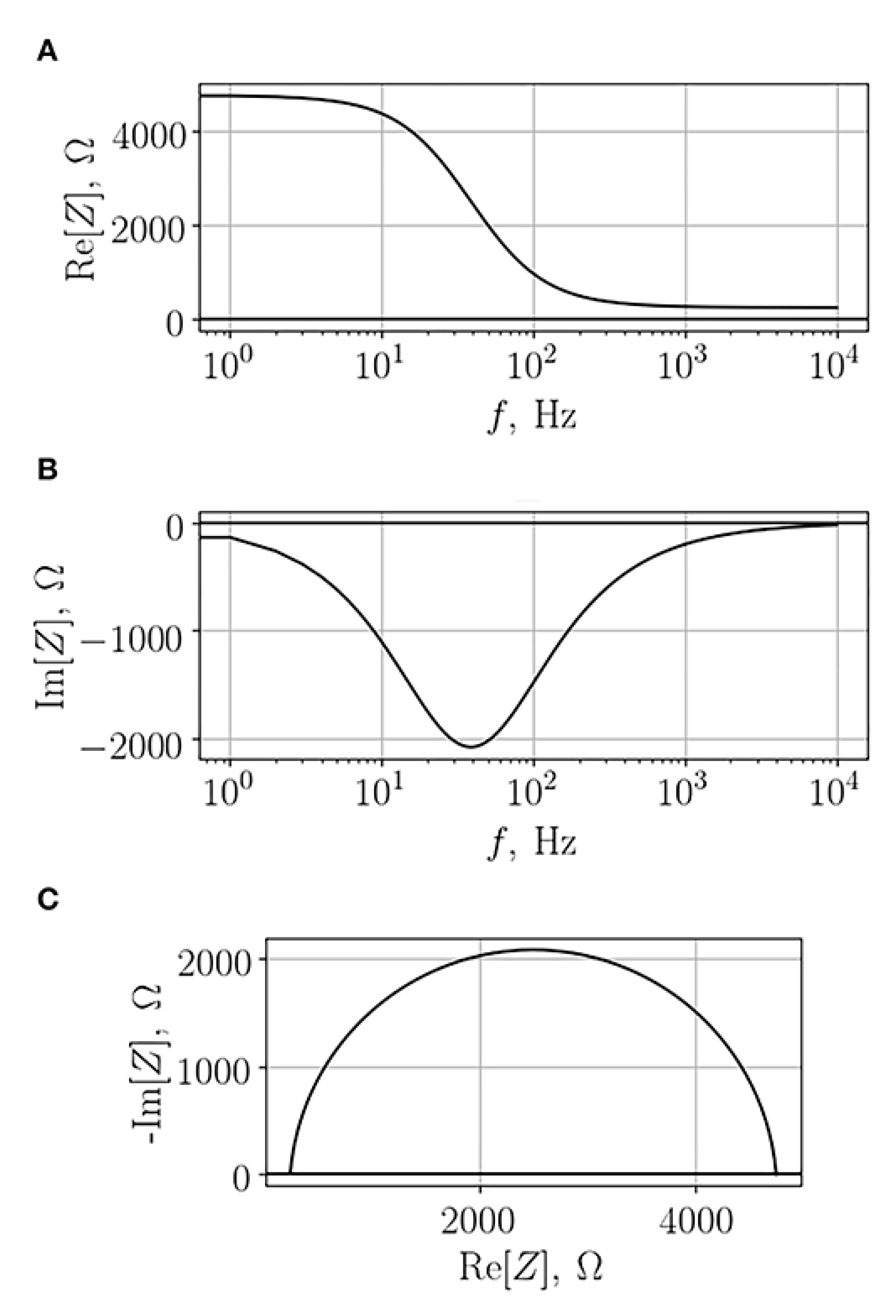
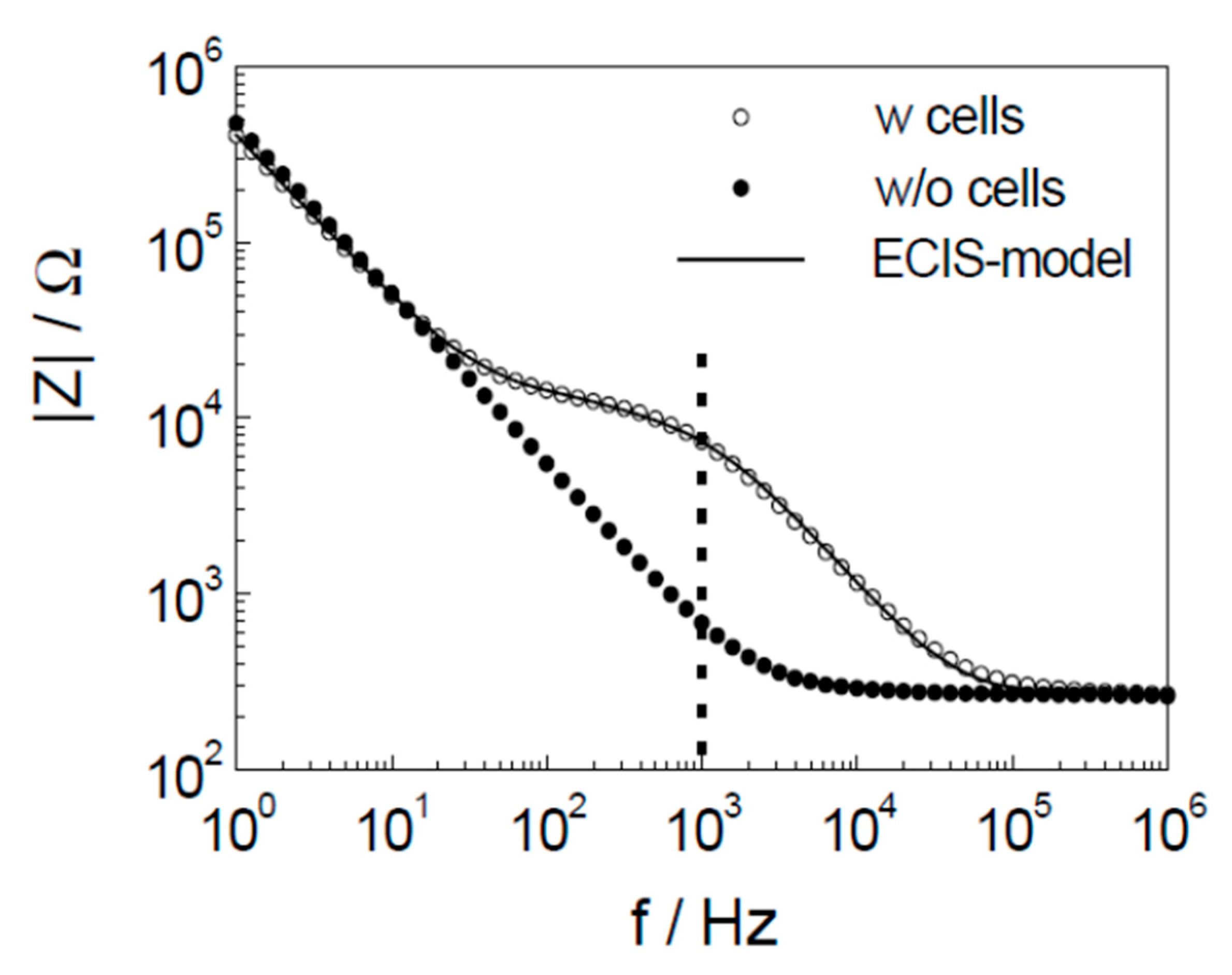
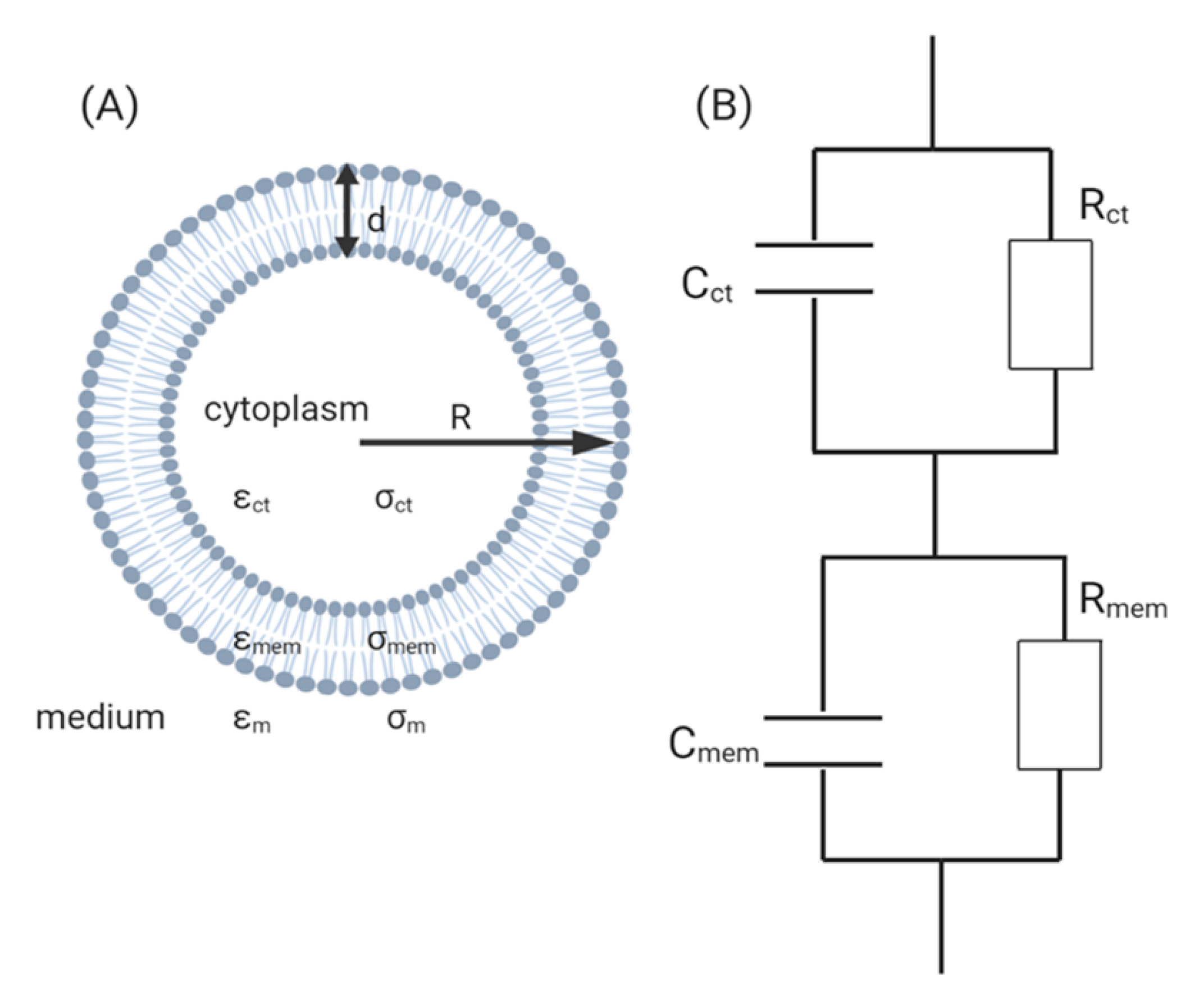
3. Theory of Electrochemical Impedance Spectroscopy for Cell Analysis
4. Applications
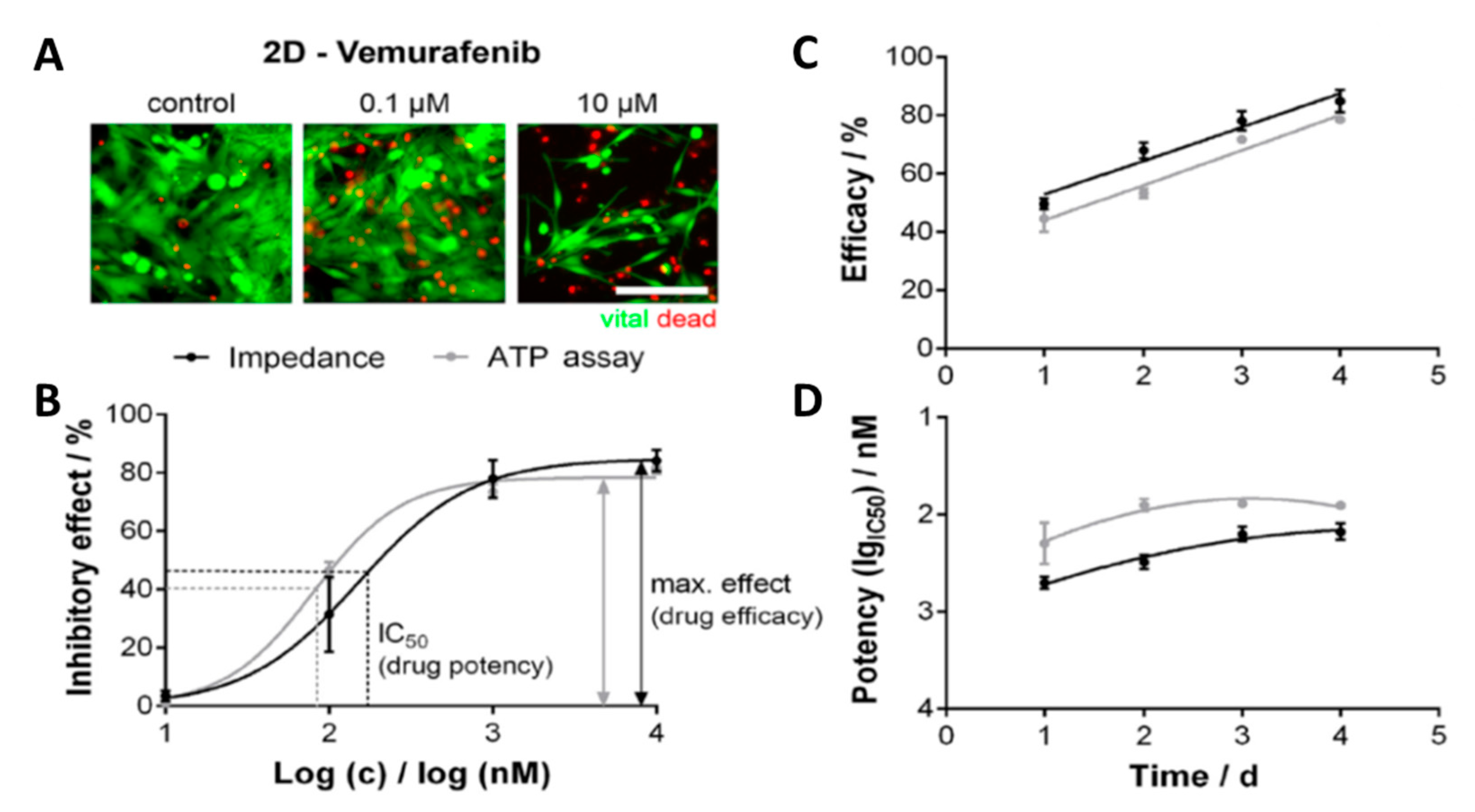
4.1. Cancer Research
4.2. Neurodegenerative Diseases
4.3. Toxicity Research
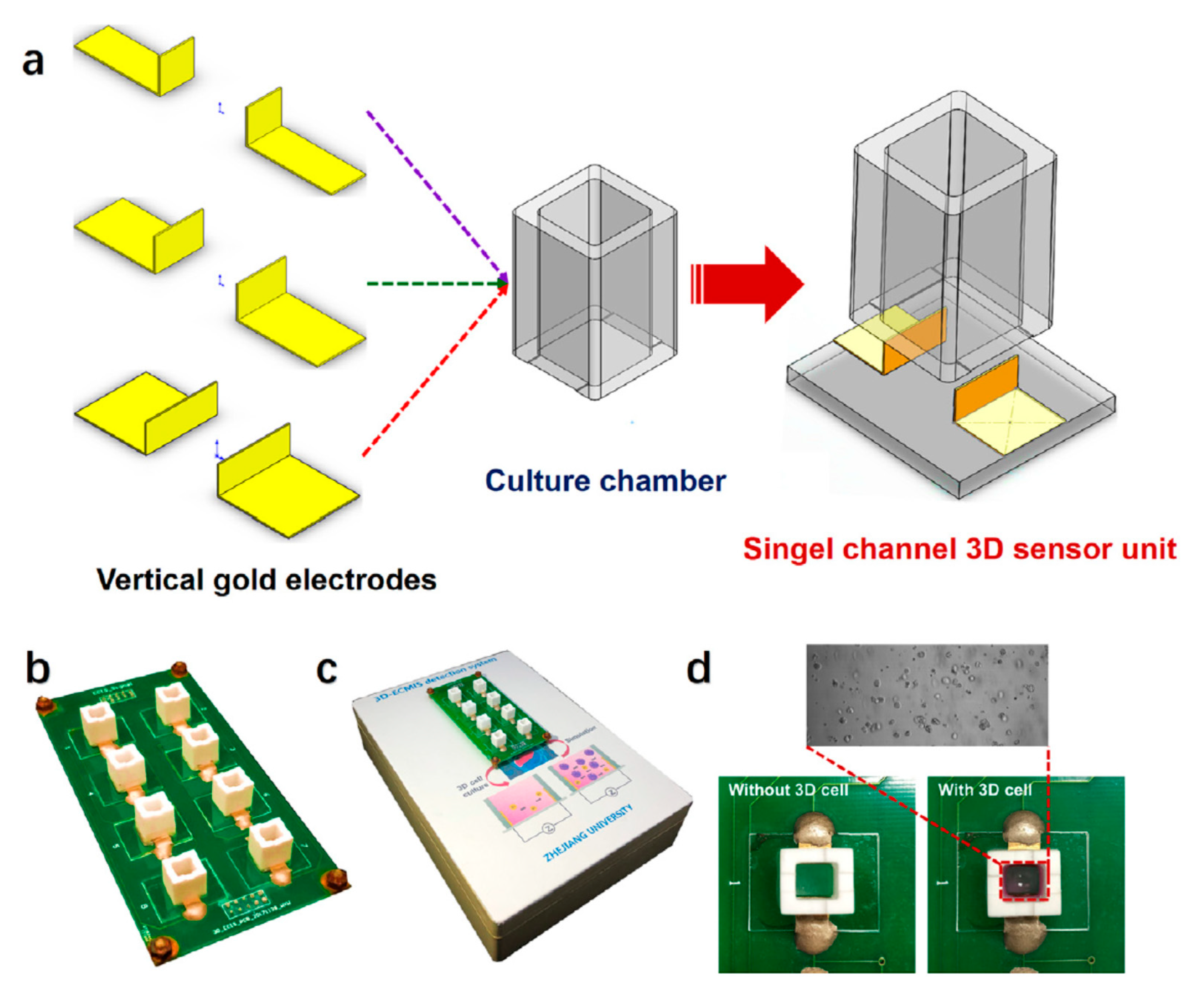
4.4. 2D and 3D In Vitro Studies
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hudu, S.A.; Alshrari, A.S.; Syahida, A.; Sekawi, Z. Cell Culture, Technology: Enhancing the Culture of Diagnosing Human Diseases. J. Clin. Diagnostic Res. 2016, 10, DE01–DE05. [Google Scholar] [CrossRef] [PubMed]
- Arango, M.-T.; Quintero-Ronderos, P.; Castiblanco, J.; Montoya-Ortíz, G. Autoimmunity from Bench to Bedside; El Rosario University Press: Bogota, Colombia, 2013; ISBN 9789587383669. [Google Scholar]
- Langhans, S.A. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Alkhilaiwi, F.; Paul, S.; Zhou, D.; Zhang, X.; Wang, F.; Palechor-Ceron, N.; Wilson, K.; Guha, R.; Ferrer, M.; Grant, N.; et al. High-throughput screening identifies candidate drugs for the treatment of recurrent respiratory papillomatosis. Papillomavirus Res. 2019, 8, 100181. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, L.; Shan, X.; Du, G.; Zhou, J.; Chen, J. High-Throughput Screening of a 2-Keto-L-Gulonic Acid-Producing Gluconobacter oxydans Strain Based on Related Dehydrogenases. Front. Bioeng. Biotechnol. 2019, 7, 1–9. [Google Scholar] [CrossRef]
- Kohara, H.; Bajaj, P.; Yamanaka, K.; Miyawaki, A.; Harada, K.; Miyamoto, K.; Matsui, T.; Okai, Y.; Wagoner, M.; Shinozawa, T. High-throughput screening to evaluate inhibition of bile acid transporters using human hepatocytes isolated from chimeric mice. Toxicol. Sci. 2020, 173, 347–361. [Google Scholar] [CrossRef]
- Liao, W.; Wang, J.; Xu, J.; You, F.; Pan, M.; Xu, X.; Weng, J.; Han, X.; Li, S.; Li, Y.; et al. High-throughput three-dimensional spheroid tumor model using a novel stamp-like tool. J. Tissue Eng. 2019, 10. [Google Scholar] [CrossRef]
- Ock, J.; Li, W. A high-throughput three-dimensional cell culture platform for drug screening. Bio-Design Manuf. 2020, 3, 40–47. [Google Scholar] [CrossRef]
- Kenny, H.A.; Lal-Nag, M.; Shen, M.; Kara, B.; Nahotko, D.A.; Wroblewski, K.; Fazal, S.; Chen, S.; Chiang, C.Y.; Chen, Y.J.; et al. Quantitative high-throughput screening using an organotypic model identifies compounds that inhibit ovarian cancer metastasis. Mol. Cancer Ther. 2020, 19, 52–62. [Google Scholar] [CrossRef]
- Chen, C.; Li, J.; Micko, C.J.; Pierce, G.F.; Cunningham, M.R.; Lumsden, A.B. Cytotoxic effects of basic FGF and heparin binding EGF conjugated with cytotoxin saporin on vascular cell cultures. J. Surg. Res. 1998, 75, 35–41. [Google Scholar] [CrossRef]
