Abstract
Toxin-antitoxin (TA) systems are small genetic elements that are ubiquitous in prokaryotes. Most studies on TA systems have focused on commensal and pathogenic bacteria; yet very few studies have focused on TAs in marine bacteria, especially those isolated from a deep sea environment. Here, we characterized a type II VapC/VapB TA system from the deep-sea derived Streptomyces sp. SCSIO 02999. The VapC (virulence-associated protein) protein belongs to the PIN (PilT N-terminal) superfamily. Overproduction of VapC strongly inhibited cell growth and resulted in a bleb-containing morphology in E. coli. The toxicity of VapC was neutralized through direct protein–protein interaction by a small protein antitoxin VapB encoded by a neighboring gene. Antitoxin VapB alone or the VapB/VapC complex negatively regulated the vapBC promoter activity. We further revealed that three conserved Asp residues in the PIN domain were essential for the toxic effect of VapC. Additionally, the VapC/VapB TA system stabilized plasmid in E. coli. Furthermore, VapC cross-activated transcription of several TA operons via a partially Lon-dependent mechanism in E. coli, and the activated toxins accumulated more preferentially than their antitoxin partners. Collectively, we identified and characterized a new deep sea TA system in the deep sea Streptomyces sp. and demonstrated that the VapC toxin in this system can cross-activate TA operons in E. coli.
1. Introduction
Toxin-antitoxin (TA) systems are widely distributed in archaea and bacteria. Recent studies have proven that activated toxins inhibit essential cellular processes, including DNA replication [1], mRNA stability [2], translation [3], cytoskeleton formation [4], membrane integrity [5,6] and cell wall synthesis [7]. TA-elicited alteration of cellular processes results in important physiological changes, such as the formation of metabolically dormant cells [8,9,10], higher tolerance to antibiotics [11], and increased phage inhibition [12]. Based on the nature and mode of action of antitoxin, TA systems have been divided into five different types [4,5,13,14]. The toxin components in TAs are all small proteins, while the antitoxins are either labile proteins or untranslated antisense RNAs. In type I TA systems, antitoxins bind to 5′ untranslated regions (UTR) or coding regions of toxin mRNAs through a complementary mechanism, and resulting in translation inhibition or degradation of toxin transcripts. In type II systems, antitoxin proteins bind to toxin proteins directly and inhibit the bioactivity of the toxins. In type III TA systems, an RNA antitoxin directly interacts with the toxin protein. Different from type I to type III TA systems, the components of type IV TA systems do not interact with each other, but instead, they have the same cellular target and the antitoxin suppresses the toxicity of toxin by stabilizing its target [4]. In the type V system, the antitoxin GhoS protein specifically cleaves the mRNA of the toxin ghoT, and thus prevents the translation of the toxin [5].
Type II TA systems have been the most extensively studied in the past, in part due to their abundance in bacterial genomes [15] and the development of bioinformatics tools to locate these loci in the sequenced genome based on their genetic features [16,17]. Currently, about 39 TA systems have been identified in E. coli K-12, including 18 type I, 19 type II, one type IV and one type V [4,5,18,19]. The type II toxins are RNases or DNA gyrase inhibitors [2,20,21,22,23,24,25,26]. Type II TA loci have been further classified into evolutionary independent gene families according to similarities of the toxins at the amino acid sequence level [27,28]. Among these families, VapC/VapB is the most common and represents more than 30% of all TA systems [16,27,29,30]. In Mycobacterium tuberculosis, 45 out of 88 type II TA loci are vapBC homologs [31]. However, in E. coli K-12, VapC/VapB homologs have not been identified.
Previous studies in TA systems have mostly focused on commensal and pathogenic bacteria. The marine ecosystem represents the largest ecosystem on earth and harbors the highest abundance and diversity of microorganisms [32]. Recently, the similarities and differences of core genes of the microbiomes between the marine ecosystems and the human gut have been compared using 243 ocean microbiome samples of the Tara Oceans Project [33,34] and microbiome samples from the human gut [35]. Despite large physicochemical differences between the two ecosystems, most of the prokaryotic gene abundance (73% in the ocean; 63% in the gut) can be attributed to a shared functional core [34]. Differences in the core gene abundance between the two ecosystems have also been revealed, including those involved with defense mechanisms, signal transduction, and energy production [34]. However, functional studies of TA systems in marine bacteria have been rarely explored. Streptomyces sp. SCSIO 02999 (SCSIO 02999) was isolated from South China Sea sediment at a depth of 880 m [36], and has been found to produce a variety of biologically active compounds with antivirus, antitumor or antibacterial activities [36,37]. The taxonomy of the strain was analyzed based on sequence of a 16S rRNA gene (GenBank accession No. JQ815089), and it is close to Streptomyces sp. VTT E-062988, ACT-40 and 1A01691. Here, we searched the genome of SCSIO 02999 and demonstrated that two neighboring genes (00087 and 00088) encode a type II VapC/VapB TA system. We further reveal that the VapC/VapB TA system confers plasmid stability and that the toxin VapC cross-actives many TA operons via a partially Lon-dependent mechanism in E. coli. To the best of our knowledge, this is the first report on characterizing a TA system in deep sea microorganisms.
2. Results
2.1. Identification of VapC/VapB TA
The putative TA loci in the deep sea Streptomyces sp. SCSIO 02999 genome were predicted with a web based tool RASTA-Bacteria [17], and several potential TA pairs were identified. Two neighboring genes, orf00087 and orf00088, encode two small proteins of 136 aa and 73 aa, respectively (Figure 1A and Figure S1); Orf00087 was predicted to be homologous to the toxin VapC of the VapC/VapB type II TA pair (Figure S2). To test the toxicity of the two-gene cassette, we cloned the coding regions into the pCA24N plasmid with a lac promoter to construct pCA24N-orf00087 (pCA24N-vapC) and pCA24N-orf00088 (pCA24N-vapB) using genomic DNA of Streptomyces sp. SCSIO 02999 as the template. After transformation into E. coli K-12 BW25113 and induction with isopropyl beta-d-thiogalactopyranoside (IPTG), cells expressing VapC using pCA24N-vapC exhibited a notable decrease in cell growth as shown by the reduction in turbidity at 600 nm (OD600) and colony forming units (CFUs) (Figure 1B–D). In contrast, overexpression of VapB using pCA24N-vapB did not affect cell growth (Figure 1B–D). Phase-contrast microscopic examination revealed that expression of the toxin led to formation of morphologically altered, non-replicating “bleb-containing” cells in E. coli. Almost all of the cells contained one to three “blebs” in the middle or near the poles (Figure 1E). The 4’, 6-Diamidino-2-Phenylindole (DAPI) staining showed that the “blebs” did not contain nucleoids (Figure 1E), and these membrane protrusions might be formed by the membrane damage generated in the cytoplasm when toxin VapC was overproduced. When IPTG was removed from cells overexpressing VapC, the size of the “blebs” was reduced in 3 h (Figure 1F, upper panel) and the number of cells having “blebs” greatly decreased in 6 h (Figure 1F, lower panel).
