Changes in Spirulina’s Physical and Chemical Properties during Submerged and Solid-State Lacto-Fermentation
Abstract
1. Introduction
2. Results and Discussion
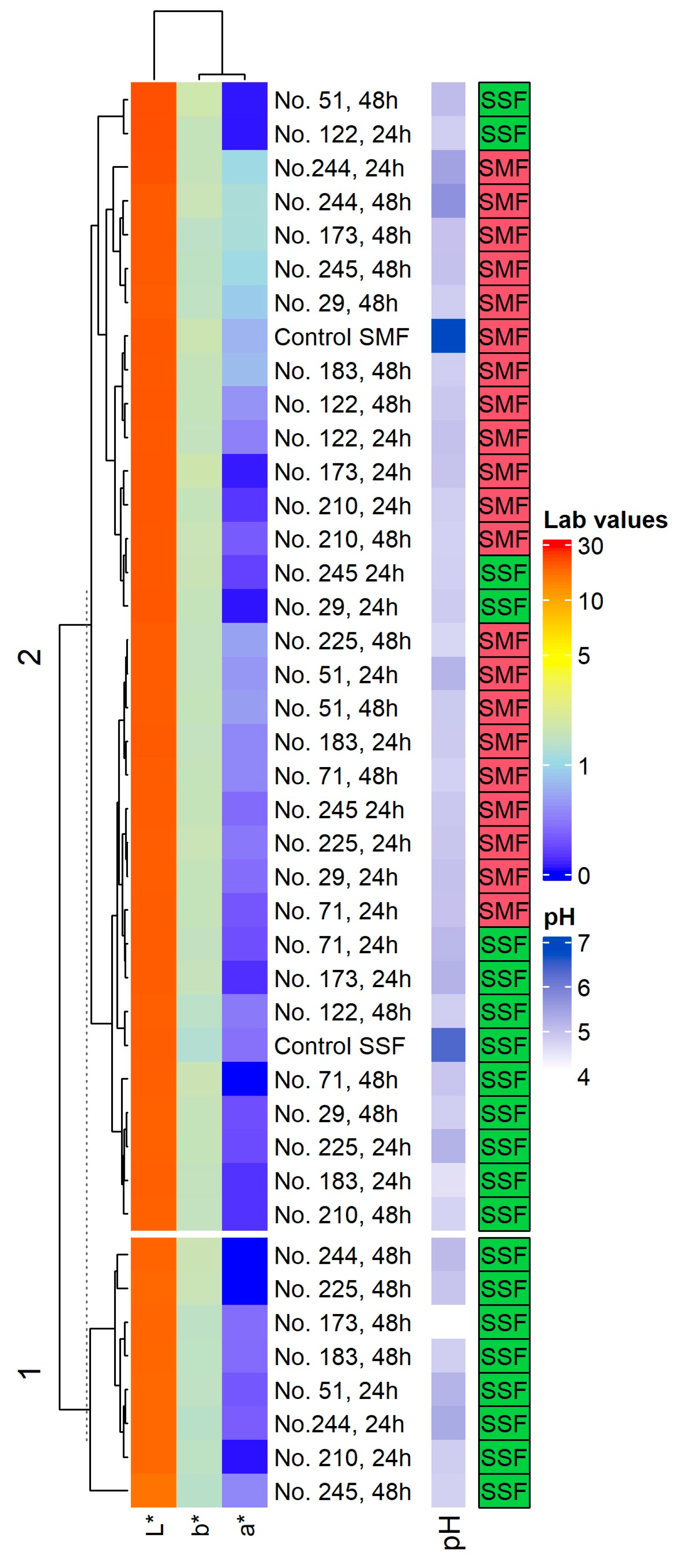
2.1. pH Values and Color Coordinates (L*, a* and b*) in the Spirulina Samples
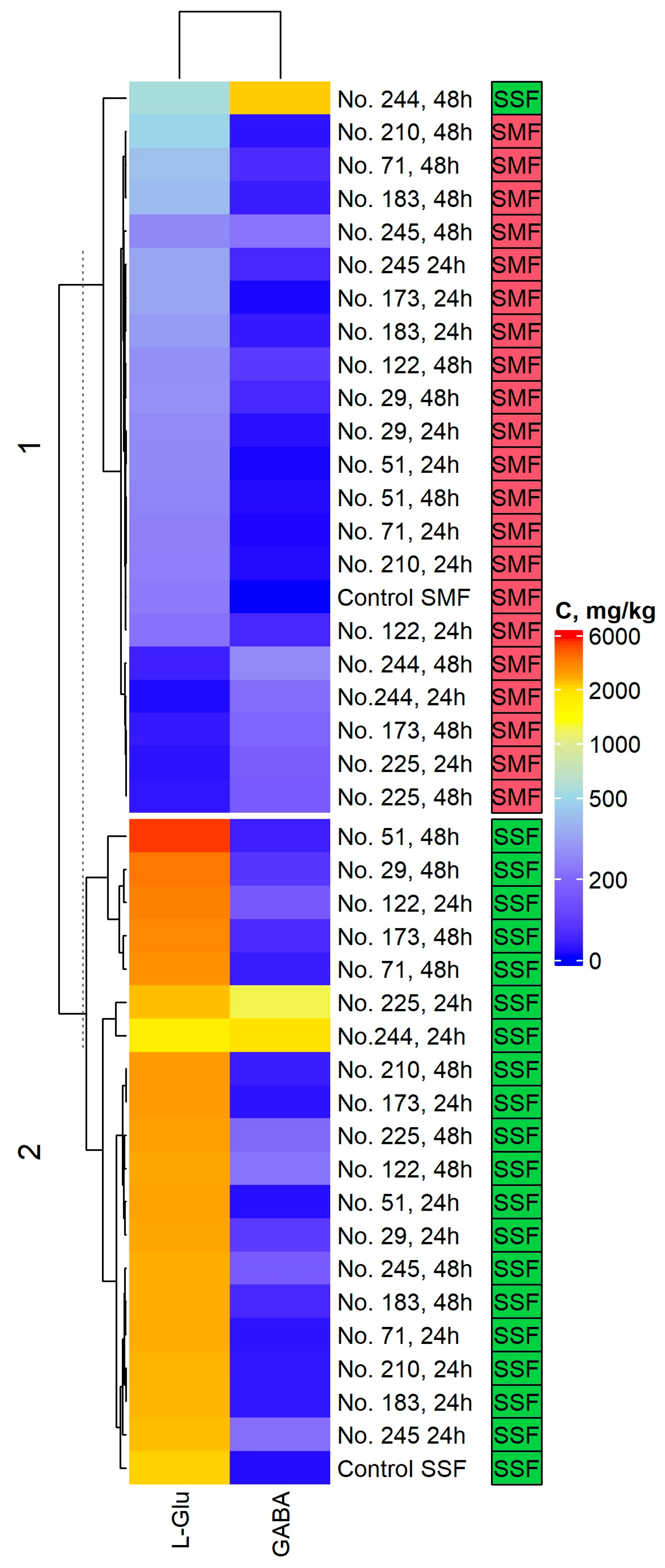
2.2. l-Glutamic Acid (L-Glu) and Gamma-Aminobutyric Acid (GABA) Concentration in the Spirulina Samples
2.3. Biogenic Amine (BA) Content in the Spirulina Samples
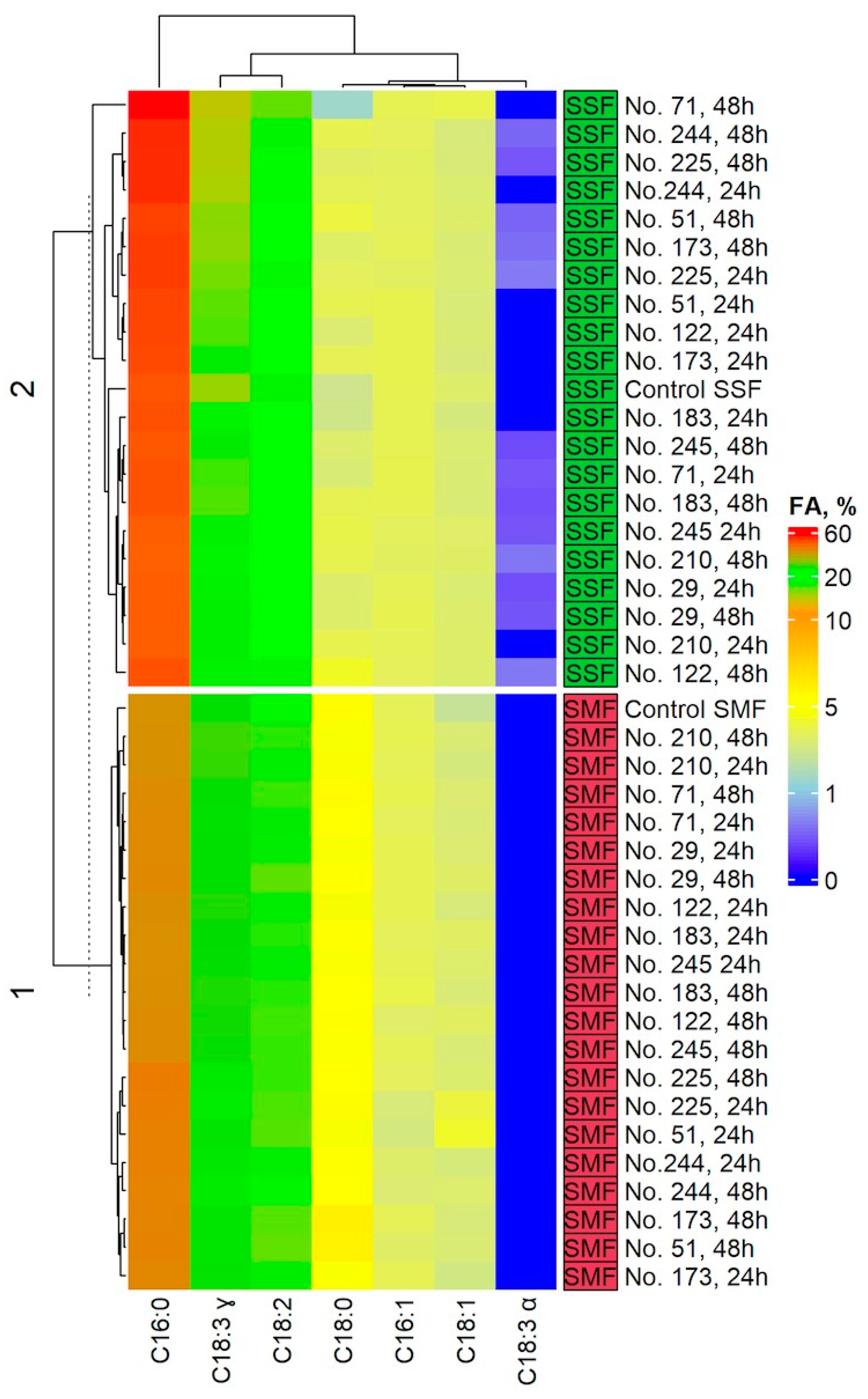
2.4. Fatty Acid (FA) Profile in the Spirulina Samples
3. Conclusions
4. Materials and Methods
4.1. Spirulina and Lactic Acid Bacteria Strains Used in Experiments and Fermentation Conditions
4.2. Analysis of pH and Color Coordinates (L*, a* and b*) in the Spirulina Samples
4.3. Evaluation of l-Glutamic Acid (L-Glu) and Gamma-Aminobutyric Acid (GABA) Concentration in Spirulina Samples
4.4. Analysis of Biogenic Amine (BA) Concentration in the Spirulina Samples
4.5. Analysis of Fatty Acid (FA) Profile in the Spirulina Samples
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yogeswara, I.B.A.; Maneerat, S.; Haltrich, D. Glutamate decarboxylase from lactic acid bacteria—A key enzyme in GABA synthesis. Microorganisms 2020, 8, 1923. [Google Scholar] [CrossRef] [PubMed]
- Aoki, H.; Uda, I.; Tagami, K.; Furuya, Y.; Endo, Y.; Fujimoto, K. The production of a new tempeh like fermented soybean containing a high level of γ-aminobutyric acid by anaerobic incubation with Rhizopus. Biosci. Biotechnol. Biochem. 2003, 67, 1018–1023. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lee, C.; Pan, T. Improvement of monacolin K, gamma-aminobutyric acid and citrinin productionratio as a function of environmental conditions of Monascus purpureus NTU 601. J. Ind. Microbiol. Biot. 2003, 30, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Miao, K.; Niyaphorn, S.; Qu, X. Production of gamma-aminobutyric acid from lactic acid bacteria: A systematic review. Int. J. Mol. Sci. 2020, 21, 995. [Google Scholar] [CrossRef]
- Sorrenti, V.; Castagna, D.A.; Fortinguerra, S.; Buriani, A.; Scapagnini, G.; Willcox, D.C. Spirulina Microalgae and Brain Health: A Scoping Review of Experimental and Clinical Evidence. Mar. Drugs 2021, 19, 293. [Google Scholar] [CrossRef] [PubMed]
- Miranda, M.; Cintra, R.; Barros, S.D.M.; Mancini-Filho, J. Antioxidant activity of the microalga Spirulina maxima. Braz. J. Med. Biol. Res. 1998, 31, 1075–1079. [Google Scholar] [CrossRef]
- Khan, Z.; Bhadouria, P.; Bisen, P. Nutritional and therapeutic potential of Spirulina. Curr. Pharm. Biotechnol. 2005, 6, 373–379. [Google Scholar] [CrossRef]
- Pratama, A.I.; Lioe, H.N.; Yuliana, N.D.; Ogawa, M. Umami compounds present in umami fraction of acid-hydrolyzed Spirulina (Spirulina platensis). Algal Res. 2022, 66, 102764. [Google Scholar] [CrossRef]
