Single Cell Analysis of Neutrophils NETs by Microscopic LSPR Imaging System
Abstract
1. Introduction
2. Materials and Methods
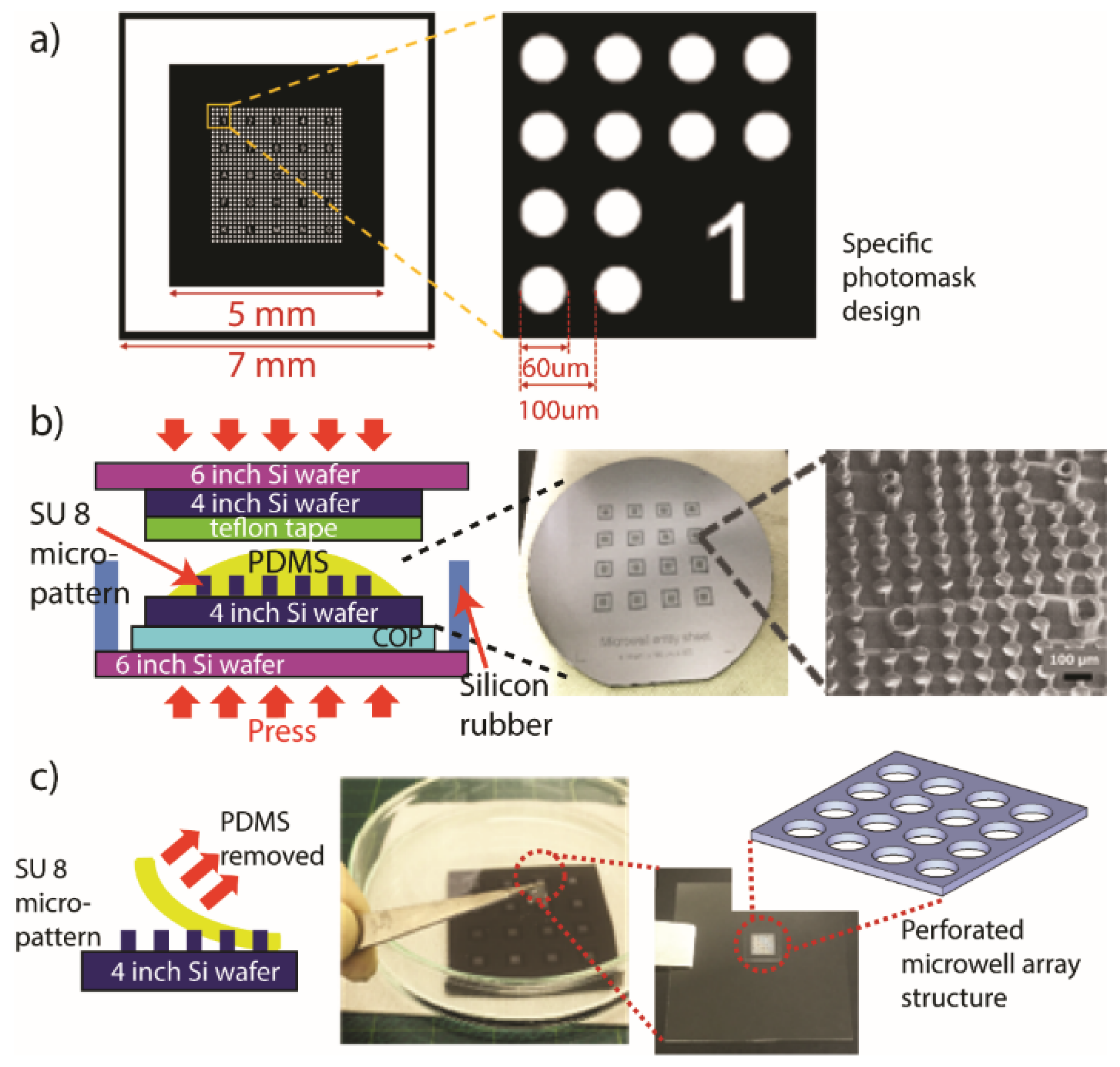
2.1. Fabrication of the Gold Sputtered Plasmonic Substrate
2.2. Fabrication of PDMS Microwell Array (MWA) Sheet
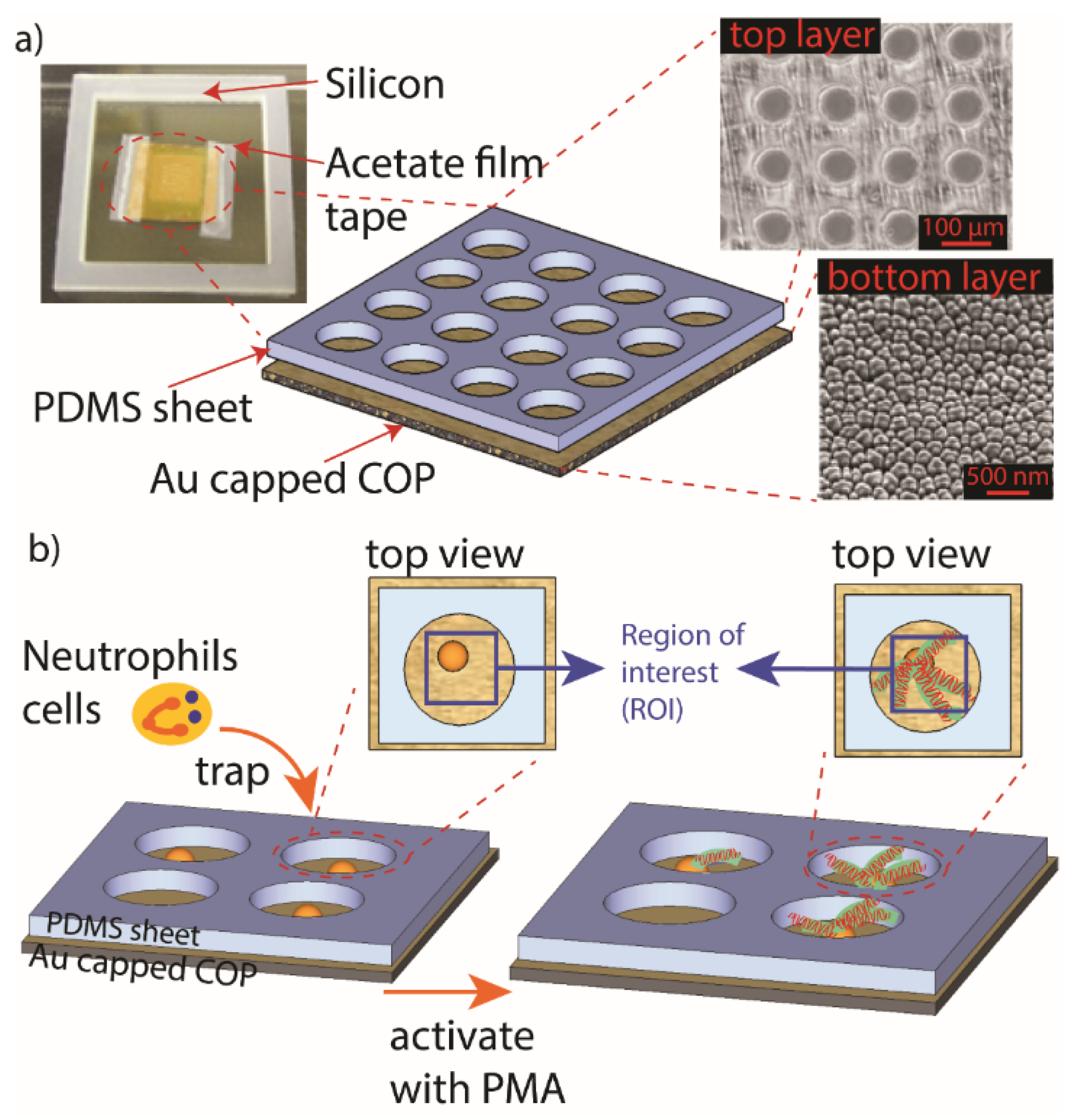
2.3. Assembly of MWA Plasmon Chips
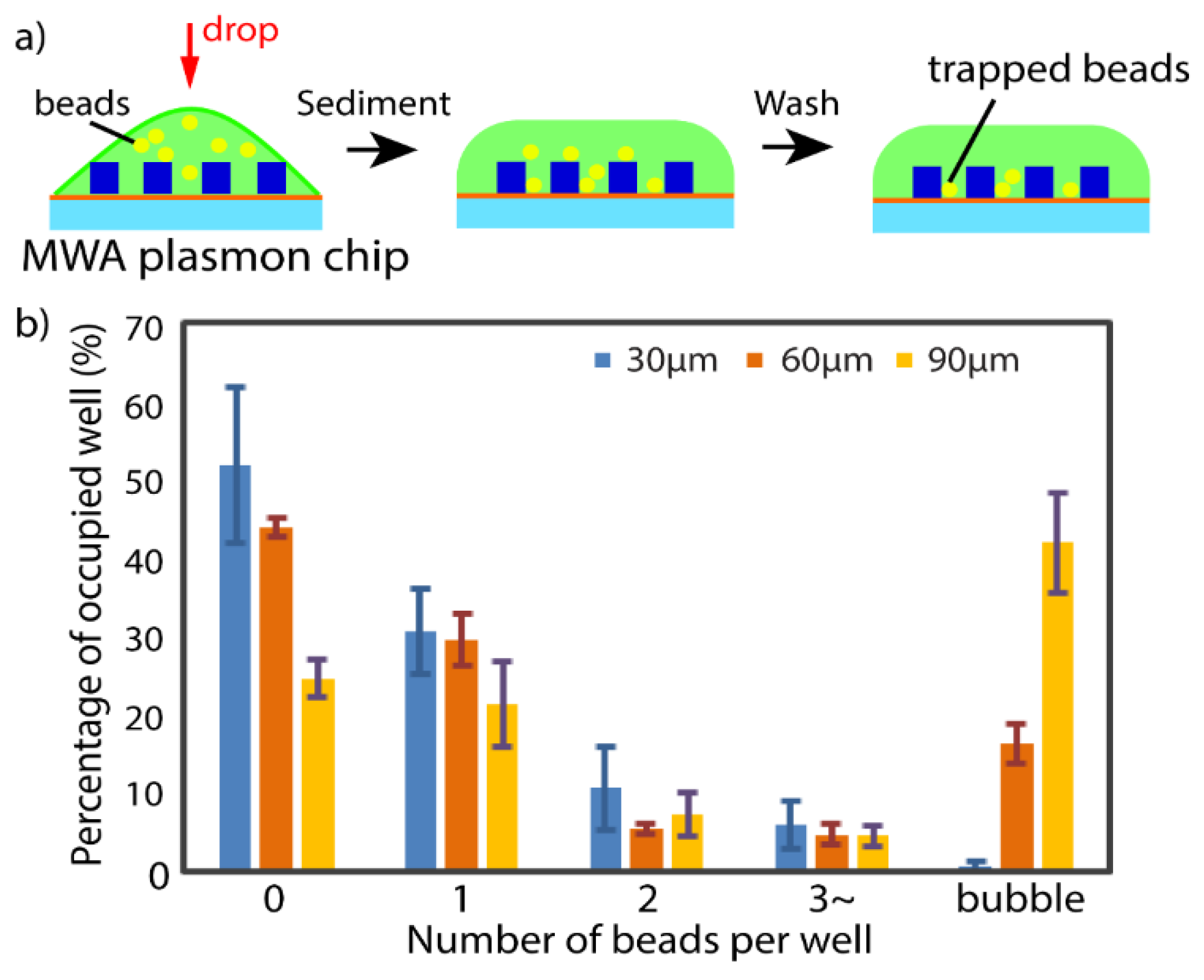
2.4. Optimization of MWA Sheet Thickness
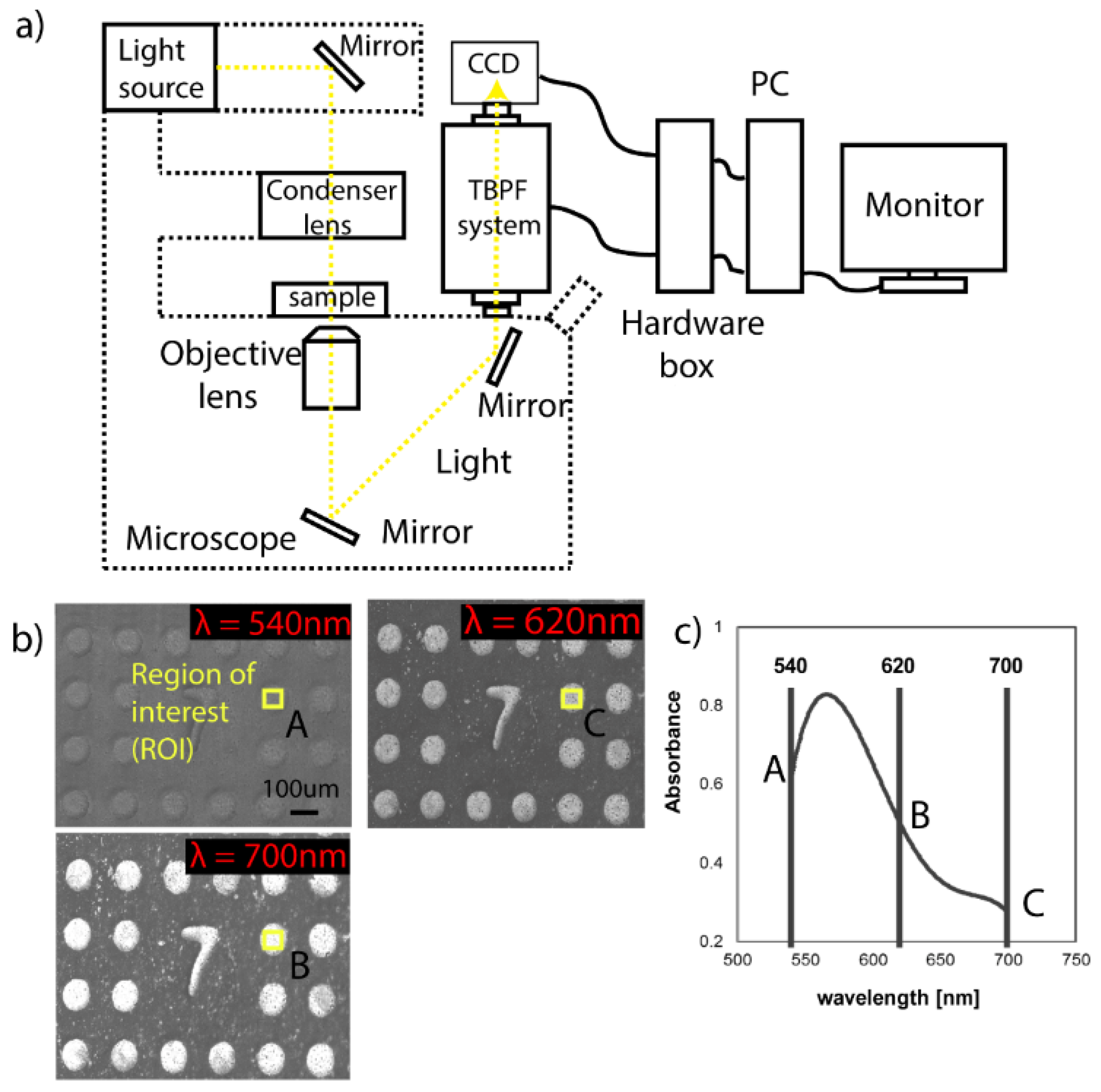
2.5. Hyperspectral Imaging System
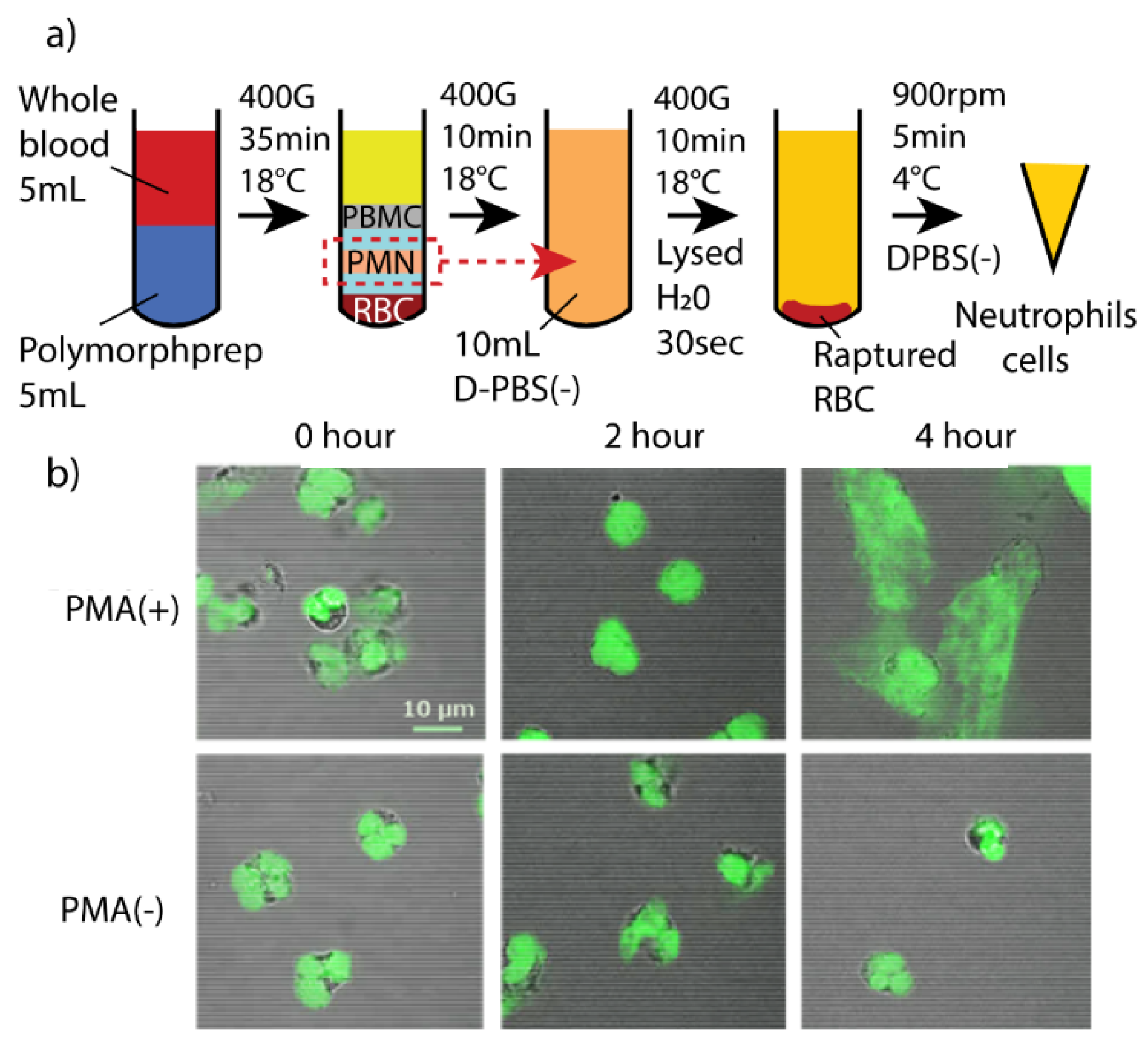
2.6. Extraction of Neutrophils from Raw Blood
2.7. Neutrophils Isolation Using MWA Plasmon Chips
2.8. Verification of PMA Induced Neutrophils of Fibril Release
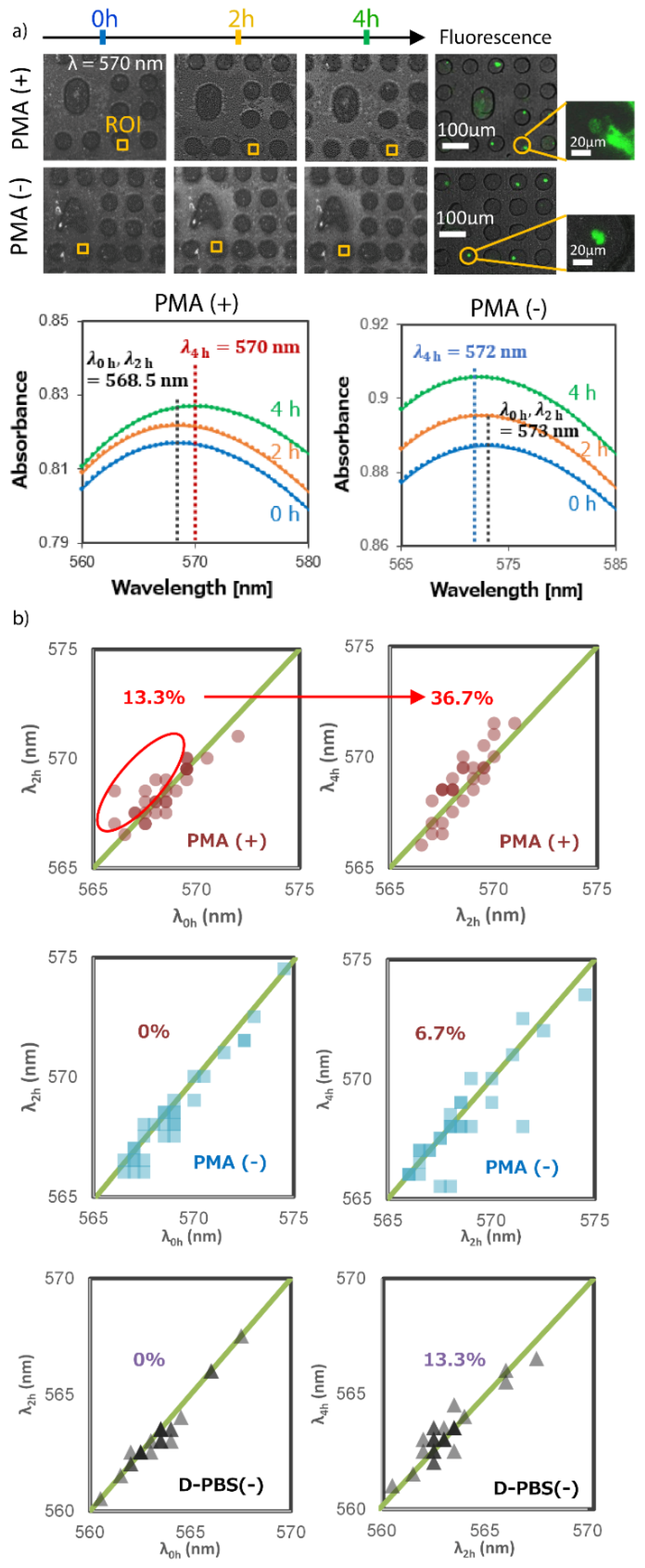
2.9. Real-Time LSPR Observation of Neutrophil Extracellular Traps (NETs)
3. Results and Discussion
3.1. Optimization of MWA Plasmon Chips
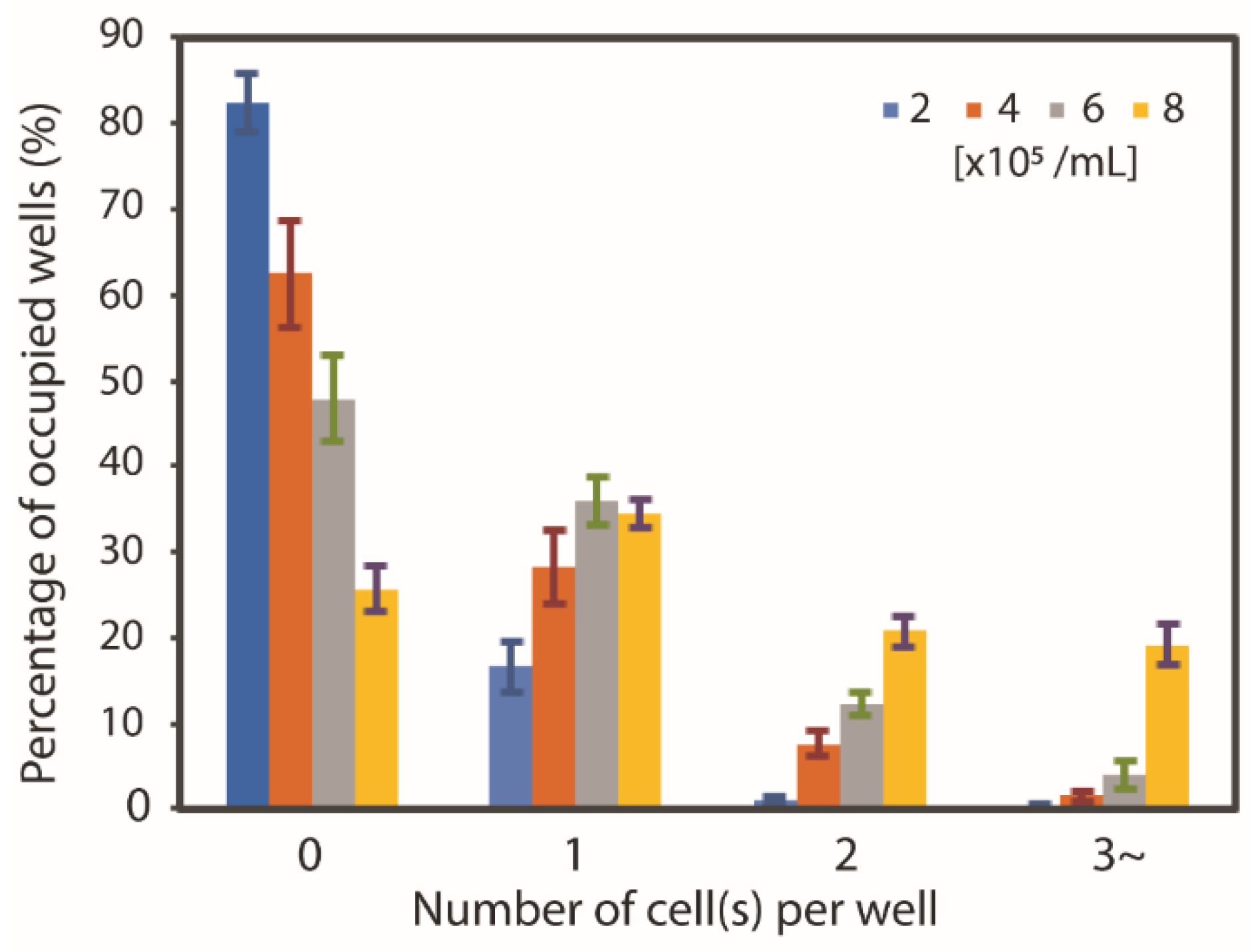
3.2. Neutrophil Isolation Capability
3.3. PMA Induced Fibril Release from Neutrophils
3.4. Real-Time LSPR Imaging Observation of Neutrophils
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Delgado-Rizo, V.; Martinez-Guzman, M.A.; Iniguez-Gutierrez, L.; Garcia-Orozco, A.; Alvarado-Navarro, A.; Fafutis-Morris, M. Neutrophil extracellular traps and its implications in inflammation: An overview. Front. Immunol. 2017, 8, 81. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, M.J.; Radic, M. Neutrophil extracellular traps: Double-edged swords of innate immunity. J. Immunol. 2012, 189, 2689–2695. [Google Scholar] [CrossRef] [PubMed]