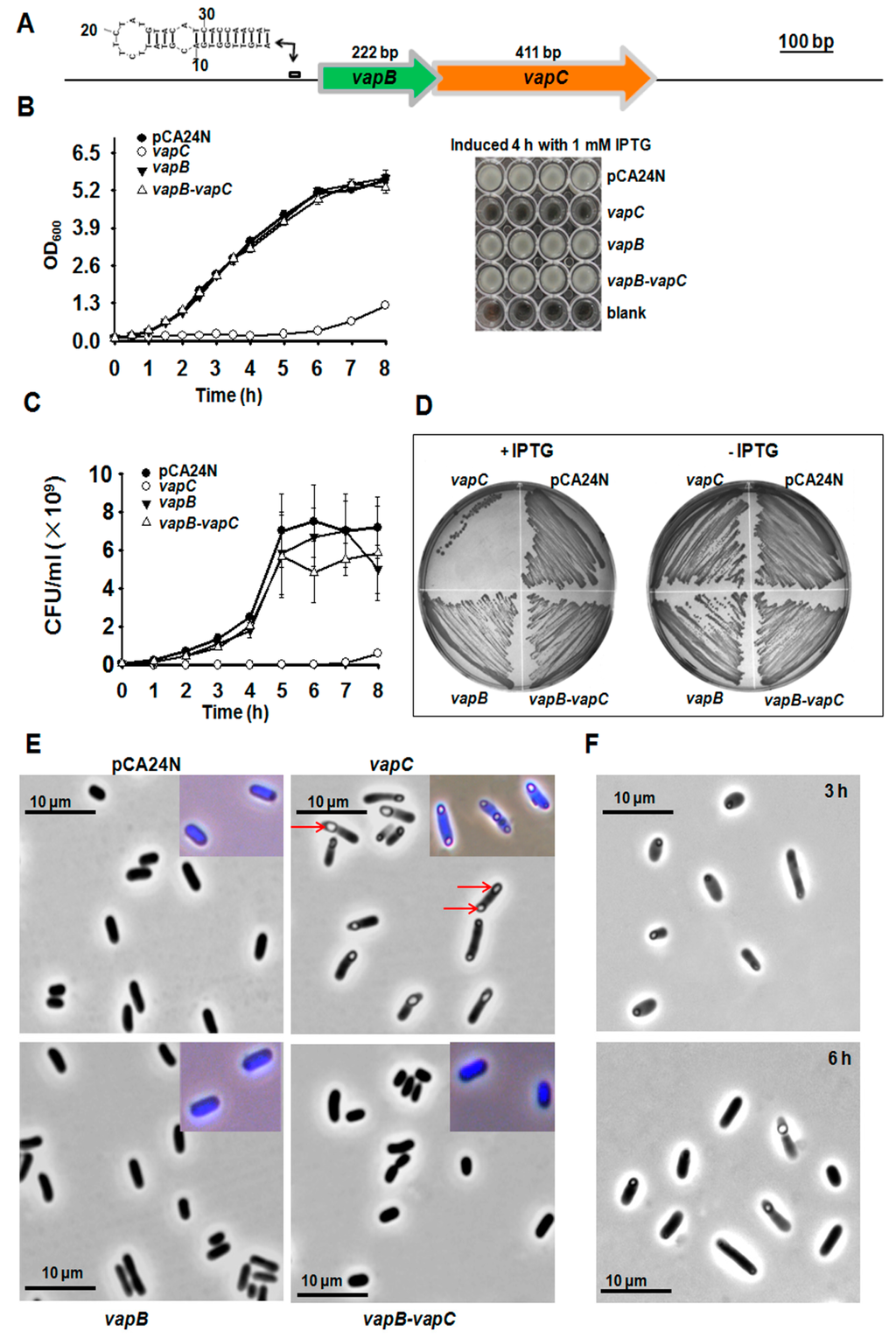
Figure 1.
VapC (00087) is toxic and VapB (00088) neutralizes the toxicity of VapC. (A) Chromosomal loci of vapBC operon. The secondary structure of the palindrome near the Ribosomal Binding Site (RBS) is also shown. (B) Growth of E. coli K-12 BW25113 harboring pCA24N-based constructs that were induced with 0.5 mM IPTG at OD600 0.1. Cell growth was tested at the time points indicated (left panel) and images were taken after 4 h induction (right panel). (C) Cell viability (CFU/mL) were determined at the time points indicated. (D) E. coli hosts harboring the above-mentioned plasmids were streaked onto LB plates supplemented with 30 μg/mL chloramphenicol with or without 0.5 mM IPTG, and were incubated for 16 h. (E) Morphology of BW25113 cells overproducing VapC, VapB and VapB-VapC TA. Red arrows point to the “blebs” of cells. Cells were grown in LB and induced with 0.5 mM IPTG at OD600 1.0 for 3 h. Induced cells stained with 4’, 6-Diamidino-2-Phenylindole (DAPI) were shown in the right corners. (F) BW25113 cells in (E) were washed with PBS to remove isopropyl beta-d-thiogalactopyranoside (IPTG) and re-cultured for another 3 h and 6 h, respectively. Data are from three independent cultures and standard deviations are shown in (B) and (C). At least two independent cultures were used and representative images were shown in (D–F).
To determine whether the neighboring protein VapB can neutralize the toxicity of VapC, we constructed the pCA24N-00088-00087 (pCA24N-vapB-vapC) plasmid to co-express VapB and VapC in E. coli cells. As expected, VapB completely neutralized the toxicity of VapC (Figure 1B–D). Moreover, co-expression of VapB effectively inhibited the formation of blebs caused by VapC overproduction (Figure 1E). Thus, expression of VapC from the deep sea Streptomyces sp. resulted in growth inhibition of E. coli, and VapB serves as the antitoxin partner for VapC.
2.2. VapB and VapC Form a Complex in Vivo
A typical feature of type II TA systems is that the toxin interacts with antitoxin directly and they form a protein complex in vivo [38]. Here, we performed a pull-down assay using pET28b-vapB-vapC-Chis to express a C-terminal hexahistidine tagged (His-tagged) VapC along with an untagged antitoxin VapB. Affinity purification using Ni-NTA agarose beads and subsequent Tricine-SDS-PAGE revealed that a small protein could be pulled down together with His-tagged VapC (Figure 2, Lane 2–4), and this small protein was found to be VapB by mass spectrometry. As a control, we constructed a pET28b-vapB-vapC to express untagged VapB and untagged VapC, and neither of them could bind to Ni-NTA beads (Figure 2, lane 5–7). Hence, we show that VapB interacts with VapC and they formed a complex in vivo.
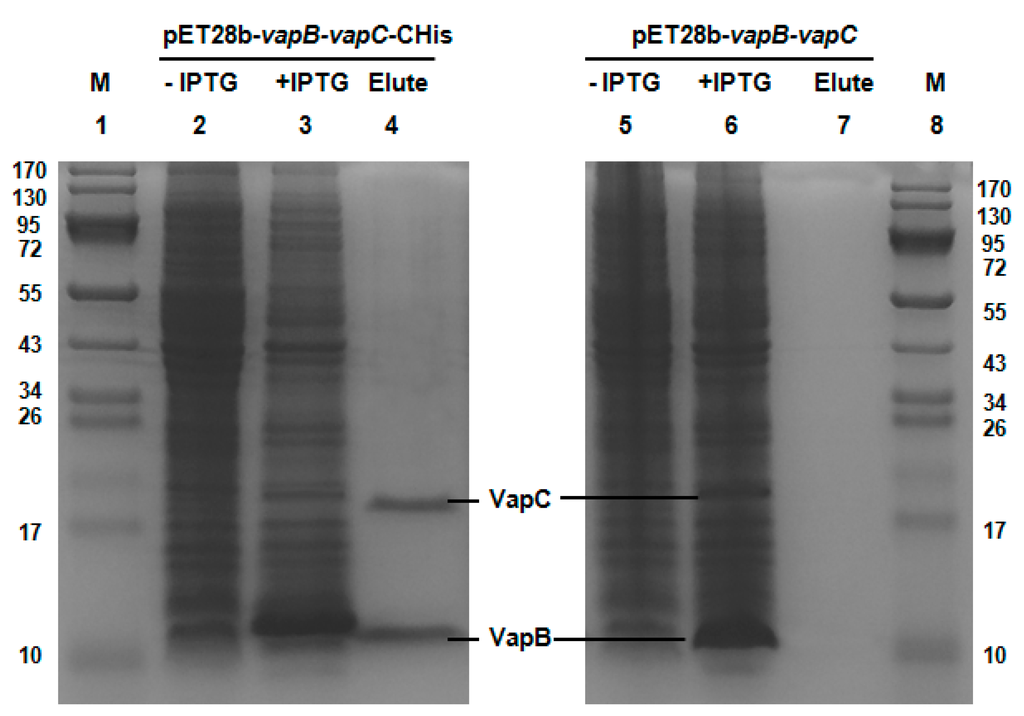
Figure 2.