- Altmann, B.A.; Rosenau, S. Spirulina as Animal Feed: Opportunities and Challenges. Foods 2022, 11, 965. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Daim, M.M.; Ahmed, A.; Ijaz, H.; Abushouk, A.I.; Ahmed, H.; Negida, A.; Aleya, L.; Bungau, S.G. Influence of Spirulina platensis and ascorbic acid on amikacin-induced nephrotoxicity in rabbits. Env. Sci. Pollut. Res. 2019, 26, 8080–8086. [Google Scholar] [CrossRef]
- Abdel-Daim, M.M.; Abushouk, A.I.; Alkhalf, M.I.; Toraih, E.A.; Fawzy, M.S.; Ijaz, H.; Aleya, L.; Bungau, S.G. Antagonistic effects of Spirulina platensis on diazinon-induced hemato-biochemical alterations and oxidative stress in rats. Environ. Sci. Pollut. Res. 2018, 25, 27463–27470. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.J.; Nam, Y.D.; Lee, S.Y.; Park, S.L.; Yi, S.H.; Lim, S.I. Expression and characterization of a glutamate decarboxylase from Lactobacillus brevis 877G producing gamma-aminobutyric acid. Biosci. Biotechnol. Biochem. 2013, 77, 853–856. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Qiu, T.; Gao, D.; Cao, Y. Medium optimization for production of gamma-aminobutyric acid by Lactobacillus brevis NCL912. Amino Acids 2010, 38, 1439–1945. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Qiu, T.; Huang, G.; Cao, Y. Production of gamma-aminobutyric acid by Lactobacillus brevis NCL912 using fed-batch fermentation. Microb. Cell Fact. 2010, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Park, K.B.; Oh, S.H. Cloning, sequencing and expression of a novel glutamate decarboxylase gene from a newly isolated lactic acid bacterium, Lactobacillus brevis OPK-3. Bioresource Technol. 2007, 98, 312–319. [Google Scholar] [CrossRef]
- Siragusa, S.; de Angelis, M.; di Cagno, R.; Rizzello, C.G.; Coda, R.; Gobbetti, M. Synthesis of γ-aminobutyric acid by lactic acid bacteria isolated from a variety of Italian cheeses. Appl. Environ. Microb. 2007, 73, 7283–7290. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, X.; Fu, J.; Wang, S.; Chen, Y.; Chang, K.; Li, H. Substrate sustained release-based high efficacy biosynthesis of GABA by Lactobacillus brevis NCL912. Micro. Cell Fact. 2018, 17, 80. [Google Scholar] [CrossRef]
- Diana, M.; Tres, A.; Quílez, J.; Llombart, M.; Rafecas, M. Spanish cheese screening and selection of lactic acid bacteria with high gamma-aminobutyric acid production. LWT-Food Sci. Technol. 2014, 56, 351–355. [Google Scholar] [CrossRef]
- Park, K.B.; Oh, S.H. Isolation and characterization of Lactobacillus buchneri strains with high gamma-aminobutyric acid producing capacity from naturally aged cheese. Food Sci. Biotechnol. 2006, 15, 86–90. [Google Scholar]
- Cho, Y.R.; Chang, J.Y.; Chang, H.C. Production of γ-aminobutyric acid (GABA) by Lactobacillus buchneri isolated from Kimchi and its neuroprotective effect on neuronal cells. J. Microbiol. Biotechnol. 2007, 17, 104–109. [Google Scholar]