- Rosales, C.; Demaurex, N.; Lowell, C.A.; Uribe-Querol, E. Neutrophils: Their role in innate and adaptive immunity. J. Immunol. Res. 2016, 2016, 1469780. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef] [PubMed]
- Takei, H.; Araki, A.; Watanabe, H.; Ichinose, A.; Sendo, F. Rapid killing of human neutrophils by the potent activator phorbol 12-myristate 13-acetate (PMA) accompanied by changes different from typical apoptosis or necrosis. J. Leukoc. Biol. 1996, 59, 229–240. [Google Scholar] [CrossRef]
- Lee, K.H.; Kronbichler, A.; Park, D.D.Y.; Park, Y.; Moon, H.; Kim, H.; Choi, J.H.; Choi, Y.; Shim, S.; Lyu, I.S.; et al. Neutrophil extracellular traps (NETs) in autoimmune diseases: A comprehensive review. Autoimmun. Rev. 2017, 16, 1160–1173. [Google Scholar] [CrossRef]
- Fritzler, M.J.; Wiik, A.; Fritzler, M.L.; Barr, S.G. The use and abuse of commercial kits used to detect autoantibodies. Arthritis Res. Ther. 2003, 5, 192–201. [Google Scholar] [CrossRef][Green Version]
- Bendall, S.C.; Nolan, G.P.; Roederer, M.; Chattopadhyay, P.K. A deep profiler’s guide to cytometry. Trends Immunol. 2012, 33, 323–332. [Google Scholar] [CrossRef]
- Bendall, S.C.; Simonds, E.F.; Qiu, P.; El-ad, D.A.; Krutzik, P.O.; Finck, R.; Bruggner, R.V.; Melamed, R.; Trejo, A.; Ornatsky, O.I.; et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011, 332, 687–696. [Google Scholar] [CrossRef]