VapC and VapB form a complex in vivo. Plasmid pET28b-vapB-vapC-Chis was constructed to produce a His-tagged VapC and untagged VapB with IPTG induction, 15.74 kDa VapC-Chis and 8.59 kDa VapB were induced (lane 3). During purification, VapB was co-purified (lane 4). Cells that were not induced with IPTG were served as control (lane 2). Additionally, as a negative control (lane 5–7), pET28b-vapB-vapC was also constructed to produce untagged VapB and untagged VapC, and neither VapB nor VapC bound to the Ni-NTA agarose beads (lane 7). The protein marker (M) was loaded in lanes 1 and 8.
2.3. VapB and VapC/VapB Negatively Autoregulate the VapBC Operon
An in vivo promoter activity assay was used to study the autoregulation of the vapBC operon. We amplified three different fragments containing a 260 bp upstream region of vapBC followed by the full coding region of vapBC (vapB-vapC), the full coding region of VapB followed by the first 45 bp of the coding region of VapC (vapB-vapC’), and the first 45 bp of the coding region of VapB (vapB’). These three fragments were fused to the lacZ gene and ligated into the pHGEI01 plasmid (Figure 3A). The promoter activity was decreased from 645.0 ± 13.6 miller units (MU) in E. coli WM3064 cells carrying the pHGEI01-vapB’ plasmid to 426.3 ± 17.9 MU in cells harboring the pHGEI01-vapB-vapC’(Figure 3B), suggesting that the presence of VapB repressed the promoter activity. Moreover, the cells carrying the pHGEI01-vapB-vapC showed much lower promoter activity (117.8 ± 7.5 MU), suggesting that the VapC helped the VapB to repress the promoter activity. One palindrome of 14 bp near the ribosome binding site (RBS) was found (Figure 3A and Figure S1), probably serving as the binding site for VapB since most type II antitoxins bind to DNA at palindromic regions.
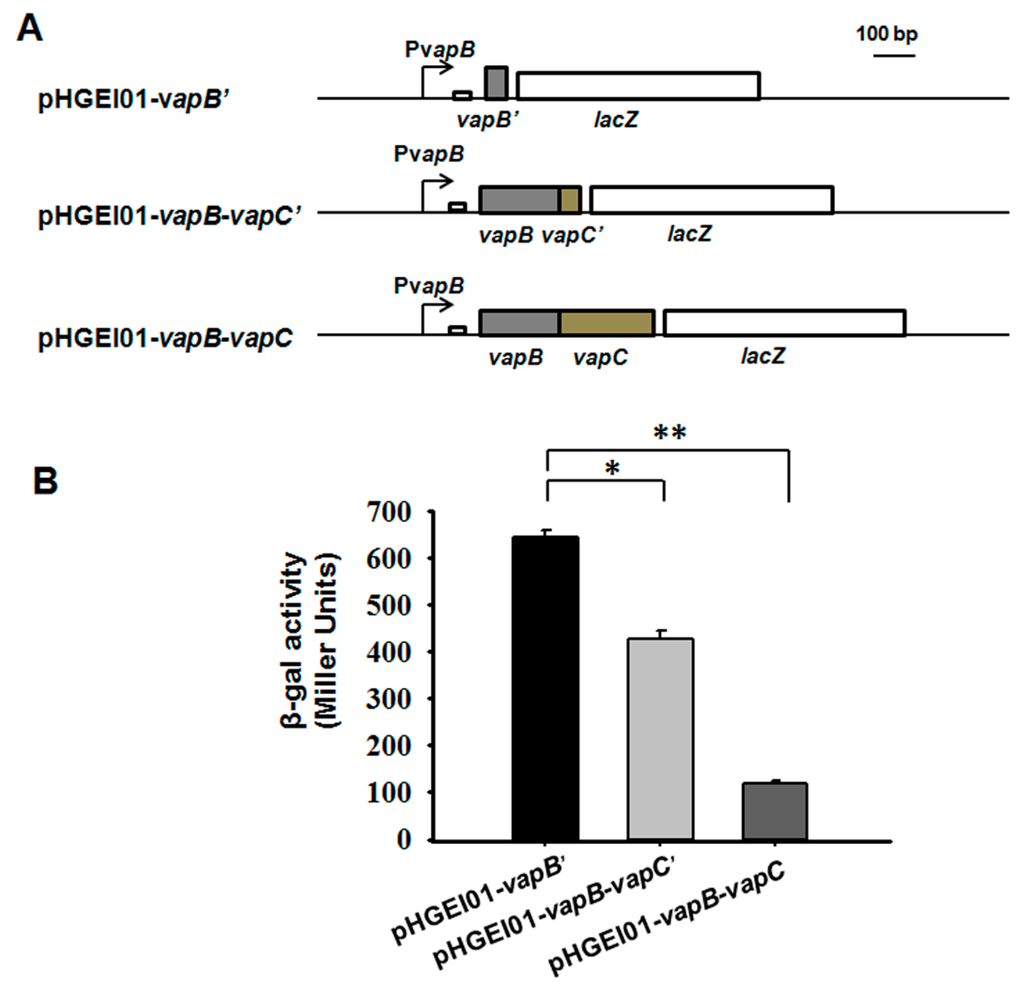
Figure 3.
VapB and VapB-VapC complex both regulate vapBC operon. (A) Schematic diagram of the constructed reporter systems for the promoter activity assay. (B) Mid-log-phase E. coli WM3064 cells harboring the reporter systems in (A) were collected and tested for β-galactosidase activity. Three independent cultures for each strain were used and the data are shown as means ± standard deviations. Asterisks represent a statistically significant difference (p < 0.01 was shown in * and p < 0.001 was shown in **; n = 3).
2.4. Key Residues for the Toxicity of VapC
Phyre2 [39] was applied to predict the 3D structure of VapC (Figure 4A), which belongs to PIN (PilT N-terminal) domain superfamily. Proteins of the PIN superfamily are small RNases. Three strictly conserved acidic residues (Asp, D) commonly found in the RNase active site of PIN domain were also identified in VapC (at position 6, 96 and 114) (Figure 4A). To investigate the importance of the conserved acidic residues in determining the toxicity of VapC, we performed site-directed mutagenesis on the D residues at positions 6, 96 and 114, separately. The results suggested that any mutation of the three D residues completely abolished the toxicity of VapC (Figure 4B–D). The 3D structure predicted here indicated that the conserved active site formed a negatively charged pocket near the center of the molecule, and they might be essential for single-stranded ribonuclease activity (Figure 4A). Furthermore, the three mutated VapCs no longer induced “bleb-containing” cells (Figure S3), further confirming the key role of the three residues in determining VapC toxicity.