- Gangaraju, D.S.; Murty, V.R.; Prapulla, S.G. Probiotic-mediated biotransformation of monosodium glutamate to γ-aminobutyric acid: Differential production in complex and minimal media and kinetic modelling. Ann. Microbiol. 2014, 64, 229–237. [Google Scholar] [CrossRef]
- Woraharn, S.; Lailerd, N.; Sivamaruthi, B.S.; Wangcharoen, W.; Sirisattha, S.; Peerajan, S.; Chaiyasut, C. Evaluation of factors that influence the L-glutamic and γ-aminobutyric acid production during Hericium erinaceus fermentation by lactic acid bacteria. CyTA-J. CyTA-J. Food 2016, 14, 47–54. [Google Scholar] [CrossRef]
- Lin, Q.; Li, D.; Qin, H. Molecular cloning, expression, and immobilization of glutamate decarboxylase from Lactobacillus fermentum YS2. Electron. J. Biotechnol. 2017, 27, 8–13. [Google Scholar] [CrossRef]
- Sun, T.S.; Zhao, S.P.; Wang, H.K.; Cai, C.K.; Chen, Y.F.; Zhang, H.P. ACE-inhibitory activity and gamma-aminobutyric acid content of fermented skim milk by Lactobacillus helveticus isolated from Xinjiang koumiss in China. Eur. Food Res. Technol. 2009, 228, 607–612. [Google Scholar] [CrossRef]
- Franciosi, E.; Carafa, I.; Nardin, T.; Schiavon, S.; Poznanski, E.; Cavazza, A.; Larcher, R.; Tuohy, K.M. Biodiversity and γ-aminobutyric acid production by lactic acid bacteria isolated from traditional alpine raw cow’s milk cheeses. BioMed Res. Int. 2015, 625740. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, K.; Jiang, Y.; Feng, X.; Li, L.; Dang, F.; Zhang, W.; Man, C. Transcriptomic response to GABA-producing Lactobacillus plantarum CGMCC 1.2437T induced by L-MSG. PLoS ONE 2018, 13, e0199021. [Google Scholar] [CrossRef] [PubMed]
- Bartkiene, E.; Lele, V.; Ruzauskas, M.; Domig, K.J.; Starkute, V.; Zavistanaviciute, P.; Bartkevics, V.; Pugajeva, I.; Klupsaite, D.; Juodeikiene, G.; et al. Lactic acid bacteria isolation from spontaneous sourdough and their characterization including antimicrobial and antifungal properties evaluation. Microorganisms 2019, 8, 64. [Google Scholar] [CrossRef]
- Zokaityte, E.; Lele, V.; Starkute, V.; Zavistanaviciute, P.; Klupsaite, D.; Bartkevics, V.; Pugajeva, I.; Bērziņa, Z.; Gruzauskas, R.; Sidlauskiene, S.; et al. The influence of combined extrusion and fermentation processes on the chemical and biosafety parameters of wheat bran. LWT 2021, 146, 111498. [Google Scholar] [CrossRef]
- Mokoena, M.P. Lactic Acid Bacteria and Their Bacteriocins: Classification, Biosynthesis and Applications against Uropathogens: A Mini-Review. Molecules 2017, 22, 1255. [Google Scholar] [CrossRef]
- Hernández-González, J.C.; Martínez-Tapia, A.; Lazcano-Hernández, G.; García-Pérez, B.E.; Castrejón-Jiménez, N.S. Bacteriocins from Lactic Acid Bacteria. A Powerful Alternative as Antimicrobials, Probiotics, and Immunomodulators in Veterinary Medicine. Animals 2021, 11, 979. [Google Scholar] [CrossRef]