- Magun, R.; Boone, D.L.; Tsang, B.K.; Sorisky, A. The effect of adipocyte differentiation on the capacity of 3T3-L1 cells to undergo apoptosis in response to growth factor deprivation. Int. J. Obes. 1998, 22, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Nixon, A.J.; Lillich, J.T.; Burton-Wurster, N.; Lust, G.; Mohammed, H.O. Differentiated cellular function in fetal chondrocytes cultured with insulin-like growth factor-I and transforming growth factor-β. J. Orthop. Res. 1998, 16, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Byrne, K.M.; Cheng, X.; Vierck, J.; Erickson, S.; Greene, E.A.; Duckett, S.; Dodson, M.V. Use of a 96-well plate reader to evaluate proliferation of equine satellite cell clones in vitro. Methods Cell Sci. 1998, 19, 311–316. [Google Scholar] [CrossRef]
- Dodson, M.V.; McFarland, D.C.; Grant, A.L.; Doumit, M.E.; Velleman, S.G. Extrinsic regulation of domestic animal-derived satellite cells. Domest. Anim. Endocrinol. 1996, 13, 107–126. [Google Scholar] [CrossRef]
- Lindsay, C.D.; Hambrook, J.L. Diisopropylglutathione ester protects A549 cells from the cytotoxic effects of sulphur mustard. Hum. Exp. Toxicol. 1998, 17, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, R.; Abe, M. Unfused C2C12 mouse skeletal muscle cells express neurofilament 140K protein. Cell Struct. Funct. 1997, 22, 117–121. [Google Scholar] [CrossRef]
- Tatsumi, R.; Anderson, J.E.; Nevoret, C.J.; Halevy, O.; Allen, R.E. HGF/SF is present in normal adult skeletal muscle and is capable of activating satellite cells. Dev. Biol. 1998, 194, 114–128. [Google Scholar] [CrossRef]
- Kumar, N.; Afjei, R.; Massoud, T.F.; Paulmurugan, R. Comparison of cell-based assays to quantify treatment effects of anticancer drugs identifies a new application for Bodipy-L-cystine to measure apoptosis. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Stewart, N.T.; Byrne, K.M.; Hosick, H.L.; Vierck, J.L.; Dodson, M.V. Traditional and emerging methods for analyzing cell activity in cell culture. Methods Cell Sci. 2000, 22, 67–78. [Google Scholar] [CrossRef]
- Cornelison, D.D.W.; Wold, B.J. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev. Biol. 1997, 191, 270–283. [Google Scholar] [CrossRef]
- Frank, A.J.; Proctor, S.J.; Tilby, M.J. Detection and quantification of melphalan-DNA adducts at the single cell level in hematopoietic tumor cells. Blood 1996, 88, 977–984. [Google Scholar] [CrossRef] [PubMed]
- Florini, R. Assay of Creatine Kinase in Microtiter Plates Using. Anal. Biochem. 1989, 182, 399–404. [Google Scholar] [CrossRef]
- Watson, J.; Ebra, E. Animal Cell Culture: A Practical Approach; Freshney, R., Ed.; Oxford University Press: Oxford, UK, 1992; ISBN 9786468600. [Google Scholar]
- Tajima, S.; Tsuji, K.; Ebihara, Y.; Su, X.; Tanaka, R.; Muraoka, K.; Yoshida, M.; Yamada, K.; Yasukawa, K.; Taga, T.; et al. Summary for Policymakers. J. Exp. Med. 1996, 184, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Morgan, H.; Sun, T.; Holmes, D.; Gawad, S.; Green, N.G. Single cell dielectric spectroscopy. J. Phys. D Appl. Phys. 2007, 40, 61–70. [Google Scholar] [CrossRef]
- Lei, K.F. Review on impedance detection of cellular responses in micro/nano environment. Micromachines 2014, 5, 1–12. [Google Scholar] [CrossRef]