- Sharma, R.; Roychoudhury, S.; Singh, N.; Sarda, Y. Methods to measure Reactive Oxygen Species (ROS) and Total Antioxidant Capacity (TAC) in the reproductive system. In Oxidative Stress in Human Reproduction; Springer International Publishing AG: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Pavelescu, L.A. On reactive oxygen species measurement in living systems. J. Med. Life 2015, 8, 38–42. [Google Scholar]
- Kippner, L.E.; Kim, J.; Gibson, G.; Kemp, M.L. Single cell transcriptional analysis reveals novel innate immune cell types. PeerJ 2014, 2, e452. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, D.; Bodovitz, S. Single cell analysis: The new frontier in ‘omics’. Trends Biotechnol. 2010, 28, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, P.K.; Gierahn, T.M.; Roederer, M.; Love, J.C. Single-cell technologies for monitoring immune systems. Nat. Immunol. 2014, 15, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Heath, J.R.; Ribas, A.; Mischel, P.S. Single-cell analysis tools for drug discovery and development. Nat. Rev. Drug Discov. 2016, 15, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, M.; Ståhlberg, A.; Rorsman, P.; Kubista, M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 2005, 15, 1388–1392. [Google Scholar] [CrossRef]
- Endo, T.; Yamamura, S.; Nagatani, N.; Morita, Y.; Takamura, Y.; Tamiya, E. Localized surface plasmon resonance based optical biosensor using surface modified nanoparticle layer for label-free monitoring of antigen–antibody reaction. Sci. Technol. Adv. Mater. 2005, 6, 491–500. [Google Scholar] [CrossRef]