- He, Q.; Peng, H.; Sheng, M.; Hu, S.; Qiu, J.; Gu, J. Humidity control strategies for solid-state fermentation: Capillary water supply by water-retention materials and negative-pressure auto-controlled irrigation. Front. Bioeng. Biotechnol. 2019, 7, 263. [Google Scholar] [CrossRef]
- Banat, I.M.; Carboué, Q.; Saucedo-Castañeda, G.; Cázares-Marinero, J.J. Biosurfactants: The green generation of speciality chemicals and potential production using Solid-State fermentation (SSF) technology. Bioresour. Technol. 2021, 320, 124222. [Google Scholar] [CrossRef]
- Sadh, P.K.; Duhan, S.; Duhan, J.S. Agro-industrial wastes and their utilization using solid state fermentation: A review. Bioresour. Bioprocess 2018, 5, 1. [Google Scholar] [CrossRef]
- Venil, C.K.; Yusof, N.Z.B.; Ahmad, W.A. Solid State Fermentation Utilizing Agroindustrial Waste for Microbial Pigment Production. In Sustainable Agriculture towards Food Security; Dhanarajan, A., Ed.; Springer Singapore: Singapore, 2017; pp. 375–381. [Google Scholar] [CrossRef]
- Amaya-Chantaca, D.; Flores-Gallegos, A.C.; Iliná, A.; Aguilar, C.N.; Sepúlveda-Torre, L.; Ascacio-Vadlés, J.A.; Chávez-González, M.L. Comparative extraction study of grape pomace bioactive compounds by submerged and solid-state fermentation. J. Chem. Technol. Biotechnol. 2022, 97, 1494–1505. [Google Scholar] [CrossRef]
- Yilmaz, N.; Özogul, F.; Moradi, M.; Fadiloglu, E.E.; Simat, V.; Rocha, J.M. Reduction of biogenic amines formation by foodborne pathogens using postbiotics in lysine-decarboxylase broth. J. Biotechnol. 2022, 7, 118–127. [Google Scholar] [CrossRef]
- Ryu, I.S.; Kim, O.H.; Kim, J.S.; Sohn, S.; Choe, E.S.; Lim, R.N.; Kim, T.W.; Seo, J.W.; Jang, E.Y. Effects of β-Phenylethylamine on Psychomotor, Rewarding, and Reinforcing Behaviors and Affective State: The Role of Dopamine D1 Receptors. Int. J. Mol. Sci 2021, 22, 9485. [Google Scholar] [CrossRef] [PubMed]
- Rutigliano, G.; Accorroni, A.; Zucchi, R. The Case for TAAR1 as a Modulator of Central Nervous System Function. Front. Pharmacol. 2018, 8, 987. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, J.; Kishino, S.; Ando, A.; Sugimoto, S.; Mihara, K.; Shimizu, S. Production of conjugated fatty acids by lactic acid bacteria. J. Biosci. Bioeng. 2005, 100, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Gunasekaran, Y.K.; Lele, V.; Sakiene, V.; Zavistanaviciut, P.; Zokaityte, E.; Klupsaite, D.; Bartkevics, V.; Guiné, R.P.; Bartkiene, E. Plant-based proteinaceous snacks: Effect of fermentation and ultrasonication on end-product characteristics. Food Sci. Nutr. 2020, 8, 4746–4756. [Google Scholar] [CrossRef]
- Jamnik, P.; Mahnič, N.; Mrak, A.; Pogačnik, L.; Jeršek, B.; Niccolai, A.; Masten Rutar, J.; Ogrinc, N.; Dušak, L.; Ferjančič, B.; et al. Fermented Biomass of Arthrospira platensis as a Potential Food Ingredient. Antioxidants 2022, 11, 216. [Google Scholar] [CrossRef]