- Qiu, Y.; Liao, R.; Zhang, X. Real-time monitoring primary cardiomyocyte adhesion based on electrochemical impedance spectroscopy and electrical cell-substrate impedance sensing. Anal. Chem. 2008, 80, 990–996. [Google Scholar] [CrossRef]
- Qiu, Y.; Liao, R.; Zhang, X. Impedance-based monitoring of ongoing cardiomyocyte death induced by tumor necrosis factor-α. Biophys. J. 2009, 96, 1985–1991. [Google Scholar] [CrossRef]
- Xu, T.; Lizarralde, M.; Nemer, W.E.; Pioufle, B.L.; Français, O. Monitoring Biological Cell Flow within a Mimicking Capillary Device with Impedance Measurement. Proceedings 2017, 1, 517. [Google Scholar] [CrossRef]
- Giaever, I.; Keese, C.R. A morphological biosensor for mammalian cells. Nature 1993, 366, 591–592. [Google Scholar] [CrossRef]
- Doijen, J.; Van Loy, T.; Landuyt, B.; Luyten, W.; Schols, D.; Schoofs, L. Advantages and shortcomings of cell-based electrical impedance measurements as a GPCR drug discovery tool. Biosens. Bioelectron. 2019, 137, 33–44. [Google Scholar] [CrossRef]
- Solly, K.; Wang, X.; Xu, X.; Strulovici, B.; Zheng, W. Application of real-time cell electronic sensing (RT-CES) technology to cell-based assays. Assay Drug Dev. Technol. 2004, 2, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Xie, X.; Duan, Y.; Wang, L.; Cheng, Z.; Cheng, J. A review of impedance measurements of whole cells. Biosens. Bioelectron. 2016, 77, 824–836. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wu, C.; Cai, H.; Hu, N.; Zhou, J.; Wang, P. Cell-based biosensors and their application in biomedicine. Chem. Rev. 2014, 114, 6423–6461. [Google Scholar] [CrossRef] [PubMed]
- Gerasimenko, T.; Nikulin, S.; Zakharova, G.; Poloznikov, A.; Petrov, V.; Baranova, A.; Tonevitsky, A. Impedance Spectroscopy as a Tool for Monitoring Performance in 3D Models of Epithelial Tissues. Front. Bioeng. Biotechnol. 2020, 7. [Google Scholar] [CrossRef] [PubMed]
- Stolwijk, J.A.; Wegener, J. Impedance-Based Assays Along the Life Span of Adherent Mammalian Cells In Vitro: From Initial Adhesion to Cell Death. In Bioanalytical Reviews; Wegener, J., Ed.; Springer: Berlin/Heidelberg, Germany, 2019; Volume 2, pp. 1–75. [Google Scholar]
- De León, S.E.; Pupovac, A.; McArthur, S.L. Three-Dimensional (3D) cell culture monitoring: Opportunities and challenges for impedance spectroscopy. Biotechnol. Bioeng. 2020, 117, 1230–1240. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wu, C.; Hu, N.; Hsia, K.J. Micro/nano cell and molecular sensors. In Micro/Nano Cell and Molecular Sensors; Springer: Singapore, 2016; pp. 1–243. ISBN 9789811016585. [Google Scholar]
- Pänke, O.; Weigel, W.; Schmidt, S.; Steude, A.; Robitzki, A.A. A cell-based impedance assay for monitoring transient receptor potential (TRP) ion channel activity. Biosens. Bioelectron. 2011, 26, 2376–2382. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.; Gawad, S.; Renaud, P. Impedance spectroscopy flow cytometry: On-chip label-free cell differentiation. Cytom. Part. A 2005, 65, 124–132. [Google Scholar] [CrossRef]
- Abdolahad, M.; Janmaleki, M.; Taghinejad, M.; Taghnejad, H.; Salehi, F.; Mohajerzadeh, S. Single-cell resolution diagnosis of cancer cells by carbon nanotube electrical spectroscopy. Nanoscale 2013, 5, 3421–3427. [Google Scholar] [CrossRef]