- Acimovic, S.S.; Ortega, M.A.; Sanz, V.; Berthelot, J.; Garcia-Cordero, J.L.; Renger, J.; Maerkl, S.J.; Kreuzer, M.P.; Quidant, R. LSPR chip for parallel, rapid, and sensitive detection of cancer markers in serum. Nano Lett. 2014, 14, 2636–2641. [Google Scholar] [CrossRef]
- Oh, B.R.; Chen, P.; Nidetz, R.; McHugh, W.; Fu, J.; Shanley, T.P.; Cornell, T.T.; Kurabayashi, K. Multiplexed nanoplasmonic temporal profiling of T-cell response under immunomodulatory agent exposure. ACS Sens. 2016, 1, 941–948. [Google Scholar] [CrossRef]
- Chen, P.; Chung, M.T.; McHugh, W.; Nidetz, R.; Li, Y.; Fu, J.; Cornell, T.T.; Shanley, T.P.; Kurabayashi, K. Multiplex serum cytokine immunoassay using nanoplasmonic biosensor microarrays. ACS Nano 2015, 9, 4173–4181. [Google Scholar] [CrossRef]
- Oh, B.R.; Huang, N.T.; Chen, W.; Seo, J.H.; Chen, P.; Cornell, T.T.; Shanley, T.P.; Fu, J.; Kurabayashi, K. Integrated nanoplasmonic sensing for cellular functional immunoanalysis using human blood. ACS Nano 2014, 8, 2667–2676. [Google Scholar] [CrossRef]
- Yoshikawa, H.; Murahashi, M.; Saito, M.; Jiang, S.; Iga, M.; Tamiya, E. Parallelized label-free detection of protein interactions using a hyper-spectral imaging system. Anal. Methods 2015, 7, 5157–5161. [Google Scholar] [CrossRef]
- Amirjani, A.; Koochak, N.N.; Haghshenas, D.F. Investigating the shape and size-dependent optical properties of silver nanostructures using UV–vis spectroscopy. J. Chem. Educ. 2019, 11, 2584–2589. [Google Scholar] [CrossRef]