- Bao, J.; Zhang, X.; Zheng, J.H.; Ren, D.F.; Lu, J. Mixed fermen tation of Spirulina platensis with Lactobacillus plantarum and Bacillus subtilis by random-centroid optimization. Food Chem. 2018, 264, 64–72. [Google Scholar] [CrossRef]
- Park, W.S.; Kim, H.J.; Li, M.; Lim, D.H.; Kim, J.; Kwak, S.S.; Kang, M.; Ferruzzi, M.G.; Ahn, M.-J. Two Classes of Pigments, Carotenoids and C-Phycocyanin, in Spirulina Powder and Their Antioxidant Activities. Molecules 2018, 23, 2065. [Google Scholar] [CrossRef] [PubMed]
- Careri, M.; Furlattini, L.; Mangia, A.; Musci, M.; Anklam, E.; Theobald, A.; Von Holst, C. Supercritical fluid extraction for liquid chromatographic determination of carotenoids in Spirulina Pacifica algae: A chemometric approach. J. Chromatogr. A 2001, 912, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Chaiklahan, R.; Chirasuwan, N.; Bunnag, B. Stability of phycocyanin extracted from Spirulina sp.: Influence of temperature, pH and preservatives. Process Biochem. 2012, 47, 659–664. [Google Scholar] [CrossRef]
- İlter, I.; Akyıl, S.; Demirel, Z.; Koç, M.; Conk-Dalay, M.; Kaymak-Ertekin, F. Optimization of phycocyanin extraction from Spirulina platensis using different techniques. J. Food Compos. Anal. 2018, 70, 78–88. [Google Scholar] [CrossRef]
- Fithriani, D.; Sinurat, E. Utilization of spirulina as functional food: Phytosterol and amino acid profiles study. IOP Conf. Ser. Earth Environ. Sci. 2019, 278, 012028. [Google Scholar] [CrossRef]
- Cian, R.E.; Drago, S.R.; De Medina, F.S.; Martínez-Augustin, O. Proteins and carbohydrates from red seaweeds: Evidence for beneficial effects on gut function and microbiota. Mar. Drugs 2015, 13, 5358–5383. [Google Scholar] [CrossRef]
- de Marco Castro, E.; Shannon, E.; Abu-Ghannam, N. Effect of fermentation on enhancing the nutraceutical properties of Arthrospira platensis (Spirulina). Fermentation 2019, 5, 28. [Google Scholar] [CrossRef]
- An, J.; Feng, Y.; Zheng, J.; Addy, M.; Zhang, L.; Ren, D. The immune-enhancing potential of peptide fractions from fermented Spirulina platensis by mixed probiotics. J. Food Biochem 2020, 44, e13245. [Google Scholar] [CrossRef]
- Mamuad, L.L.; Lee, S.S. The Role of Glutamic Acid-producing Microorganisms in Rumen Microbial Ecosystems. J. Life Sci. 2021, 31, 520–526. [Google Scholar] [CrossRef]
- Dhakal, R.; Bajpai, V.K.; Baek, K.H. Production of GABA (γ-aminobutyric acid) by microorganisms: A review. Brazilian J. Microbiol. 2012, 43, 1230–1241. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Hwang, C.; Do, H.K.; Kwon, G.S.; Kang, S.C. Anti-oxidizing Functional Effect of Polyamine as a Bioconversion Starter Using Microbes Isolated from Fluke babsikhae. J. Korean Soc. Appl. Biol. Chem. 2013, 56, 83–86. [Google Scholar] [CrossRef]
- Sahay, N.; Chaudhari, N.; Trivedi, R.; Patel, T.; Kommu, S. Spirulina the super food; regulation of foul-fishy smell. Acad. Lett. 2022, 2. [Google Scholar] [CrossRef]