- Wang, J.; Wu, C.; Hu, N.; Zhou, J.; Du, L.; Wang, P. Microfabricated Electrochemical Cell-Based Biosensors for Analysis of Living Cells In Vitro. Biosensors 2012, 2, 127–170. [Google Scholar] [CrossRef]
- Wang, W.; Foley, K.; Shan, X.; Wang, S.; Eaton, S.; Nagaraj, V.J.; Wiktor, P.; Patel, U.; Tao, N. Single cells and intracellular processes studied by a plasmonic-based electrochemical impedance microscopy. Nat. Chem. 2011, 3, 249–255. [Google Scholar] [CrossRef]
- Arndt, S.; Seebach, J.; Psathaki, K.; Galla, H.J.; Wegener, J. Bioelectrical impedance assay to monitor changes in cell shape during apoptosis. Biosens. Bioelectron. 2004, 19, 583–594. [Google Scholar] [CrossRef]
- Asami, K. Characterization of biological cells by dielectric spectroscopy. J. Non. Cryst. Solids 2002, 305, 268–277. [Google Scholar] [CrossRef]
- Gawad, S.; Cheung, K.; Seger, U.; Bertsch, A.; Renaud, P. Dielectric spectroscopy in a micromachined flow cytometer: Theoretical and practical considerations. Lab Chip 2004, 4, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Asami, K.; Yonezawa, T.; Wakamatsu, H.; Dielectric, N.K. Spectroscopy of biological cells. Bioelectrochemistry Bioenerg. 1996, 40, 141–145. [Google Scholar] [CrossRef]
- Benson, K.; Cramer, S.; Galla, H.J. Impedance-based cell monitoring: Barrier properties and beyond. Fluids Barriers CNS 2013, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Q.; Ren, S.; Dong, F. A complete sensor model for miniscopic electrical impedance tomography. In Proceedings of the 2018 IEEE International Instrumentation and Measurement Technology Conference (I2MTC), Houston, TX, USA, 14–17 May 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Wu, H.; Zhou, W.; Yang, Y.; Jia, J.; Bagnaninchi, P. Exploring the potential of electrical impedance tomography for tissue engineering applications. Materials 2018, 11, 930. [Google Scholar] [CrossRef]
- Yang, Y.; Jia, J.; Smith, S.; Jamil, N.; Gamal, W.; Bagnaninchi, P.O. A miniature electrical impedance tomography sensor and 3-D Image Reconstruction for Cell Imaging. IEEE Sens. J. 2017, 17, 514–523. [Google Scholar] [CrossRef]
- Wu, H.; Yang, Y.; Bagnaninchi, P.O.; Jia, J. Electrical impedance tomography for real-time and label-free cellular viability assays of 3D tumour spheroids. Analyst 2018, 143, 4189–4198. [Google Scholar] [CrossRef]
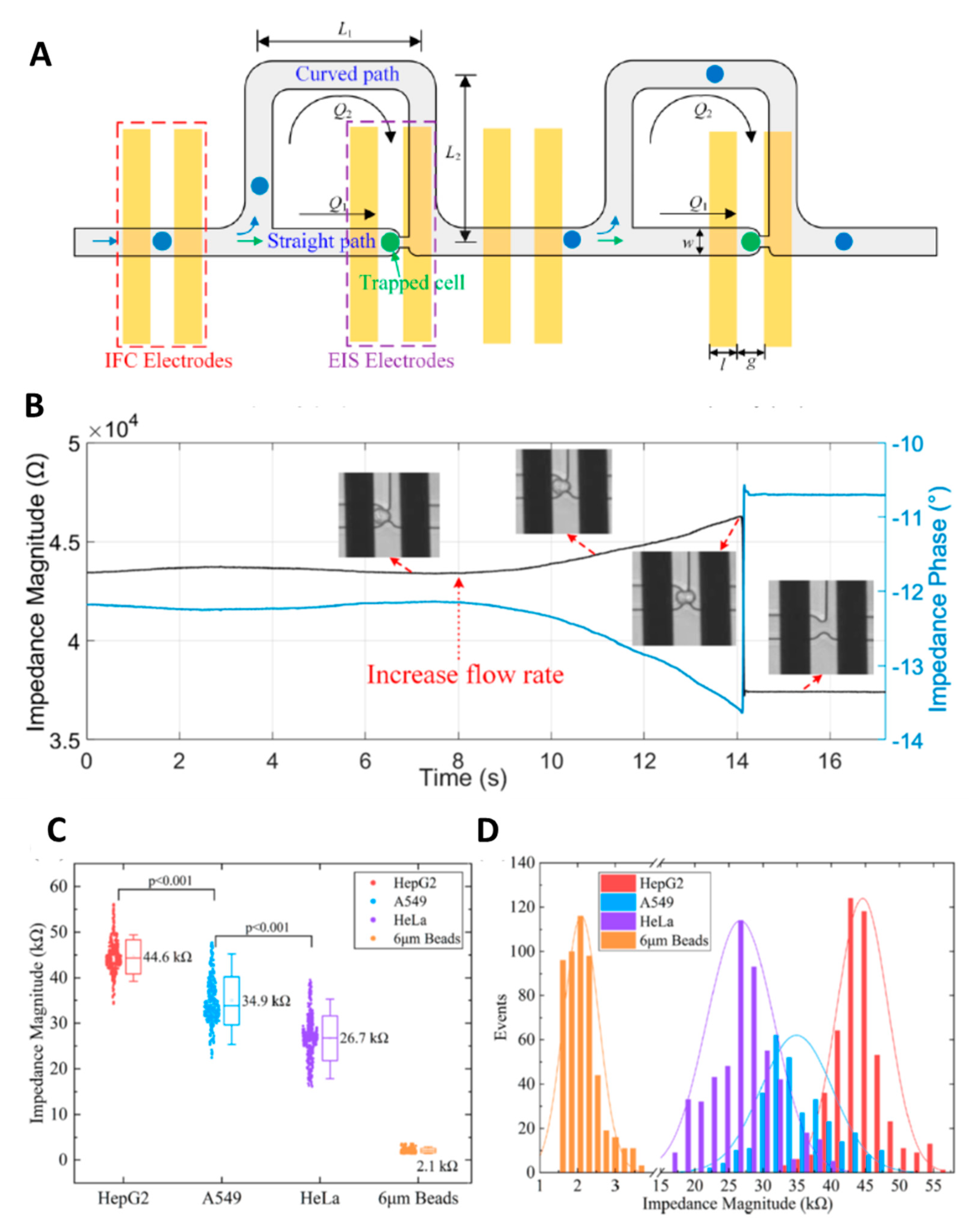
- Feng, Y.; Huang, L.; Zhao, P.; Liang, F.; Wang, W. A Microfluidic Device Integrating Impedance Flow Cytometry and Electric Impedance Spectroscopy for High-Efficiency Single-Cell Electrical Property Measurement. Anal. Chem. 2019, 91, 15204–15212. [Google Scholar] [CrossRef]
- Nguyen, T.A.; Yin, T.I.; Reyes, D.; Urban, G.A. Microfluidic chip with integrated electrical cell-impedance sensing for monitoring single cancer cell migration in three-dimensional matrixes. Anal. Chem. 2013, 85, 11068–11076. [Google Scholar] [CrossRef]