- Amirjani, A.; Haghshenas, D.F. Facile and on-line colorimetric detection of Hg2+ based on localized surface plasmon resonance (LSPR) of Ag nanotriangles. Talanta 2019, 192, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Amirjani, A.; Haghshenas, D.F. Ag nanostructures as the surface plasmon resonance (SPR)-based sensors: A mechanistic study with an emphasis on heavy metallic ions detection. Sens. Actuators B Chem. 2018, 273, 1768–1779. [Google Scholar] [CrossRef]
- Bhagawati, M.; You, C.; Piehler, J. Quantitative real-time imaging of protein–protein interactions by LSPR detection with micropatterned gold nanoparticles. Anal. Chem. 2013, 85, 9564–9571. [Google Scholar] [CrossRef] [PubMed]
- Fernández, F.; Garcia-Lopez, O.; Tellechea, E.; Asensio, A.C.; Cornago, I. LSPR Cuvette for Real-Time Biosensing by using a common spectrophotometer. IEEE Sens. J. 2016, 16, 4158–4165. [Google Scholar] [CrossRef]
- Mayer, K.M.; Lee, S.; Liao, H.; Rostro, B.C.; Fuentes, A.; Scully, P.T.; Nehl, C.L.; Hafner, J.H. A label-free immunoassay based upon localized surface plasmon resonance of gold nanorods. ACS Nano 2008, 2, 687–692. [Google Scholar] [CrossRef]
- Bellapadrona, G.; Tesler, A.B.; Grünstein, D.; Hossain, L.H.; Kikkeri, R.; Seeberger, P.H.; Vaskevich, A.; Rubinstein, I. Optimization of localized surface plasmon resonance transducers for studying carbohydrate–protein interactions. Anal. Chem. 2012, 84, 232–240. [Google Scholar] [CrossRef]
- Endo, T.; Kerman, K.; Nagatani, N.; Takamura, Y.; Tamiya, E. Label-free detection of peptide nucleic acid-DNA hybridization using localized surface plasmon resonance based optical biosensor. Anal. Chem. 2005, 77, 6976–6984. [Google Scholar] [CrossRef]