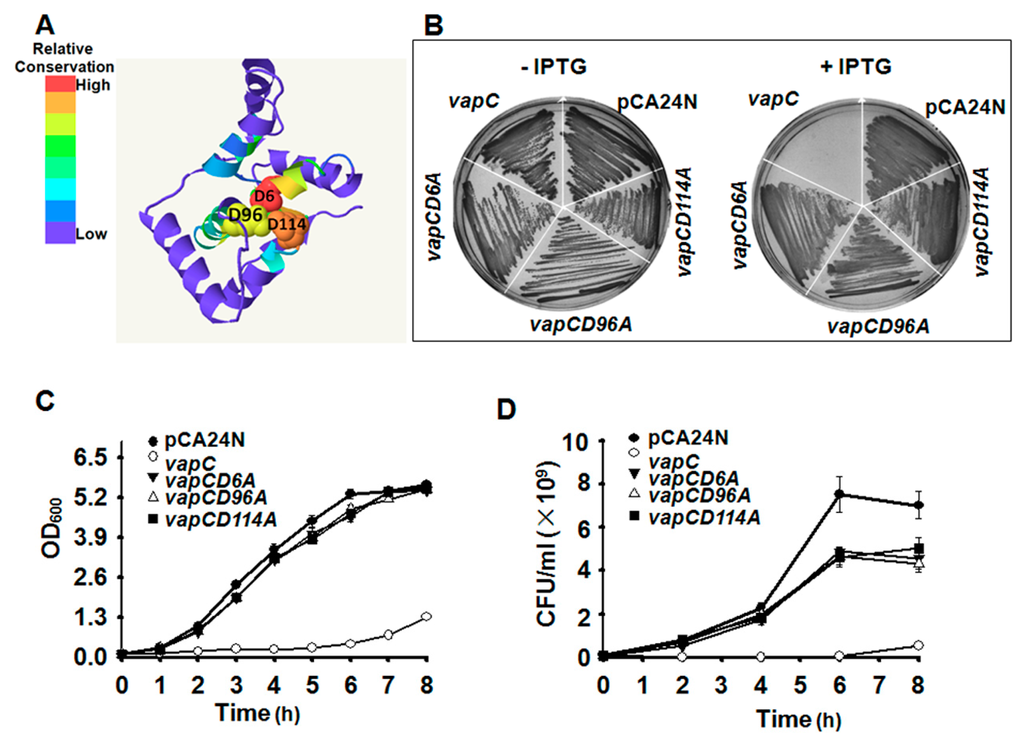
Figure 4.
Key residues for determining VapC toxicity. (A) Predicted 3D structure of VapC. The three conserved D in PilT N-terminal (PIN) domain were indicated. The number in the mutated protein indicates the position of the amino acid in VapC. (B) Toxicity after single-site mutagenesis to convert the three D residues to A was determined in E. coli BW25113 wild type. Growth (C) and colony forming units (CFU) (D) tests of E. coli cells expressing the VapC and the three mutant VapC proteins, 0.5 mM IPTG was added at OD600 0.1. Three independent cultures were evaluated for each analysis, and only one representative image is shown in (B).
2.5. VapC/VapB Stabilizes Plasmids in E. coli
Toxin-antitoxin systems are well known to stabilize plasmids in cells. The ability of VapC/VapB to stabilize plasmids was tested in E. coli. Empty plasmid pCA24N carrying a chloramphenicol resistance gene exhibited a loss of ~99% in E. coli in the absence of chloramphenicol after continuous culture for four days. In contrast, cells expressing TA pair VapB/VapC did not exhibit significant plasmid loss (Figure 5). The E. coli cells harboring pCA24N completely lost resistance to chloramphenicol after five days, whereas more than 90% of the cells harboring the pCA24N-vapB-vapC were still resistant to chloramphenicol after seven days (Figure 5). These results demonstrate that the VapC/VapB TA pair from the deep sea Streptomyces sp. SCSIO 02999 confers plasmid stabilization in E. coli.
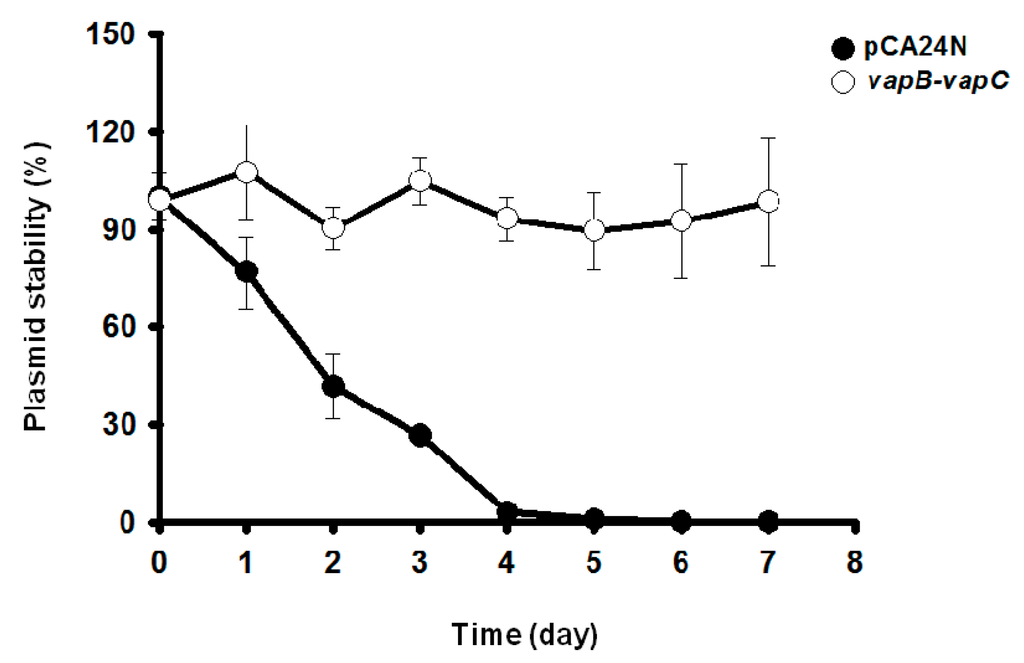
Figure 5.
VapC/VapB toxin-antitoxin (TA) system confers plasmid stability in E. coli. E. coli K-12 BW25113 harboring plasmids pCA24N and pCA24N-vapB-vapC were used for the plasmid stability assay. Overnight cultures were diluted 100-fold in LB medium without any antibiotics, then incubated at 37 °C for 12 h. This process was repeated every 12 h for seven days. Three independent cultures were conducted, and the data are shown as means ± standard deviations.
2.6. VapC Cross-Activates E. coli TA Systems in a Partially Lon-Dependent Manner
Cross-talk among TAs has been reported previously [40,41]. VapC-VapB in the deep sea microbe could be transferred into a different host if it is present on a mobile genetic element such as conjugative plasmids. Here, we explored whether deep sea derived VapC also activates transcription of reported TA systems, especially type II endoribonucleases in E. coli. The VapC was overexpressed via pCA24N-vapC after IPTG induction for 1 h in E. coli K-12 host. Changes at the transcriptional levels of toxin components of ten different type II TAs (the same 10 TA deleted in the Δ10 strain [42]) and two other toxins (RalR of type I TA and GhoT of type V TA) [5,19] were determined via qRT-PCR. Unexpectedly, transcription of all twelve toxins was induced by VapC overproduction (Figure 6). To check whether the antitoxins that are co-transcribed with the toxin components were also induced, expression levels of yefM and relB were also determined. The expression levels of these two antitoxins were both induced, but with lower fold-changes compared to their toxin components (Figure 6). VapC also induced the expression of another RNase gene rbn (Figure 6). As a negative control, expression of purA (adenylosuccinate synthase gene) was not affected by VapC overproduction (Figure 6). Moreover, the upregulation of toxin transcriptions by VapC was partially abolished in a Δlon mutant strain (Figure 6), suggesting that the cross-activation of TA systems by VapC functions is partially Lon-dependent.