- Pandya, H.J.; Dhingra, K.; Prabhakar, D.; Chandrasekar, V.; Natarajan, S.K.; Vasan, A.S.; Kulkarni, A.; Shafiee, H. A microfluidic platform for drug screening in a 3D cancer microenvironment. Biosens. Bioelectron. 2017, 94, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Chawla, K.; Modena, M.M.; Ravaynia, P.S.; Lombardo, F.C.; Leonhardt, M.; Panic, G.; Bürgel, S.C.; Keiser, J.; Hierlemann, A. Impedance-Based Microfluidic Assay for Automated Antischistosomal Drug Screening. Sensors 2018, 3, 2613–2620. [Google Scholar] [CrossRef] [PubMed]
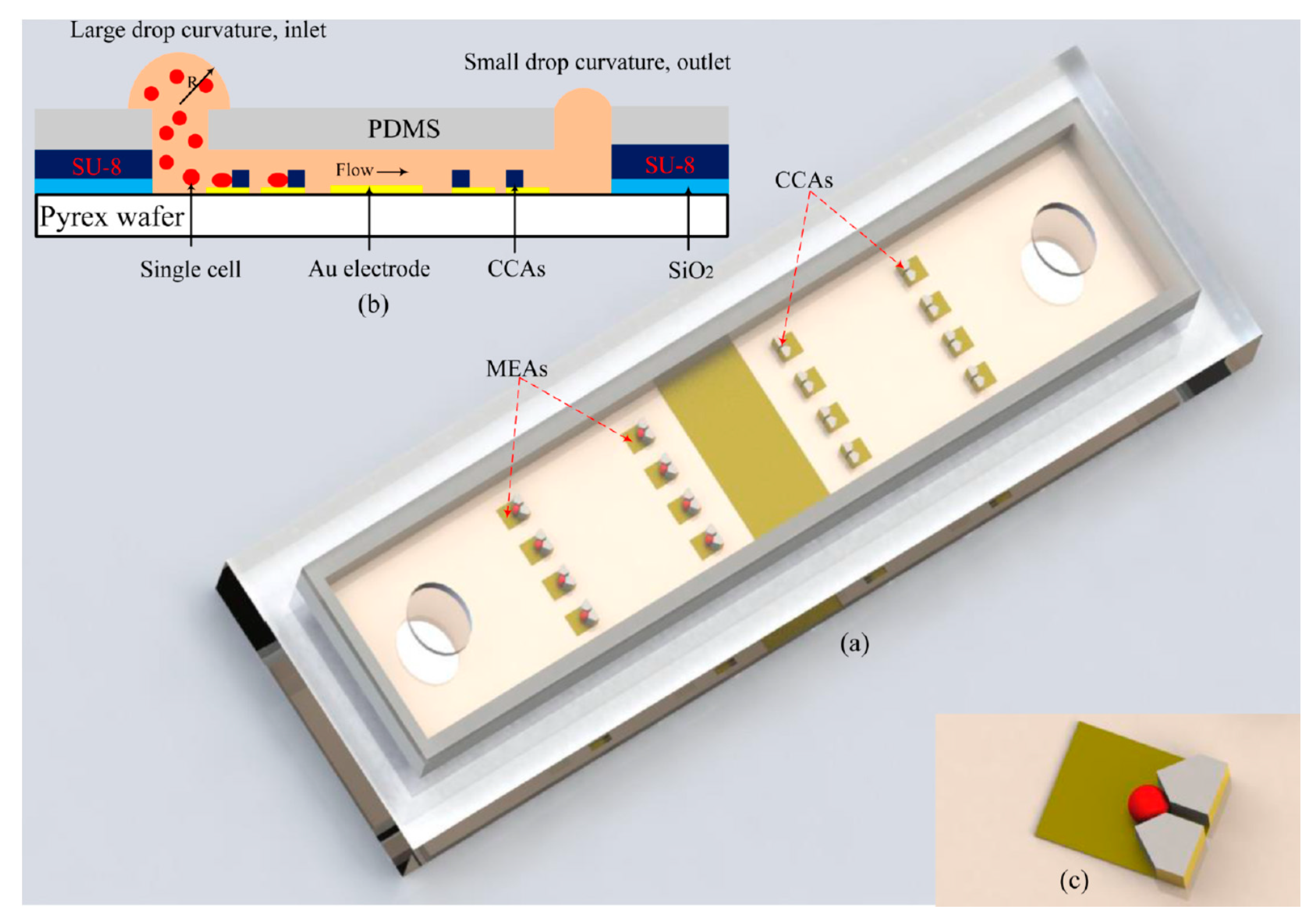
- Panwar, J.; Roy, R. Integrated Field’s metal microelectrodes based microfluidic impedance cytometry for cell-in-droplet quantification. Microelectron. Eng. 2019, 215, 111010. [Google Scholar] [CrossRef]
- Cheung, K.C.; Berardino, M.D.; Schade-Kampmann, G.; Hebeisen, M.; Pierzchalski, A.; Bocsi, J.; Mittag, A.; Tárnok, A. Microfluidic impedance-based flow cytometry. Cytom. Part A 2010, 77, 648–666. [Google Scholar] [CrossRef]
- Claudel, J.; Nadi, M.; El Mazria, O.; Kourtiche, D. High reliability microfluidic biosensor for single cell impedance cytometry. In Proceedings of the 2017 Eleventh International Conference on Sensing Technology (ICST), Sydney, NSW, Australia, 4–6 December 2017; pp. 1–5. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA. Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Andrade, E.L.; Bento, A.F.; Cavalli, J.; Oliveira, S.K.; Freitas, C.S.; Marcon, R.; Schwanke, R.C.; Siqueira, J.M.; Calixto, J.B. Non-clinical studies required for new drug development—Part I: Early in silico and in vitro studies, new target discovery and validation, proof of principles and robustness of animal studies. Braz. J. Med. Biol. Res. 2016, 49, 1–9. [Google Scholar] [CrossRef]
- Ediriweera, M.K.; Tennekoon, K.H.; Samarakoon, S.R. In vitro assays and techniques utilized in anticancer drug discovery. J. Appl. Toxicol. 2019, 39, 38–71. [Google Scholar] [CrossRef]
- Korpal, M.; Feala, J.; Puyang, X.; Zou, J.; Ramos, A.H.; Wu, J.; Baumeister, T.; Yu, L.; Warmuth, M.; Zhu, P. Implementation of in vitro drug resistance assays: Maximizing the potential for uncovering clinically relevant resistance mechanisms. J. Vis. Exp. 2015, 106, e52879. [Google Scholar] [CrossRef]
- Polacheck, W.J.; Zervantonakis, I.K.; Kamm, R.D. Tumor cell migration in complex microenvironments. Cell. Mol. Life Sci. 2013, 70, 1335–1356. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Wyckoff, J.; Condeelis, J. Cell migration in tumors. Curr. Opin. Cell Biol. 2005, 17, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Hulkower, K.I.; Herber, R.L. Cell migration and invasion assays as tools for drug discovery. Pharmaceutics 2011, 3, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Stolwijk, J.A.; Wegener, J. Impedance-Based Assays Along the Life Span of Adherent Mammalian Cells In Vitro: From Initial Adhesion to Cell Death. In Label-Free Monitoring of Cells In Vitro; Matysik, F.-M., Wegener, J., Eds.; Springer Nature Switzerland AG.: Cham, Switzerland, 2019; ISBN 9783030324322. [Google Scholar]