- Huang, T.; Nallathamby, P.D.; Xu, X.H.N. Photostable single-molecule nanoparticle optical biosensors for real-time sensing of single cytokine molecules and their binding reactions. J. Am. Chem. Soc. 2008, 130, 17095–17105. [Google Scholar] [CrossRef]
- Breault-Turcot, J.; Masson, J.F. Nanostructured substrates for portable and miniature SPR biosensors. Anal. Bioanal. Chem. 2012, 403, 1477–1484. [Google Scholar] [CrossRef] [PubMed]
- Cetin, A.E.; Coskun, A.F.; Galarreta, B.C.; Huang, M.; Herman, D.; Ozcan, A.; Altug, H. Handheld high-throughput plasmonic biosensor using computational on-chip imaging. Light Sci. Appl. 2014, 3, e122. [Google Scholar] [CrossRef]
- Coskun, A.F.; Cetin, A.E.; Galarreta, B.C.; Alvarez, D.A.; Altug, H.; Ozcan, A. Lensfree optofluidic plasmonic sensor for real-time and label-free monitoring of molecular binding events over a wide field-of-view. Sci. Rep. 2014, 4, 6789. [Google Scholar] [CrossRef] [PubMed]
- Kinpara, T.; Mizuno, R.; Murakami, Y.; Kobayashi, M.; Yamaura, S.; Hasan, Q.; Morita, Y.; Nakano, H.; Yamane, T.; Tamiya, E. A picoliter chamber array for cell-free protein synthesis. J. Biochem. 2004, 136, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Sulka, G.D. Nanostructured Materials in Electrochemistry; Eftekhari, A., Ed.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2008; Chapter 1; pp. 1–116. [Google Scholar]