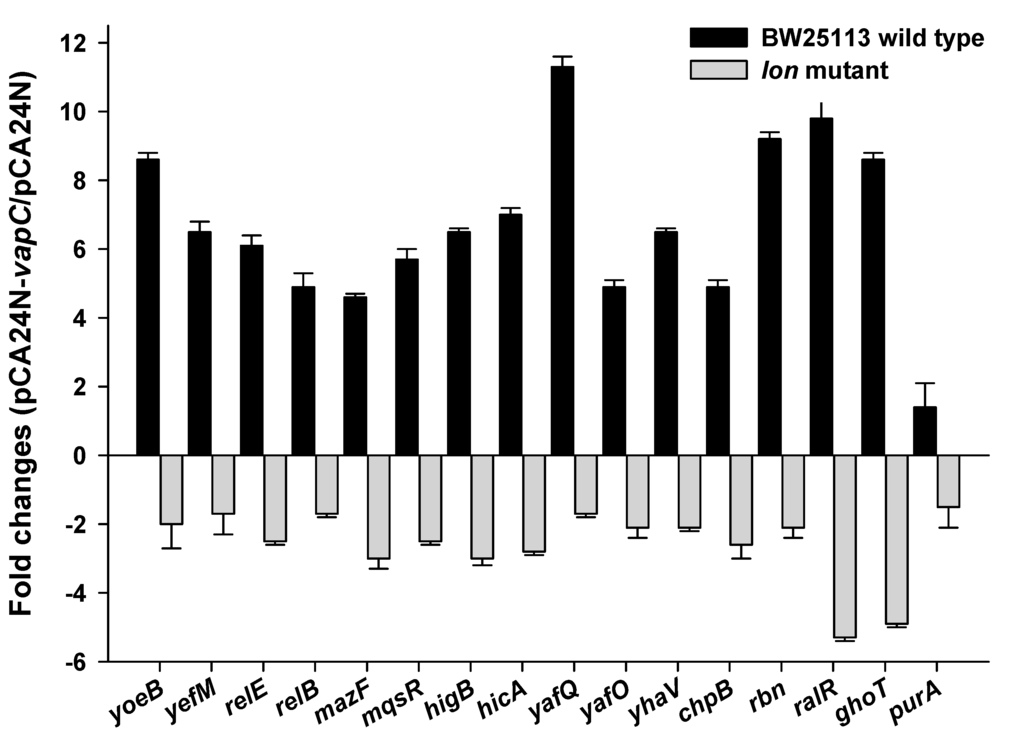
Figure 6.
VapC cross-activates the toxins in E. coli in a Lon-dependent manner. Fold changes of 14 TA transcripts and 1 RNase gene (rbn) in cells overexpressing VapC via pCA24N-vapC as compared to empty vector pCA24N in E. coli were quantified by qRT-PCR. The purA was used as negative control. Two independent cultures were used for the assay, and standard errors are indicated.
3. Discussion
Our findings strongly support the hypothesis that VapC and VapB in deep sea Streptomyces sp. SCSIO 02999 form a type II TA pair. The evidences are as follows: (i) both proteins are small; (ii) VapC is a toxin that inhibits cell growth and induces a bleb-containing phenotype in E. coli; (iii) the cognate VapB counteracts the toxicity of VapC through direct protein–protein interaction; (iv) the antitoxin VapB and VapB-VapC complex negatively regulates promoter activity of the vapBC operon; (v) the VapC/VapB TA stabilizes plasmid in E. coli; and (vi) ectopic production of VapC cross-activates many TA operons via a partially Lon-dependent mechanism in E. coli. These observations fit the features of type II TA system.
The VapC toxin identified in SCSIO 02999 belongs to the PIN domain (PF01850) superfamily, which contains 3673 proteins from 721 different species across the three domains of life [43]. Based on the secondary structure, 130 residues of the deep sea VapC (96% coverage) shared the highest similarity with VapC from Mycobacterium tuberculosis [44] (Figure S2). In eukaryotes, PIN domains are found in proteins involved in nonsense mediated mRNA decay [45], and in processing of 18S ribosomal RNA [46]. The majority of PIN-domain containing proteins identified in prokaryotes encodes the VapC toxins of VapC/VapB TA systems [47,48]. Recently, the PIN domain was identified in the ocean microbes by hologenome analysis of marine sponges microbiomes [49]. The VapC proteins are RNases targeting cellular mRNAs or tRNAfMet, showing substrates sequence specific or non-specific activities [50,51,52,53,54,55]. The cell morphology induced by VapC overproduction is different from that induced by other known toxins in E. coli. For example, the “ghost” cells were caused by overproduction of the lytic membrane toxin GhoT of type V GhoT/GhoS TA pair [5], the “filamentous growing” cells were induced by the toxin ParE of ParE/ParD TA pair [56,57], and the “swollen” cells were induced by higher eukaryotes and prokaryotes nucleotide-binding (HEPN) family toxin [58]. These results suggest that VapC in SCSIO 02999 should have different cellular targets from other known toxins in E. coli. Additionally, the “ovoid cells” morphology was reported for VapC overproduction in Mycobacterium smegmatis [59]. These previous results indicate that VapC from different species may have different targets and may be involved in distinct biological processes. Thus, the marine derived TAs may have potential application for treating pathogenic bacteria infection due to their similarity and differences to those in commensal and pathogenic bacteria.
Structural analysis has revealed that the antitoxin VapB tightly wrapped around toxin VapC to neutralize its toxicity [50,60,61]. Four DNA binding domains in VapB have also been characterized previously, including helix–turn–helix (HTH), ribbon–helix–helix (RHH), AbrB, and Phd/YefM domains [28,62]. VapB of SCSIO 02999 shares a similar RHH domain with VapB3 in M. Tuberculosis, and we demonstrated that both VapB and VapB-VapC are capable of auto-regulating the transcription of the TA operon. In this study, the attempt to purify the VapC was unsuccessful due to its high toxicity, which is consistent with previous studies [63,64]. In addition, co-purification of VapB and VapC resulted in a strong interaction between VapB and VapC, which made it difficult to separate them without affecting the activity of the toxin. New approaches are in needed to purify the toxin for in vitro characterization of the cellular targets of the toxin.
Studies on cross-activation among TA systems or other toxins mainly focus on type II TA system in which the toxin components are endoribonucleases. The well-studied type II TA MqsR/MqsA induces or represses various toxin genes in E. coli. For example, titrating antitoxin MqsA with MqsR or degrading MqsA through proteases Lon and ClpXP represses the expression of small toxic gene cspD [65]. In contrast, the deletion of toxin gene mqsR represses small toxic polypeptides encoding genes hokA and hokE [65]. In addition, MqsR overproduction induced expression of the relBEF operon and relF encodes a hok-like toxin targeting the inner membrane [65,66]. Furthermore, MqsR degrades ghoS mRNA and enriches toxin ghoT mRNA of the GhoT/GhoS type V TA pair during stress conditions [40]. In turn, HipA toxin activates the mqsR/mqsA operon [22], and RelE toxin induces the transcription of several TA operons including mqsR/mqsA [66,67]. Expression of VapC from Salmonella and Shigella activates toxin gene yoeB expression in E. coli, and the activation depends on Lon protease [41]. However, cross-activation among TAs in E. coli can also occur in protease deficient strains such as in lon, ppk, clpP, and hslV deficient strains [66]. Collectively, these results suggest that cross-talk among TAs is rather complex, and the exact mechanism remains to be elucidated.
4. Experimental Procedures
4.1. Bacterial Strains, Plasmids and Growth Conditions
The Streptomyces sp. SCSIO 02999 and E. coli strains and plasmids used in this study are listed in Table 1, and the sequences of all primers used in this study are listed in Table S1. The E. coli strains were grown in luria-bertani (LB) medium at 37 °C. The marine-derived Streptomyces sp. SCSIO 02999 was isolated from a South China Sea sediment (E 109° 153.171′, N 116° 103.576′) at a depth of 880 m [36], the strain was grown in AM6 culture [37] at 28 °C, and the strain was deposited in the type culture collection of the Center for Marine Microbiology, Research Network of Applied Microbiology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China. The taxonomy of the strain was analyzed based on a 16S rRNA gene and was deposited in GenBank (accession No. JQ815089) Chloramphenicol (30 µg/mL) was used for maintaining the pCA24N based plasmids, and kanamycin (50 µg/mL) was used for maintaining pET28b and pHGEI01 based plasmids.

Table 1.
Bacterial strains and plasmids used in this study. CmR and KmR indicate chloramphenicol and kanamycin resistance, respectively. The aa indicates amino acids.