- Kivirand, K.; Somerik, H.; Oldekop, M.L.; Rebane, R.; Rinken, T. Effect of spermidine and its metabolites on the activity of pea seedlings diamine oxidase and the problems of biosensing of biogenic amines with this enzyme. Enzym. Microb. Technol. 2016, 82, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Glória, M.B.A.; Tavares-Neto, J.; Labanca, R.A.; Carvalho, M.S. Influence of cultivar and germination on bioactive amines in soybeans (Glycine max L. Merril). J. Agric. Food Chem. 2005, 53, 7480–7485. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xin, S.; Zhang, C.; Ran, X.; Tang, H.; Cao, D. Development of a novel chromophore reaction-based fluorescent probe for biogenic amines detection. J. Mater. Chem. B. 2021, 9, 9383–9394. [Google Scholar] [CrossRef]
- Müller, D.G.; Oreste, E.Q.; Heinemann, M.G.; Dias, D.; Kessler, F. Biogenic amine sensors and its building materials: A review. Eur. Polym. J. 2022, 111221. [Google Scholar] [CrossRef]
- Bartkiene, E.; Krungleviciute, V.; Juodeikiene, G.; Vidmantiene, D.; Maknickiene, Z. Solid state fermentation with lactic acid bacteria to improve the nutritional quality of lupin and soya bean. J. Sci. Food Agric. 2015, 95, 1336–1342. [Google Scholar] [CrossRef]
- AlFadhly, N.K.; Alhelfi, N.; Altemimi, A.B.; Verma, D.K.; Cacciola, F.; Narayanankutty, A. Trends and Technological Advancements in the Possible Food Applications of Spirulina and Their Health Benefits: A Review. Molecules 2022, 27, 5584. [Google Scholar] [CrossRef]
- Diraman, H.; Koru, E.; Dibeklioglu, H. Fatty Acid Profile of Spirulina platensis Used as a Food Supplement. Isr. J. Aquac. Bamidgeh 2009, 61, 134–142. [Google Scholar] [CrossRef]
- Ramírez-Rodrigues, M.M.; Estrada-Beristain, C.; Metri-Ojeda, J.; Pérez-Alva, A.; Baigts-Allende, D.K. Spirulina platensis Protein as Sustainable Ingredient for Nutritional Food Products Development. Sustainability 2021, 13, 6849. [Google Scholar] [CrossRef]
- Jung, F.; Krüger-Genge, A.; Waldeck, P.; Küpper, J.H. Spirulina platensis, a super food? J. Cell Biotechnol. 2019, 5, 43–54. [Google Scholar] [CrossRef]
- Choopani, A.; Poorsoltan, M.; Fazilati, M.; Latifi, A.M.; Salavati, H. Spirulina: A Source of Gamma-linoleic Acid and Its Applications. J. Appl. Biotechnol. Rep. 2016, 3, 483–488. [Google Scholar]
- Dewi, E.N.; Amalia, U. Nutritional comparison of Spirulina sp powder by solid-state fermentation using Aspergillus sp (FNCL 6088) and Lactobacillus plantarum (FNCL 0127). IOP Conf. Ser. Earth Environ. Sci. 2018, 102, 012024. [Google Scholar] [CrossRef]
- Hosseini, S.M.; Shahbazizadeh, S.; Khosravi-Darani, K.; Reza Mozafari, M. Spirulina paltensis: Food and function. Curr. Nutr. Food Sci. 2013, 9, 189–193. [Google Scholar] [CrossRef]
- Falquet, J.; Hurni, J.P. The Nutritional Aspects of Spirulina. Antenna Found. 1997. Available online: https://www.antenna.ch/wp-content/uploads/2017/03/AspectNut_UK (accessed on 10 August 2022).