- Lundstrom, K. Cell-impedance-based label-free technology for the identification of new drugs. Expert Opin. Drug Discov. 2017, 12, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Jiang, D.; Gu, C.; Qiu, Y.; Wan, H.; Wang, P. 3D microgroove electrical impedance sensing to examine 3D cell cultures for antineoplastic drug assessment. Microsystems Nanoeng. 2020, 6. [Google Scholar] [CrossRef]
- Jahnke, H.G.; Mewes, A.; Zitzmann, F.D.; Schmidt, S.; Azendorf, R.; Robitzki, A.A. Electrochemical live monitoring of tumor cell migration out of micro-tumors on an innovative multiwell high-dense microelectrode array. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Seidel, D.; Rothe, R.; Kirsten, M.; Jahnke, H.G.; Dumann, K.; Ziemer, M.; Simon, J.C.; Robitzki, A.A. A multidimensional impedance platform for the real-time analysis of single and combination drug pharmacology in patient-derived viable melanoma models. Biosens. Bioelectron. 2019, 123, 185–194. [Google Scholar] [CrossRef]
- Ahuja, K.; Rather, G.M.; Lin, Z.; Sui, J.; Xie, P.; Le, T.; Bertino, J.R.; Javanmard, M. Toward point-of-care assessment of patient response: A portable tool for rapidly assessing cancer drug efficacy using multifrequency impedance cytometry and supervised machine learning. Microsyst. Nanoeng. 2019, 5, 1–11. [Google Scholar] [CrossRef]
- Kumar, M.; Yadav, S.; Kumar, A.; Sharma, N.N.; Akhtar, J.; Singh, K. MEMS impedance flow cytometry designs for effective manipulation of micro entities in health care applications. Biosens. Bioelectron. 2019, 142, 111526. [Google Scholar] [CrossRef]
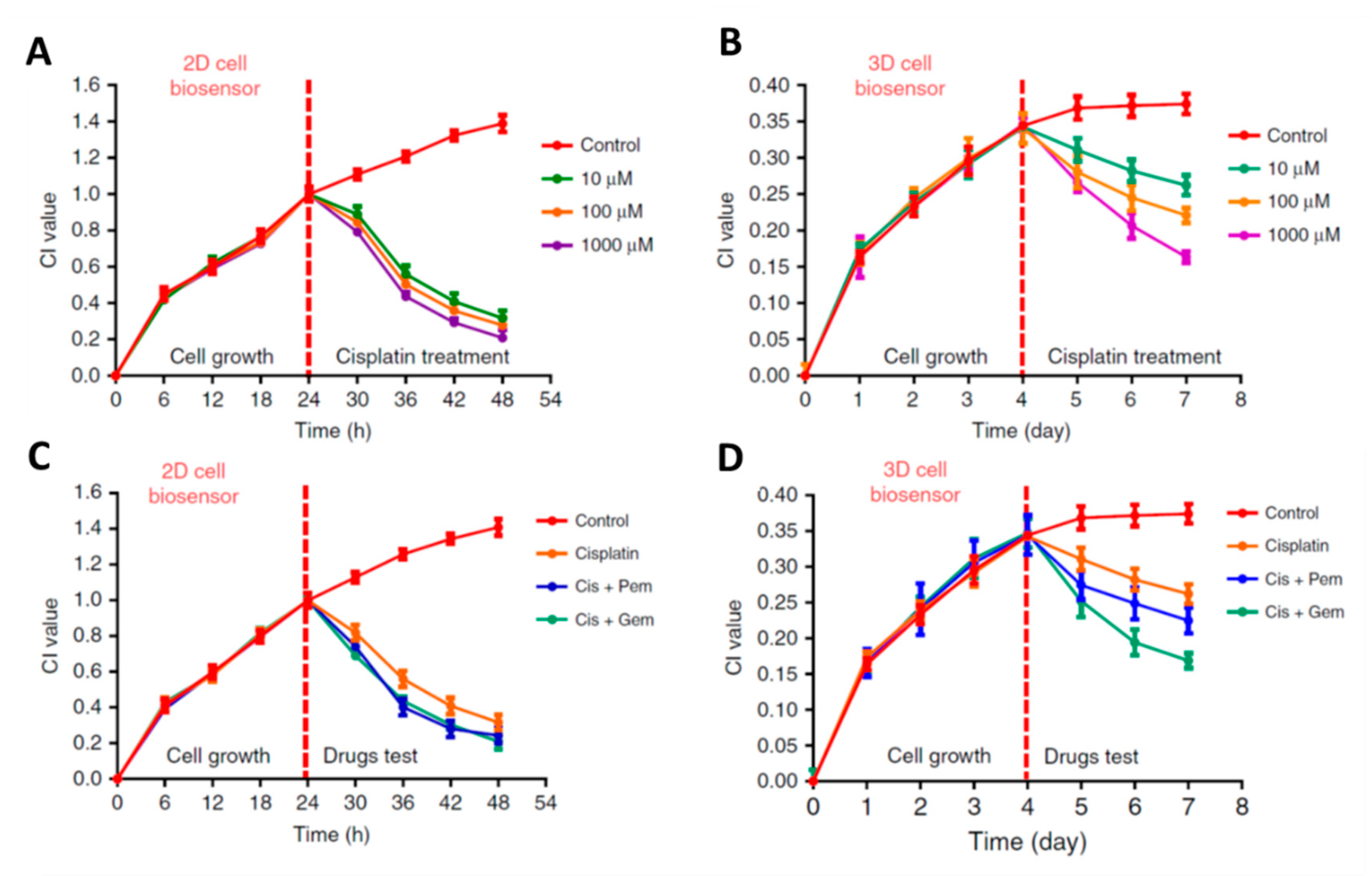
- Pan, Y.; Hu, N.; Wei, X.; Gong, L.; Zhang, B.; Wan, H.; Wang, P. 3D cell-based biosensor for cell viability and drug assessment by 3D electric cell/matrigel-substrate impedance sensing. Biosens. Bioelectron. 2019, 130, 344–351. [Google Scholar] [CrossRef]
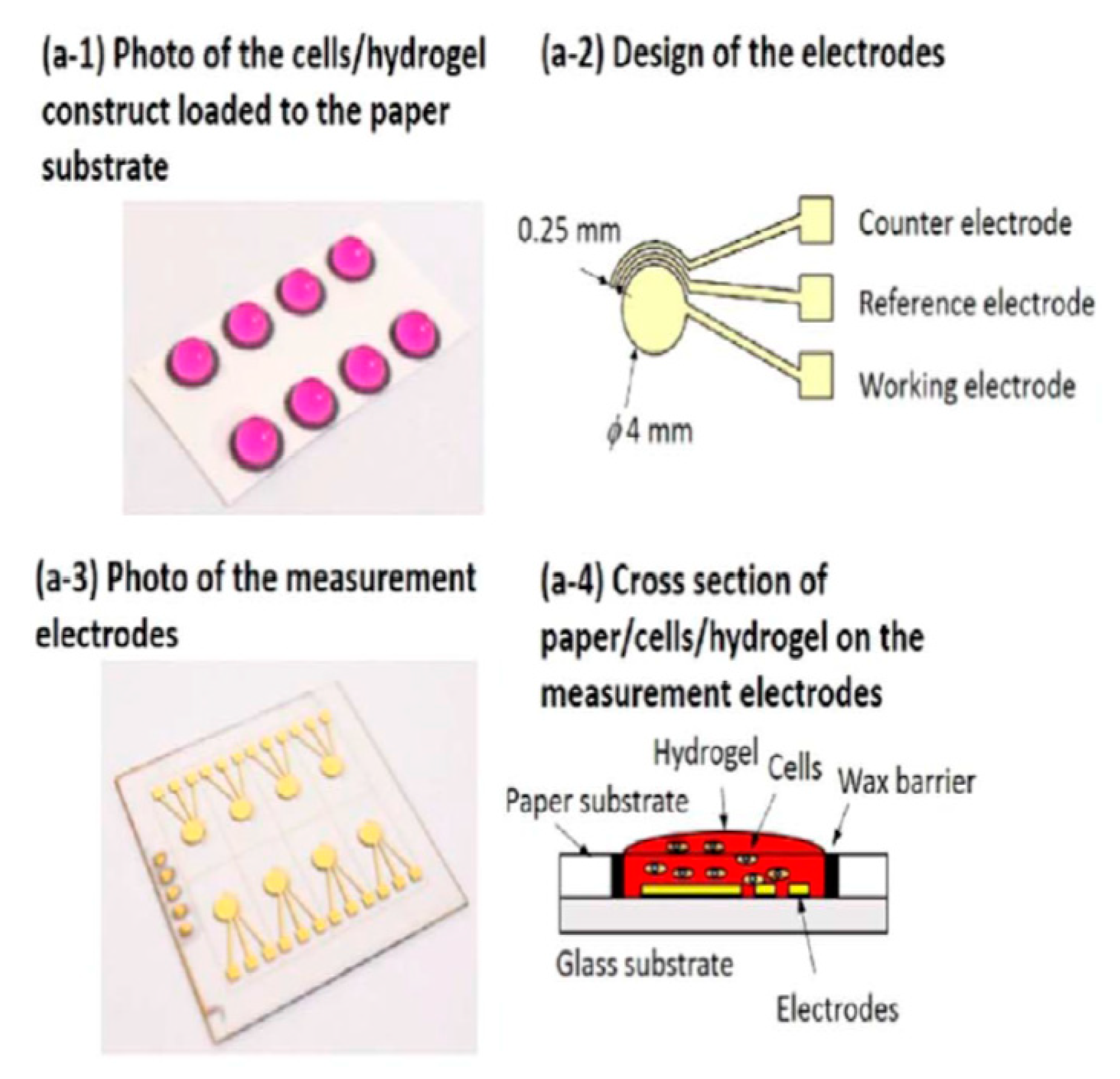
- Lei, K.F.; Liu, T.K.; Tsang, N.M. Towards a high throughput impedimetric screening of chemosensitivity of cancer cells suspended in hydrogel and cultured in a paper substrate. Biosens. Bioelectron. 2018, 100, 355–360. [Google Scholar] [CrossRef]
- Adcock, A.F.; Agbai, C.O.; Yang, L. Application of electric cell-substrate impedance sensing toward personalized anti-cancer therapeutic selection. J. Anal. Sci. Technol. 2018, 9. [Google Scholar] [CrossRef]
- Bilican, I.; Guler, M.T.; Serhatlioglu, M.; Kirindi, T.; Elbuken, C. Focusing-free impedimetric differentiation of red blood cells and leukemia cells: A system optimization. Sens. Actuators B Chem. 2020, 307. [Google Scholar] [CrossRef]
- Lin, Z.; Lin, S.Y.; Xie, P.; Lin, C.Y.; Rather, G.M.; Bertino, J.R.; Javanmard, M. Rapid Assessment of Surface Markers on Cancer Cells Using Immuno-Magnetic Separation and Multi-frequency Impedance Cytometry for Targeted Therapy. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Steeg, P.S. Tumor metastasis: Mechanistic insights and clinical challenges. Nat. Med. 2006, 12, 895–904. [Google Scholar] [CrossRef]
- Massagué, J.; Obenauf, A.C. Metastatic colonization by circulating tumour cells. Nature 2016, 529, 298–306. [Google Scholar] [CrossRef]