4.2. Cloning of Genes
The pCA24N-based plasmids were constructed according to a previous procedure [69] using primer pairs pCA24N-vapB-f/r, pCA24N-vapC-f/r and pCA24N-vapB-f/pCA24N-vapC-r. For pET28b-based constructs, different fragments were amplified with primer pairs pET28b-vapB-f/pET28b-vapC-Chis-r and pET28b-vapB-f/pET28b-vapC-r. After digestion with NcoI and HindIII, they were ligated into the pET28b empty vectors digested with the same two enzymes digested. The pHGEI01-based plasmids were constructed according to previous procedure [70] using primer pairs pHGEI01-PvapB-vapC-f as the forward primer, pHGEI01-vapB’-r1, pHGEI01-vapC’-r1 and pHGEI01-vapB-vapC-r1 as the trans-primer, respectively, for the first-step PCR; and then add the RBS (AGATCTCACACAGGAAACAGCT) sequence between the vap genes and the lacZ gene using primer pairs pHGEI01-PvapB-vapC-f and the primer pHGEI01-vapB-vapC-r2. The PCR products of the second-step PCR was digested with EcoRI and BamHI, the purified fragments were ligated into the plasmid digested with the same enzymes. All the three pHGEI01 based constructs are transcriptional fusion other than translational fusion. Genomic DNAs isolated from Streptomyces sp. SCSIO 02999 was used as DNA templates.
4.3. Protein Expression and Purification
The VapB-VapC complex containing a six histidine tag at the C-terminus of VapC and the VapB-VapC complex without any tag were purified via BL21 (DE3) with pET28b-vapB-vapC-CHis and pET28b-vapB-vapC, respectively. The strains were induced with 0.5 mM IPTG at OD600 0.5 for 6 h. Then, the cells were collected and resuspended in 10 mL of lysis buffer (50 mM monosodium phosphate buffer (pH 8.0), 300 mM NaCl, 5 mM imidazole and protease inhibitor cocktail (Sigma-Aldrich, Shanghai, China)). The samples were lysed with the constant systems cell disruptor (Constant Systems Limited, Northants, UK) twice with 30 MPa. Nickel-nitrilotriacetic acid (Ni-NTA) agarose beads (Qiagen, Valencia, CA, USA) were used according to the manufacturer’s protocol. Purified proteins were desalted using a desalination column with 20 mM Tris-HCl buffer containing 300 mM NaCl (pH 8.0), and the protein concentration was measured using a Bi Yuntian bicinchoninic acid (BCA) assay kit (Bioteke corporation, Haimen, Jiangsu, China). Tricine-SDS-PAGE was performed as previously described [71]. A total of 20 µg of protein from each sample was loaded for SDS-PAGE.
4.4. DAPI (4′, 6-Diamidino-2-Phenylindole) Staining
Overnight cultures of E. coli K-12 BW25113 harboring pCA24N, pCA24N-vapB, pCA24N-vapC and pCA24N-vapB-vapC were diluted to OD600 0.1, and cultured at 37 °C till the turbidity reached around 1.0. Then, 0.5 mM IPTG was added to induce protein expression for 5 h. Cells (2 × 105 to 1 × 106) were collected by centrifugation at 3000 g for 5 min, and the cell pellets were resuspended in 1 mL of phosphate buffer saline (PBS, pH 7.4). DAPI staining solution (300 µL of 300 nM) was added to the cell suspensions, then the cells were incubated for 5 min and rinsed several times in PBS before re-suspending in 100 µL PBS.
4.5. Promoter Activity Assay
Three plasmids were constructed to study the auto-regulation of the antitoxin VapB and VapB-VapC complex in vivo. A DNA fragment containing 260 bp upstream of the translational start site of vapB was selected as the promoter region, and pHGEI01-vapB’ contains the promoter region and the first 45 bp coding region of VapB, pHGEI01-vapB-vapC’ contains the promoter region, the VapB coding region and the first 45 bp coding region of VapC, and pHGEI01-vapB-vapC contains the promoter region and full length vapBC operon. All three fragments were digested with EcoRI and BamHI and cloned into the promoter-less lacZ-fusion vector pHGEI01diegested with the two enzymes [71] to create plasmid pHGEI01-vapB’, pHGEI01-vapB-vapC’ and pHGEI01-vapB-vapC. The resulting plasmids were verified by sequencing in WM3064. Mid-log-phase (OD600 ~ 0.7) cells of the indicated strains carrying the reporter systems were collected by centrifugation and washed with PBS. The β-galactosidase activity was measured according to previous protocols [72].
4.6. Site-Directed Mutagenesis
Single site-directed mutagenesis [2] was used to mutate the three conserved putative active sites of VapC. Mutation of D (GAT) to A (GCT) used primer pair vapCD6A-f/-r, D (GAC) to A (GCC) used primer pair vapCD96A-f/-r and D (GAT) to A (GCT) used primer pair vapCD114A-f/-r (Table S1). The mutations were verified by DNA sequencing using primers pCA24N-f.
4.7. Plasmid Stabilization Test
Overnight cultures of E. coli BW25113 carrying the plasmids pCA24N and pCA24N-vapB-vapC were obtained with chloramphenicol selection in LB. The cultures were diluted 1% in LB medium without antibiotics and cultured for 12 h. The cells were re-inoculated into 3 mL of fresh LB without antibiotics for a further 12 h is process. The cultures were serially diluted 100–107 by 10-fold from days 1 to 7, and 10 µL was dropped onto LB plates with and without 30 µg/mL of chloramphenicol. The plates were incubated at 37 °C for 16 h, and viable colonies were counted and the CFU were analyzed. The ratio of the chloramphenicol resistance colonies versus the total viable colony counts was used to estimate the percentage of plasmid maintained in the population. The CFU assay was conducted every day up to 7 days.
4.8. RNA Isolation and qRT-PCR
Total RNAs was isolated as indicated previously [73] and to avoid contamination of DNA to during the isolation process, DNase was added to treat the RNA samples for 30 min at room temperature. Then, the isolated RNAs were used as the templates for qRT-PCR reactions using the SuperScriptTM III Platinum SYBR® Green One-Step qRT-PCR Kit (Invitrogen, Carlsbad, CA, USA). All of the primers for qRT-PCR are listed in Table S1. The level of the rrsG transcript was used as a reference to normalize the gene expression data. Exponentially growing cells (OD600 0.8) were induced with 0.5 mM IPTG for 1 h. Lower Ct values indicate higher expression levels. Fold changes in the transcription of various targets with pCA24N or pCA24N-vapC were calculated as: 2-((Ct target(pCA24N-vapC)-CtrrsG(pCA24N-vapC)) -(Cttarget(pCA24N)-CtrrsG(pCA24N))).
Supplementary Materials
The following are available online at www.mdpi.com/2072-6651/8/7/195/s1, Figure S1: Gene and protein sequence of vapBC operon in Streptomyces sp. SCSIO 02999. Figure S2: VapC in Streptomyces sp. SCSIO 02999 belongs to the PIN domain superfamily. Figure S3: VapC mutant could not induce “bubble forming like” cells. Table S1: Oligonucleotides used for plasmid construction site-directed mutagenesis (target mutated nucleotides are in red font and highlighted yellow) and DNA sequencing. If an enzymatic restriction site is included in the sequence, the enzyme restriction site is underlined. f indicates forward primer and r indicates reverse primer. P indicates promoter.
Acknowledgements
This work was supported by the Chinese Academy of Sciences (XDA11030402), by the National Science Foundation of China (31290233, 31270214 and 31500025), by National Science Foundation of Guangdong Province (2015A030310405 and 2014A030310385) and China Postdoctoral Science Foundation funded project (2013M542217 and 2014T70830). We are grateful for Changsheng Zhang from our institute for his generosity in providing us the Streptomyces sp. SCSIO 02999 and its genomic sequence. XW is a 1000-Youth Elite Program recipient in China.