- Usharani, G.; Saranraj, P.; Kanchana, D. Spirulina cultivation: A review. Int. J. Pharm. Biol. Arch. 2012, 3, 1327–1341. [Google Scholar]
- Andrade, L.M.; Andrade, C.J.; Dias, M.; Nascimento, C.; Mendes, M.A. Chlorella and spirulina microalgae as sources of functional foods. Nutraceuticals Food Suppl. 2018, 6, 45–58. [Google Scholar] [CrossRef]
- Al-Dhabi, N.A.; Valan Arasu, M. Quantification of phytochemicals from commercial Spirulina products and their antioxidant activities. Evid. Based Complement. Altern. Med. 2016, 2016, 7631864. [Google Scholar] [CrossRef] [PubMed]
- Liestianty, D.; Rodianawati, I.; Andi Arfah, R.; Asma Assa, A.; Muliadi, S.P. Nutritional Analysis of Spirulina sp. to Promote as Superfood Candidate. In Proceedings of the 13th Joint Conference on Chemistry (13th JCC), Semarang, Indonesia, 7–8 September 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Tolpeznikaite, E.; Ruzauskas, M.; Pilkaityte, R.; Bartkevics, V.; Zavistanaviciute, P.; Starkute, V.; Lele, V.; Zokaityte, E.; Mozuriene, E.; Ruibys, R.; et al. Influence of Fermentation on the Characteristics of Baltic Sea Macroalgae, Including Microbial Profile and Trace Element Content. Food Control 2021, 129, 108235. [Google Scholar] [CrossRef]
- Ben-Gigirey, B.J.M.; de Sousa, V.B.; Villa, T.G.; Barros-Velazquez, J. Changes in biogenic amines and microbiological analysis in albacore (Thunnus alalunga) muscle during frozen storage. J. Food Prot. 1998, 61, 608–615. [Google Scholar] [CrossRef]
- Pérez-Palacios, T.; Ruiz, J.; Ferreira, I.; Petisca, C.; Antequera, T. Effect of solvent to sample ratio on total lipid extracted and fatty acid composition in meat products within different fat content. Meat Sci. 2012, 91, 369–373. [Google Scholar] [CrossRef] [PubMed]



| Factors and Their Interaction | Dependent Variable | p |
|---|---|---|
| Lactic acid bacteria strain used for fermentation | L* | 0.403 |
| a* | 0.0001 | |
| b* | 0.377 | |
| pH | 0.791 | |
| Duration of fermentation | L* | 0.317 |
| a* | 0.0001 | |
| b* | 0.807 | |
| pH | 0.898 | |
| Conditions of fermentation (submerged or solid state) | L* | 0.438 |
| a* | 0.0001 | |
| b* | 0.286 | |
| pH | 0.042 | |
| Lactic acid bacteria strain used for fermentation × Duration of fermentation | L* | 0.422 |
| a* | 0.0001 | |
| b* | 0.448 | |
| pH | 0.719 | |
| Lactic acid bacteria strain used for fermentation × Conditions of fermentation (submerged or solid state) | L* | 0.398 |
| a* | 0.0001 | |
| b* | 0.112 | |
| pH | 0.439 | |
| Duration of fermentation × Conditions of fermentation (submerged or solid state) | L* | 0.307 |
| a* | 0.012 | |
| b* | 0.313 | |
| pH | 0.665 | |
| Lactic acid bacteria strain used for fermentation × Duration of fermentation × Conditions of fermentation (submerged or solid state) | L* | 0.393 |
| a* | 0.0001 | |