- Ghassemi, P.; Ren, X.; Foster, B.M.; Kerr, B.A.; Agah, M. Post-enrichment circulating tumor cell detection and enumeration via deformability impedance cytometry. Biosens. Bioelectron. 2020, 150, 111868. [Google Scholar] [CrossRef]
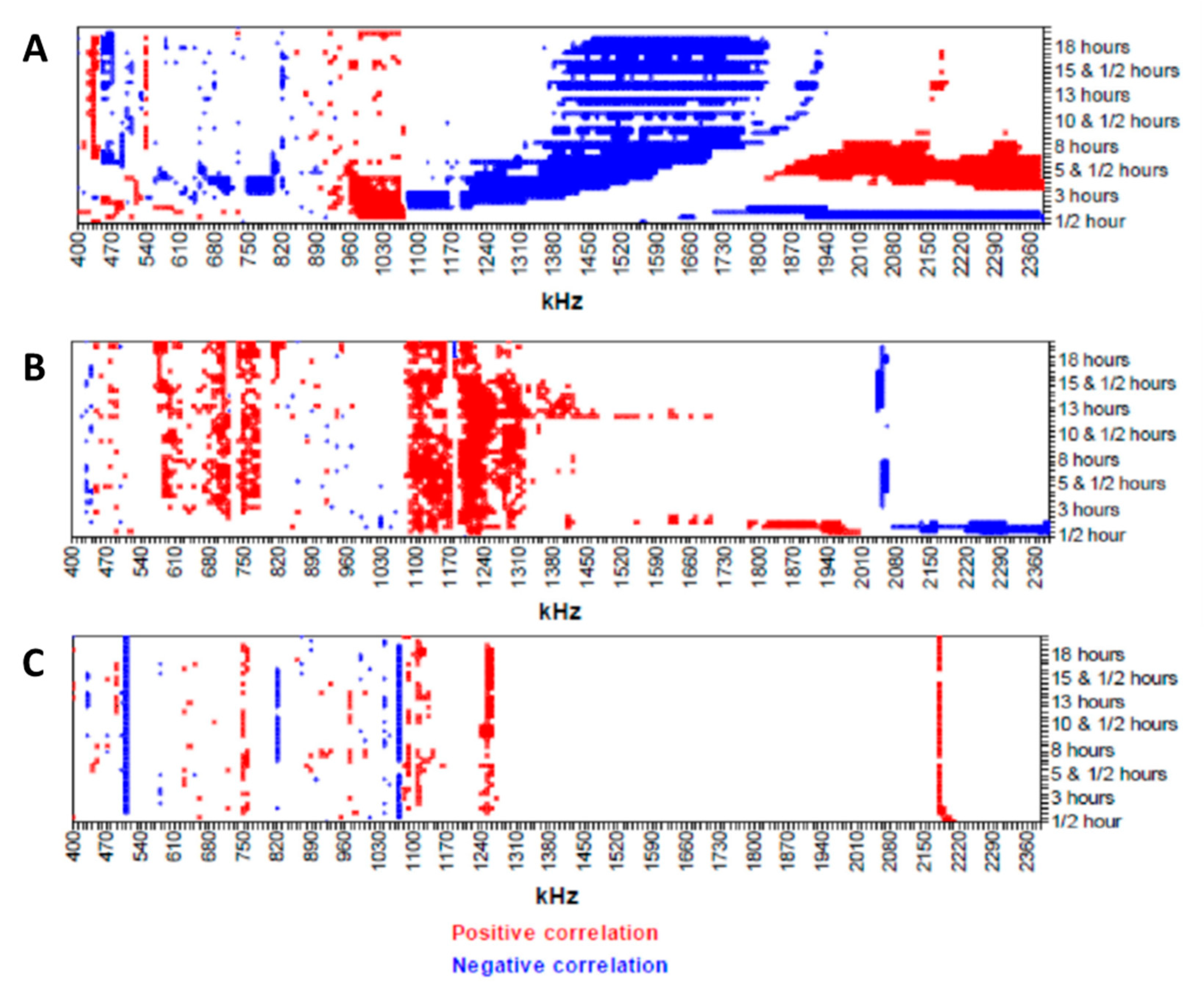
- Jahnke, H.G.; Braesigk, A.; Mack, T.G.A.; Pönick, S.; Striggow, F.; Robitzki, A.A. Impedance spectroscopy based measurement system for quantitative and label-free real-time monitoring of tauopathy in hippocampal slice cultures. Biosens. Bioelectron. 2012, 32, 250–258. [Google Scholar] [CrossRef]
- Jahnke, H.G.; Rothermel, A.; Sternberger, I.; MacK, T.G.A.; Kurz, R.G.; Pänke, O.; Striggow, F.; Robitzki, A.A. An impedimetric microelectrode-based array sensor for label-free detection of tau hyperphosphorylation in human cells. Lab Chip 2009, 9, 1422–1428. [Google Scholar] [CrossRef]
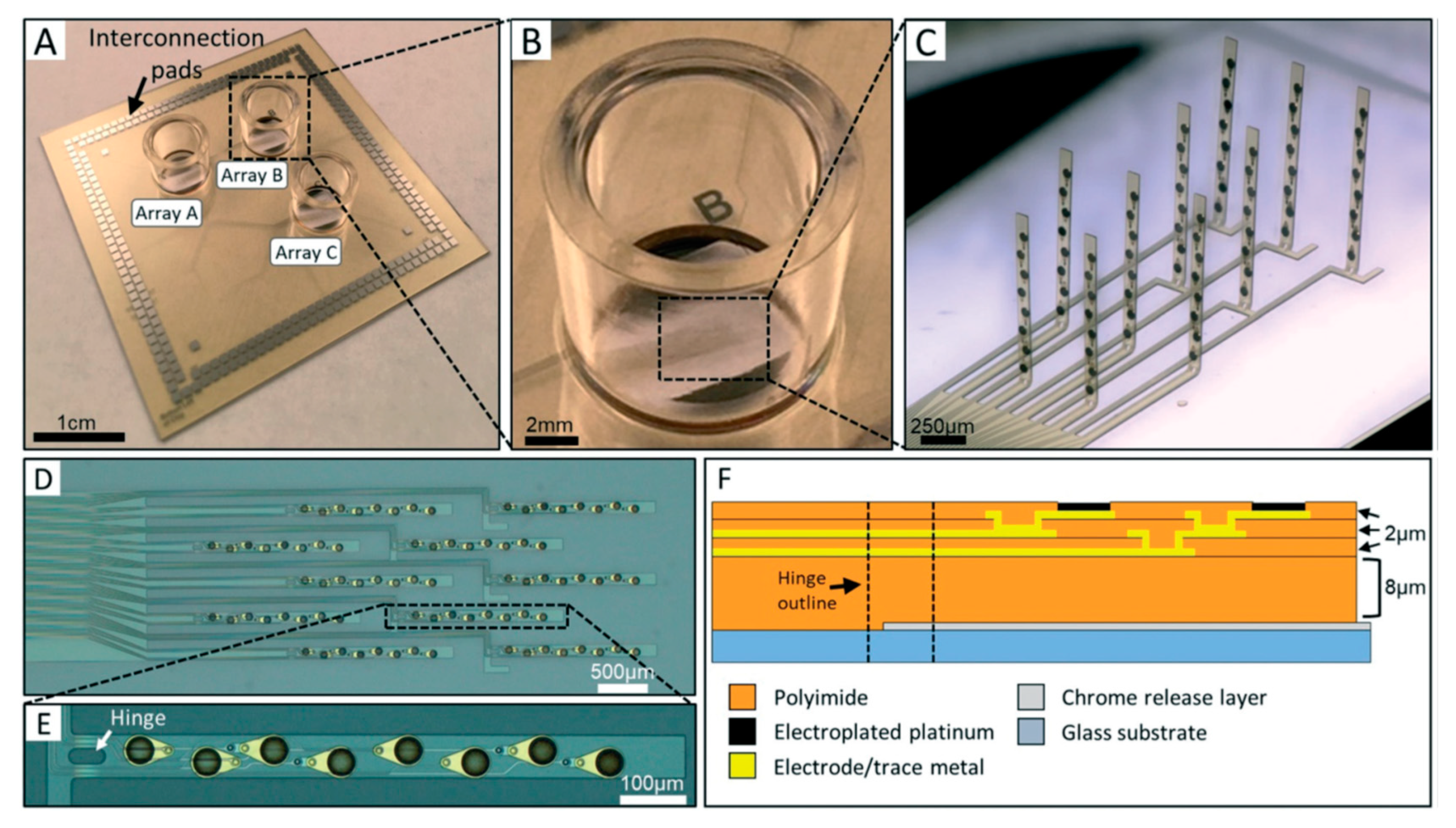
- Soscia, D.A.; Lam, D.; Tooker, A.C.; Enright, H.A.; Triplett, M.; Karande, P.; Peters, S.K.G.; Sales, A.P.; Wheeler, E.K.; Fischer, N.O. A flexible 3-dimensional microelectrode array for: In vitro brain models. Lab Chip 2020, 20, 901–911. [Google Scholar] [CrossRef]
- Zuo, L.; Yu, S.; Briggs, C.A.; Kantor, S.; Pan, J.Y. Design and fabrication of a three-dimensional multi-electrode array for neuron electrophysiology. J. Biomech. Eng. 2017, 139, 1–6. [Google Scholar] [CrossRef]
- Stamp, M.E.M.; Tong, W.; Ganesan, K.; Prawer, S.; Ibbotson, M.R.; Garrett, D.J. 3D Diamond Electrode Array for High-Acuity Stimulation in Neural Tissue. ACS Appl. Bio Mater. 2020, 3, 1544–1552. [Google Scholar] [CrossRef]
- Jahnke, H.G.; Krinke, D.; Seidel, D.; Lilienthal, K.; Schmidt, S.; Azendorf, R.; Fischer, M.; Mack, T.; Striggow, F.; Althaus, H.; et al. A novel 384-multiwell microelectrode array for the impedimetric monitoring of Tau protein induced neurodegenerative processes. Biosens. Bioelectron. 2017, 88, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Krinke, D.; Jahnke, H.G.; Mack, T.G.A.; Hirche, A.; Striggow, F.; Robitzki, A.A. A novel organotypic tauopathy model on a new microcavity chip for bioelectronic label-free and real time monitoring. Biosens. Bioelectron. 2010, 26, 162–168. [Google Scholar] [CrossRef]
- Seidel, D.; Obendorf, J.; Englich, B.; Jahnke, H.G.; Semkova, V.; Haupt, S.; Girard, M.; Peschanski, M.; Brüstle, O.; Robitzki, A.A. Impedimetric real-time monitoring of neural pluripotent stem cell differentiation process on microelectrode arrays. Biosens. Bioelectron. 2016, 86, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Arendt, T.; Stieler, J.T.; Holzer, M. Tau and tauopathies. Brain Res. Bull. 2016, 126, 238–292. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Noble, W.; Hanger, D.P. Roles of tau protein in health and disease. Acta Neuropathol. 2017, 133, 665–704. [Google Scholar] [CrossRef]
- Kroschinsky, F.; Stölzel, F.; von Bonin, S.; Beutel, G.; Kochanek, M.; Kiehl, M.; Schellongowski, P. New drugs, new toxicities: Severe side effects of modern targeted and immunotherapy of cancer and their management. Crit. Care 2017, 21, 1–11. [Google Scholar] [CrossRef]