Author Contributions
Y.G. and X.W. conceived and designed the ideas and article structure; Y.G., J.Y., C.S. and Z.W. performed experiments; Y.G. and J.Y. analyzed the data; and Y.G. and X.W. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bernard, P.; Couturier, M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J. Mol. Biol. 1992, 226, 735–745. [Google Scholar] [CrossRef]
- Wang, X.; Kim, Y.; Hong, S.H.; Ma, Q.; Brown, B.L.; Pu, M.; Tarone, A.M.; Benedik, M.J.; Peti, W.; Page, R.; et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 2011, 7, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Prysak, M.H. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol. Microbiol. 2009, 71, 1071–1087. [Google Scholar] [CrossRef] [PubMed]
- Masuda, H.; Tan, Q.; Awano, N.; Wu, K.P.; Inouye, M. YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol. Microbiol. 2012, 84, 979–989. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lord, D.M.; Cheng, H.-Y.; Osbourne, D.O.; Hong, S.H.; Sanchez-Torres, V.; Quiroga, C.; Zheng, K.; Herrmann, T.; Peti, W.; et al. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol. 2012, 8, 858–861. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.Y.; Soo, V.W.; Islam, S.; McAnulty, M.J.; Benedik, M.J.; Wood, T.K. Toxin GhoT of the GhoT/GhoS toxin/antitoxin system damages the cell membrane to reduce adenosine triphosphate and to reduce growth under stress. Environ. Microbiol. 2014, 16, 1741–1754. [Google Scholar] [CrossRef] [PubMed]
- Silvaggi, J.M.; Perkins, J.B.; Losick, R. Small untranslated RNA antitoxin in Bacillus subtilis. J. Bacteriol. 2005, 187, 6641–6650. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 2007, 5, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.; Zhang, Z.; Khodursky, A.; Kaldalu, N.; Kurg, K.; Lewis, K. Persisters: A distinct physiological state of E. coli. BMC Microbiol. 2006, 6. [Google Scholar] [CrossRef] [PubMed]
- Wood, T.K.; Knabel, S.J.; Kwan, B.W. Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 2013, 79, 7116–7121. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Persister cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Pecota, D.C.; Wood, T.K. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J. Bacteriol. 1996, 178, 2044–2050. [Google Scholar] [PubMed]
- Hayes, F. Toxins-antitoxins: Plasmid maintenance, programmed cell death, and cell cycle arrest. Science 2003, 301, 1496–1499. [Google Scholar] [CrossRef] [PubMed]
- Fineran, P.C.; Blower, T.R.; Foulds, I.J.; Humphreys, D.P.; Lilley, K.S.; Salmond, G.P. The phage abortive infection system, toxin, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. USA 2009, 106, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Unterholzner, S.J.; Poppenberger, B.; Rozhon, W. Toxin-antitoxin systems: Biology, identification, and application. Mob. Genet. Elements 2013, 3, e26219. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.C.; Harrison, E.M.; Bi, D.X.; Tai, C.; He, X.Y.; Ou, H.Y.; Rajakumar, K.; Deng, Z.X. TADB: A web-based resource for type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2011, 39, D606–D611. [Google Scholar] [CrossRef] [PubMed]
- Sevin, E.; Barloy-Hubler, F. Rasta-bacteria: A web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol. 2007, 8. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Inouye, M. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 2011, 9, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Quiroga, C.; Chen, Q.; McAnulty, M.J.; Benedik, M.J.; Wood, T.K.; Wang, X. RalR (a DNase) and RalA (a small RNA) form a type I toxin-antitoxin system in Escherichia coli. Nucleic Acids Res. 2014, 42, 6448–6462. [Google Scholar] [CrossRef] [PubMed]
- Kamada, K.; Hanaoka, F. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol. Cell 2005, 19, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.L.; Grigoriu, S.; Kim, Y.; Arruda, J.M.; Davenport, A.; Wood, T.K.; Peti, W.; Page, R. Three dimensional structure of the MqsR:MqsA complex: A novel toxin:Antitoxin pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog. 2009, 5, e1000706. [Google Scholar] [CrossRef] [PubMed]
- Kasari, V.; Kurg, K.; Margus, T.; Tenson, T.; Kaldalu, N. The Escherichia coli mqsR and ygiT genes encode a new toxin-antitoxin pair. J. Bacteriol. 2010, 192, 2908–2919. [Google Scholar] [CrossRef] [PubMed]
- Kamada, K.; Hanaoka, F.; Burley, S.K. Crystal structure of the MazE/MazF complex: Molecular bases of antidote-toxin recognition. Mol. Cell 2003, 11, 875–884. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Hara, H.; Kato, I.; Inouye, M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 2005, 280, 3143–3150. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Pogliano, J.; Helinski, D.R.; Konieczny, I. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 2002, 44, 971–979. [Google Scholar] [CrossRef] [PubMed]
- Miki, T.; Park, J.A.; Nagao, K.; Murayama, N.; Horiuchi, T. Control of segregation of chromosomal DNA by sex factor F in Escherichia coli. Mutants of DNA gyrase subunit a suppress letD (ccdB) product growth inhibition. J. Mol. Biol. 1992, 225, 39–52. [Google Scholar] [CrossRef]
- Jorgensen, M.G.; Pandey, D.P.; Jaskolska, M.; Gerdes, K. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J. Bacteriol. 2009, 191, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Comprehensive comparative- genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct 2009, 4. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 2000, 182, 561–572. [Google Scholar] [CrossRef] [PubMed]
- Pandey, D.P.; Gerdes, K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005, 33, 966–976. [Google Scholar] [CrossRef] [PubMed]
- Ramage, H.R.; Connolly, L.E.; Cox, J.S. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: Implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009, 5, e1000767. [Google Scholar] [CrossRef] [PubMed]
- Schippers, A.; Neretin, L.N.; Kallmeyer, J.; Ferdelman, T.G.; Cragg, B.A.; Parkes, R.J.; Jorgensen, B.B. Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature 2005, 433, 861–864. [Google Scholar] [CrossRef] [PubMed]
- Moran, M.A. The global ocean microbiome. Science 2015, 350. [Google Scholar] [CrossRef] [PubMed]
- Sunagawa, S.; Coelho, L.P.; Chaffron, S.; Kultima, J.R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D.R.; Alberti, A.; et al. Structure and function of the global ocean microbiome. Science 2015, 348. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.J.; Li, R.Q.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.B.; Mandi, A.; Li, S.M.; Chen, Y.C.; Zhang, W.J.; Tian, X.P.; Zhang, H.B.; Li, H.X.; Zhang, W.M.; Zhang, S.; et al. N–N-coupled indolo-sesquiterpene atropo-diastereomers from a marine-derived actinomycete. Eur. J. Org. Chem. 2012, 2012, 5256–5262. [Google Scholar] [CrossRef]
- Li, H.X.; Zhang, Q.B.; Li, S.M.; Zhu, Y.G.; Zhang, G.T.; Zhang, H.B.; Tian, X.P.; Zhang, S.; Ju, J.H.; Zhang, C.S. Identification and characterization of xiamycin A and oxiamycin gene cluster reveals an oxidative cyclization strategy tailoring indolosesquiterpene biosynthesis. J. Am.Chem. Soc. 2012, 134, 8996–9005. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.L.; Wood, T.K.; Peti, W.; Page, R. Structure of the Escherichia coli antitoxin MqsA (YgiT/b3021) bound to its gene promoter reveals extensive domain rearrangements and the specificity of transcriptional regulation. J. Biol. Chem. 2011, 286, 2285–2296. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Sternberg, M.J.E. Protein structure prediction on the web: A case study using the phyre server. Nat. Protoc. 2009, 4, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lord, D.M.; Hong, S.H.; Peti, W.; Benedik, M.J.; Page, R.; Wood, T.K. Type II toxin/antitoxin MqsR/MqsA controls type V toxin/antitoxin GhoT/GhoS. Environ. Microbiol. 2013, 15, 1734–1744. [Google Scholar] [CrossRef] [PubMed]