| b* | 0.197 | |
| pH | 0.486 |
| Factors and Their Interaction | Dependent Variable | p |
|---|---|---|
| Lactic acid bacteria strain used for fermentation | GABA | 0.0001 |
| l-Glutamic acid | 0.641 | |
| Duration of fermentation | GABA | 0.987 |
| l-Glutamic acid | 0.328 | |
| Conditions of fermentation (submerged or solid state) | GABA | 0.020 |
| l-Glutamic acid | 0.0001 | |
| Lactic acid bacteria strain used for fermentation × Duration of fermentation | GABA | 0.813 |
| l-Glutamic acid | 0.942 | |
| Lactic acid bacteria strain used for fermentation × Conditions of fermentation (submerged or solid state) | GABA | 0.0001 |
| l-Glutamic acid | 0.740 | |
| Duration of fermentation × Conditions of fermentation (submerged or solid state) | GABA | 0.499 |
| l-Glutamic acid | 0.358 | |
| Lactic acid bacteria strain used for fermentation × Duration of fermentation × Conditions of fermentation (submerged or solid state) | GABA | 0.893 |
| l-Glutamic acid | 0.957 |
| Correlations | Correlation (r) | Significance (p) | Correlation (r) | Significance (p) |
|---|---|---|---|---|
| with GABA | with l-Glutamic acid | |||
| TRP | 0.215 * | 0.016 | 0.541 ** | 0.0001 |
| PUT | 0.309 ** | 0.0001 | 0.486 ** | 0.0001 |
| CAD | 0.298 ** | 0.001 | −0.076 | 0.401 |
| HIS | 0.648 ** | 0.0001 | −0.073 | 0.414 |
| TYR | 0.681 ** | 0.0001 | −0.014 | 0.879 |
| SPRMD | 0.211 * | 0.018 | 0.627 ** | 0.0001 |
| SPRM | 0.172 | 0.054 | 0.528 ** | 0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tolpeznikaite, E.; Bartkevics, V.; Skrastina, A.; Pavlenko, R.; Mockus, E.; Zokaityte, E.; Starkute, V.; Klupsaite, D.; Ruibys, R.; Rocha, J.M.; et al. Changes in Spirulina’s Physical and Chemical Properties during Submerged and Solid-State Lacto-Fermentation. Toxins 2023, 15, 75. https://doi.org/10.3390/toxins15010075
Tolpeznikaite E, Bartkevics V, Skrastina A, Pavlenko R, Mockus E, Zokaityte E, Starkute V, Klupsaite D, Ruibys R, Rocha JM, et al. Changes in Spirulina’s Physical and Chemical Properties during Submerged and Solid-State Lacto-Fermentation. Toxins. 2023; 15(1):75. https://doi.org/10.3390/toxins15010075
Chicago/Turabian StyleTolpeznikaite, Ernesta, Vadims Bartkevics, Anna Skrastina, Romans Pavlenko, Ernestas Mockus, Egle Zokaityte, Vytaute Starkute, Dovile Klupsaite, Romas Ruibys, João Miguel Rocha, and et al. 2023. "Changes in Spirulina’s Physical and Chemical Properties during Submerged and Solid-State Lacto-Fermentation" Toxins 15, no. 1: 75. https://doi.org/10.3390/toxins15010075
APA StyleTolpeznikaite, E., Bartkevics, V., Skrastina, A., Pavlenko, R., Mockus, E., Zokaityte, E., Starkute, V., Klupsaite, D., Ruibys, R., Rocha, J. M., Santini, A., & Bartkiene, E. (2023). Changes in Spirulina’s Physical and Chemical Properties during Submerged and Solid-State Lacto-Fermentation. Toxins, 15(1), 75. https://doi.org/10.3390/toxins15010075