- Karlsson, A.; Nixon, J.B.; McPhail, L.C. McPhail, Phorbol myristate acetate induces neutrophil NADPH-oxidase activity by two separate signal transduction pathways: Dependent or independent of phosphatidylinositol 3-kinase. J. Leukoc. Biol. 2000, 67, 396–404. [Google Scholar] [CrossRef]
- Metzler, K.D.; Goosmann, C.; Lubojemska, A.; Zychlinsky, A.; Papayannopoulos, V. A myeloperoxidase-containing complex regulates neutrophil elastase release and actin dynamics during NETosis. Cell Rep. 2014, 8, 883–896. [Google Scholar] [CrossRef]
- Masuda, H.; Satoh, M. Fabrication of gold nanodot array using anodic porous alumina as an evaporation mask. Jpn. J. Appl. Phys. 1996, 35, L126. [Google Scholar] [CrossRef]
- Hiep, H.M.; Yoshikawa, H.; Taniyama, S.; Kondoh, K.; Saito, M.; Tamiya, E. Immobilization of gold nanoparticles on aluminum oxide nanoporous structure for highly sensitive plasmonic sensing. Jpn. J. Appl. Phys. 2010, 49, 06GM02. [Google Scholar] [CrossRef]
- Jiang, S.; Saito, M.; Murahashi, M.; Tamiya, E. Pressure free nanoimprinting lithography using ladder-type HSQ material for LSPR biosensor chip. Sens. Actuators B 2017, 242, 47–55. [Google Scholar] [CrossRef]
- Saito, M.; Kitamura, A.; Murahashi, M.; Yamanaka, K.; Hoa le, Q.; Yamaguchi, Y.; Tamiya, E. Novel gold-capped nanopillars imprinted on a polymer film for highly sensitive plasmonic biosensing. Anal. Chem. 2012, 84, 5494–5500. [Google Scholar] [CrossRef]
- Ali, R.A.M.; Espulgar, W.V.; Aoki, W.; Jiang, S.; Saito, M.; Ueda, M.; Tamiya, E. One-step nanoimprinted hybrid micro-/nano-structure for in situ protein detection of isolated cell array via localized surface plasmon resonance. Jpn. J. Appl. Phys. 2018, 57, 03EC03. [Google Scholar] [CrossRef]