- Winther, K.S.; Gerdes, K. Ectopic production of VapCs from Enterobacteria inhibits translation and trans-activates YoeB mRNA interferase. Mol. Microbiol. 2009, 72, 918–930. [Google Scholar] [CrossRef] [PubMed]
- Maisonneuve, E.; Shakespeare, L.J.; Jorgensen, M.G.; Gerdes, K. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. USA 2011, 108, 13206–13211. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Clemens, D.L.; Horwitz, M.A. Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagosomal maturation is inhibited. J. Exp. Med. 1995, 181, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, D.; Zenno, S.; Lee, W.C.; Saigo, K.; Tanokura, M. Crystal structure of the PIN domain of human telomerase-associated protein EST1A. Proteins Struct. Funct. Bioinform. 2007, 68, 980–989. [Google Scholar] [CrossRef] [PubMed]
- Lamanna, A.C.; Karbstein, K. Nob1 binds the single-stranded cleavage site D at the 3′-end of 18S rRNA with its PIN domain. Proc. Natl. Acad. Sci. USA 2009, 106, 14259–14264. [Google Scholar] [CrossRef] [PubMed]
- Arcus, V.L.; Rainey, P.B.; Turner, S.J. The PIN-domain toxin-antitoxin array in Mycobacteria. Trends Microbiol. 2005, 13, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Arcus, V.L.; McKenzie, J.L.; Robson, J.; Cook, G.M. The PIN-domain ribonucleases and the prokaryotic VapBC toxin-antitoxin array. Protein Eng. Des. Sel. 2011, 24, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Ryu, T.; Seridi, L.; Moitinho-Silva, L.; Oates, M.; Liew, Y.J.; Mavromatis, C.; Wang, X.; Haywood, A.; Lafi, F.F.; Kupresanin, M.; et al. Hologenome analysis of two marine sponges with different microbiomes. BMC Genom. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Miallau, L.; Faller, M.; Chiang, J.; Arbing, M.; Guo, F.; Cascio, D.; Eisenberg, D. Structure and proposed activity of a member of the VapBC family of toxin-antitoxin systems VapBC-5 from Mycobacterium tuberculosis. J. Biol. Chem. 2009, 284, 276–283. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, J.L.; Robson, J.; Berney, M.; Smith, T.C.; Ruthe, A.; Gardner, P.P.; Arcus, V.L.; Cook, G.M. A VapBC toxin-antitoxin module is a posttranscriptional regulator of metabolic flux inMycobacteria. J. Bacteriol. 2012, 194, 2189–2204. [Google Scholar] [CrossRef] [PubMed]
- Radnedge, L.; Davis, M.A.; Youngren, B.; Austin, S.J. Plasmid maintenance functions of the large virulence plasmid of Shigella flexneri. J. Bacteriol. 1997, 179, 3670–3675. [Google Scholar] [PubMed]
- Winther, K.S.; Gerdes, K. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc. Natl. Acad. Sci. USA 2011, 108, 7403–7407. [Google Scholar] [CrossRef] [PubMed]
- Sharp, J.D.; Cruz, J.W.; Raman, S.; Inouye, M.; Husson, R.N.; Woychik, N.A. Growth and translation inhibition through sequence-specific RNA binding by Mycobacterium tuberculosis VapC toxin. J. Biol. Chem. 2012, 287, 12835–12847. [Google Scholar] [CrossRef] [PubMed]
- Daines, D.A.; Wu, M.H.; Yuan, S.Y. VapC-1 of nontypeable Haemophilus influenzae is a ribonuclease. J. Bacteriol. 2007, 189, 5041–5048. [Google Scholar] [CrossRef] [PubMed]
- Fiebig, A.; Castro Rojas, C.M.; Siegal-Gaskins, D.; Crosson, S. Interaction specificity, toxicity and regulation of a paralogous set of ParE/RelE-family toxin–antitoxin systems. Mol. Microbiol. 2010, 77, 236–251. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.T.; Yeo, C.C.; Sadowy, E.; Espinosa, M. Functional validation of putative toxin-antitoxin genes from the gram-positive pathogen Streptococcus pneumoniae: Phd-doc is the fourth bona-fide operon. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Guo, Y.; Zeng, Z.; Liu, X.; Shi, F.; Wang, X. Identification and characterization of a HEPN-MNT family type II toxin-antitoxin in Shewanella oneidensis. Microb. Biotechnol. 2015, 8, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Demidenok, O.I.; Kaprelyants, A.S.; Goncharenko, A.V. Toxin-antitoxin VapBC locus participates in formation of the dormant state in Mycobacterium smegmatis. FEMS Microbiol. Lett. 2014, 352, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Mate, M.J.; Vincentelli, R.; Foos, N.; Raoult, D.; Cambillau, C.; Ortiz-Lombardia, M. Crystal structure of the DNA-bound VapBC2 antitoxin/toxin pair from Rickettsia felis. Nucleic Acids Res. 2012, 40, 3245–3258. [Google Scholar] [CrossRef] [PubMed]
- Min, A.B.; Miallau, L.; Sawaya, M.R.; Habel, J.; Cascio, D.; Eisenberg, D. The crystal structure of the Rv0301-Rv0300 VapBC-3 toxin-antitoxin complex from M. Tuberculosis reveals a Mg2+ ion in the active site and a putative RNA-binding site. Protein Sci. 2012, 21, 1754–1767. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Christensen, S.K.; Lobner-Olesen, A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005, 3, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Robson, J.; McKenzie, J.L.; Cursons, R.; Cook, G.M.; Arcus, V.L. The VapBC operon from Mycobacterium smegmatis is an autoregulated toxin–antitoxin module that controls growth via inhibition of translation. J. Mol. Biol. 2009, 390, 353–367. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, J.L.; Duyvestyn, J.M.; Smith, T.; Bendak, K.; MacKay, J.; Cursons, R.; Cook, G.M.; Arcus, V.L. Determination of ribonuclease sequence-specificity using pentaprobes and mass spectrometry. RNA 2012, 18, 1267–1278. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Wang, X.; Zhang, X.-S.; Grigoriu, S.; Page, R.; Peti, W.; Wood, T.K. Escherichia coli toxin/antitoxin pair MqsR/MqsA regulate toxin CspD. Environ. Microbiol. 2010, 12, 1105–1121. [Google Scholar] [CrossRef] [PubMed]
- Kasari, V.; Mets, T.; Tenson, T.; Kaldalu, N. Transcriptional cross-activation between toxin-antitoxin systems of Escherichia coli. BMC Microbiol. 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Pino, A.; Christensen-Dalsgaard, M.; Wyns, L.; Yarmolinsky, M.; Magnuson, R.D.; Gerdes, K.; Loris, R. Doc of prophage P1 is inhibited by its antitoxin partner Phd through fold complementation. J. Biol. Chem. 2008, 283, 30821–30827. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, M.; Ara, T.; Arifuzzaman, M.; Ioka-Nakamichi, T.; Inamoto, E.; Toyonaga, H.; Mori, H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005, 12, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Jin, M.; Ju, L.; Mao, Y.; Gao, H. Evidence for function overlapping of CymA and the cytochrome bc1 complex in the Shewanella oneidensis nitrate and nitrite respiration. Environ. Microbiol. 2014, 16, 3181–3195. [Google Scholar] [CrossRef] [PubMed]
- Schägger, H. Tricine-SDS-PAGE. Nat. Protoc. 2006, 1, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, J.; Tang, P.; Chen, H.; Gao, H. Genetic and molecular characterization of flagellar assembly in Shewanella oneidensis. PLoS ONE 2011, 6, e21479. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Bedzyk, L.A.; Thomas, S.M.; Ye, R.W.; Wood, T.K. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 2004, 64, 515–524. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).