- Jiang, H.; Qiu, Y.; Hou, W.; Cheng, X.; Yim, M.; Ching, W.K. Drug Side-effect Profiles Prediction: From Empirical Risk Minimization to Structural Risk Minimization. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 17, 402–410. [Google Scholar] [CrossRef]
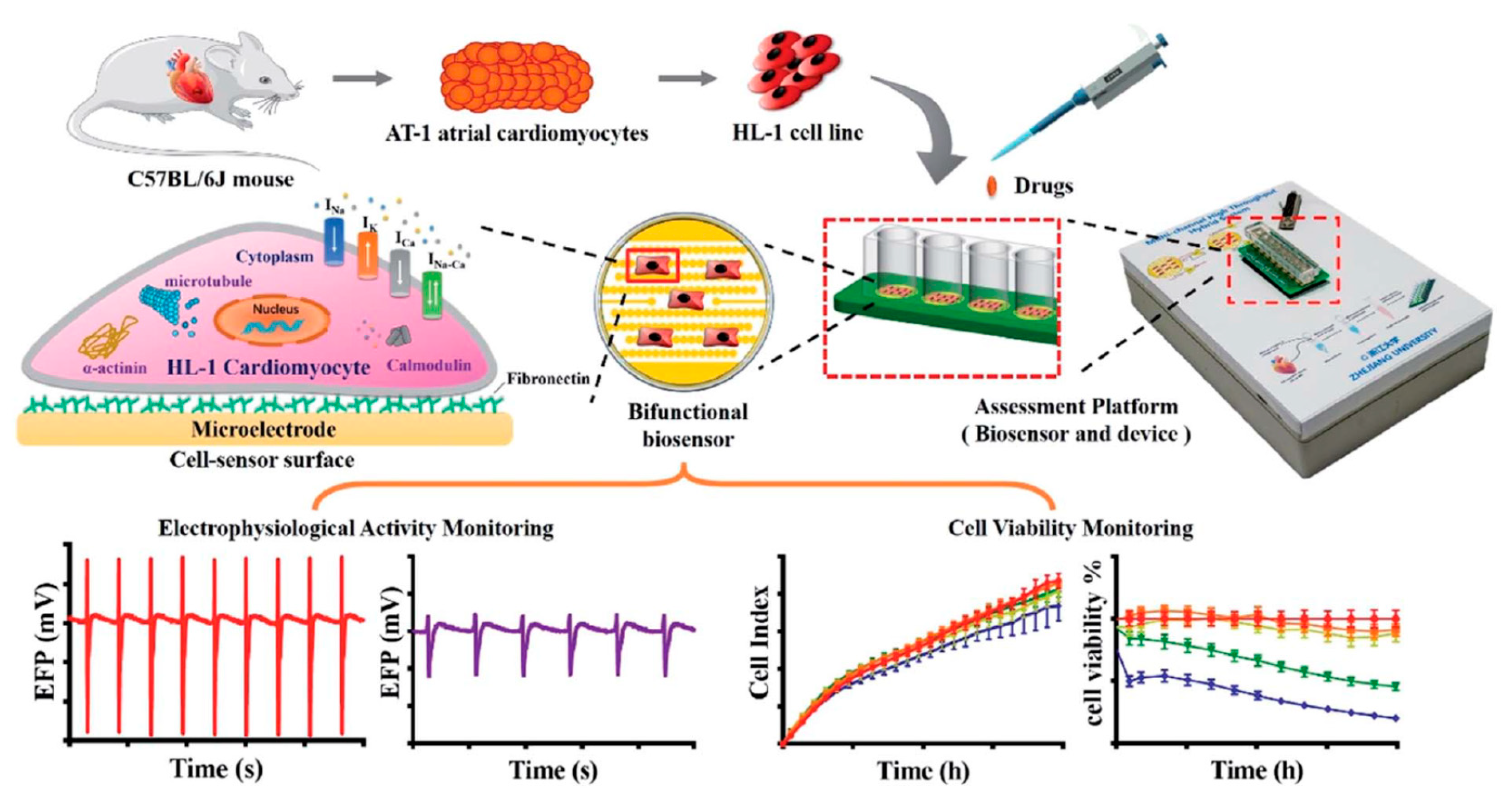
- Gu, C.; Wei, X.; Pan, Y.; Liang, T.; Gan, Y.; Gao, K.; Qiu, Y.; Wan, H.; Wang, P. High-temporal-range drug-induced cardiac side-effect evaluation using simultaneous HL-1-based impedance and long-term electrophysiology recording systems. Anal. Methods 2019, 11, 5250–5259. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, T.; Wang, P.; Hu, N. High-throughput assessment of drug cardiac safety using a high-speed impedance detection technology-based heart-on-a-chip. Micromachines 2016, 7, 122. [Google Scholar] [CrossRef]
- Kustermann, S.; Boess, F.; Buness, A.; Schmitz, M.; Watzele, M.; Weiser, T.; Singer, T.; Suter, L.; Roth, A. A label-free, impedance-based real time assay to identify drug-induced toxicities and differentiate cytostatic from cytotoxic effects. Toxicol. Vitr. 2013, 27, 1589–1595. [Google Scholar] [CrossRef] [PubMed]
- Gasser, A.; Eveness, J.; Kiely, J.; Attwood, D.; Luxton, R. A non-contact impedimetric biosensing system for classification of toxins associated with cytotoxicity testing. Bioelectrochemistry 2020, 133, 107448. [Google Scholar] [CrossRef] [PubMed]
- Breslin, S.; O’Driscoll, L. Three-dimensional cell culture: The missing link in drug discovery. Drug Discov. Today 2013, 18, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Alexander, F.; Eggert, S.; Price, D. Label-Free Monitoring of 3D Tissue Models via Electrical Impedance Spectroscopy. In Bioanalytical Reviews; Wegener, J., Ed.; Springer: Berlin/Heidelberg, Germany, 2019; Volume 2, pp. 111–134. [Google Scholar]
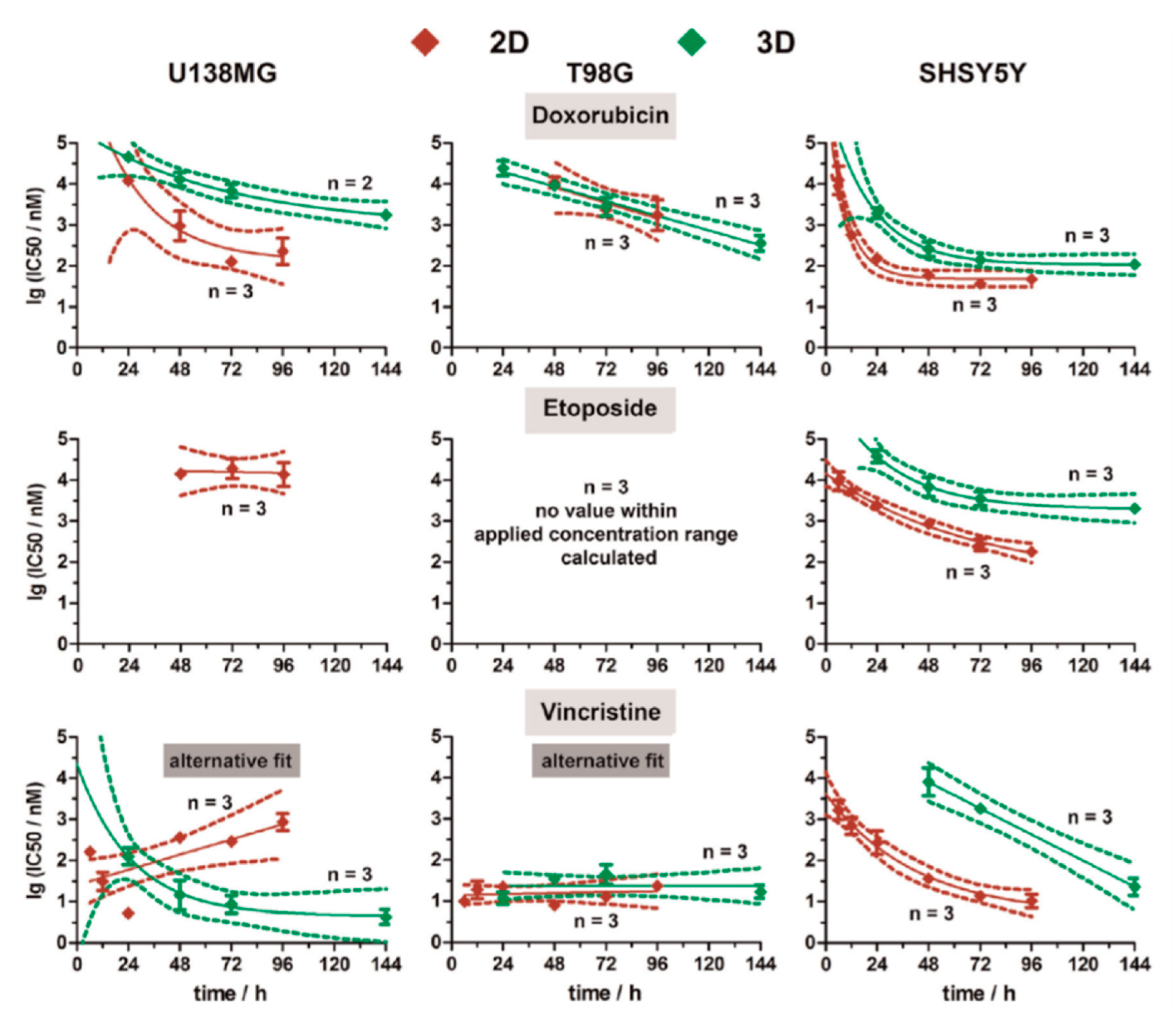
- Eichler, M.; Jahnke, H.G.; Krinke, D.; Müller, A.; Schmidt, S.; Azendorf, R.; Robitzki, A.A. A novel 96-well multielectrode array based impedimetric monitoring platform for comparative drug efficacy analysis on 2D and 3D brain tumor cultures. Biosens. Bioelectron. 2015, 67, 582–589. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassan, Q.; Ahmadi, S.; Kerman, K. Recent Advances in Monitoring Cell Behavior Using Cell-Based Impedance Spectroscopy. Micromachines 2020, 11, 590. https://doi.org/10.3390/mi11060590
Hassan Q, Ahmadi S, Kerman K. Recent Advances in Monitoring Cell Behavior Using Cell-Based Impedance Spectroscopy. Micromachines. 2020; 11(6):590. https://doi.org/10.3390/mi11060590
Chicago/Turabian StyleHassan, Qusai, Soha Ahmadi, and Kagan Kerman. 2020. "Recent Advances in Monitoring Cell Behavior Using Cell-Based Impedance Spectroscopy" Micromachines 11, no. 6: 590. https://doi.org/10.3390/mi11060590
APA StyleHassan, Q., Ahmadi, S., & Kerman, K. (2020). Recent Advances in Monitoring Cell Behavior Using Cell-Based Impedance Spectroscopy. Micromachines, 11(6), 590. https://doi.org/10.3390/mi11060590