- Kovalchuk, N.M.; Roumpea, E.; Nowak, E.; Chinaud, M.; Angeli, P.; Simmons, M.J. Effect of surfactant on emulsification in microchannels. Chem. Eng. Sci. 2018, 176, 139–152. [Google Scholar]
- Jorch, S.K.; Kubes, P. An emerging role for neutrophil extracellular traps in noninfectious disease. Nat. Med. 2017, 23, 279–287. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad Mohamed Ali, R.; Mita, D.; Espulgar, W.; Saito, M.; Nishide, M.; Takamatsu, H.; Yoshikawa, H.; Tamiya, E. Single Cell Analysis of Neutrophils NETs by Microscopic LSPR Imaging System. Micromachines 2020, 11, 52. https://doi.org/10.3390/mi11010052
Ahmad Mohamed Ali R, Mita D, Espulgar W, Saito M, Nishide M, Takamatsu H, Yoshikawa H, Tamiya E. Single Cell Analysis of Neutrophils NETs by Microscopic LSPR Imaging System. Micromachines. 2020; 11(1):52. https://doi.org/10.3390/mi11010052
Chicago/Turabian StyleAhmad Mohamed Ali, Riyaz, Daiki Mita, Wilfred Espulgar, Masato Saito, Masayuki Nishide, Hyota Takamatsu, Hiroyuki Yoshikawa, and Eiichi Tamiya. 2020. "Single Cell Analysis of Neutrophils NETs by Microscopic LSPR Imaging System" Micromachines 11, no. 1: 52. https://doi.org/10.3390/mi11010052
APA StyleAhmad Mohamed Ali, R., Mita, D., Espulgar, W., Saito, M., Nishide, M., Takamatsu, H., Yoshikawa, H., & Tamiya, E. (2020). Single Cell Analysis of Neutrophils NETs by Microscopic LSPR Imaging System. Micromachines, 11(1), 52. https://doi.org/10.3390/mi11010052





