Screening of TNFR1 Binding Peptides from Deinagkistrodon acutus Venom through Phage Display
Abstract
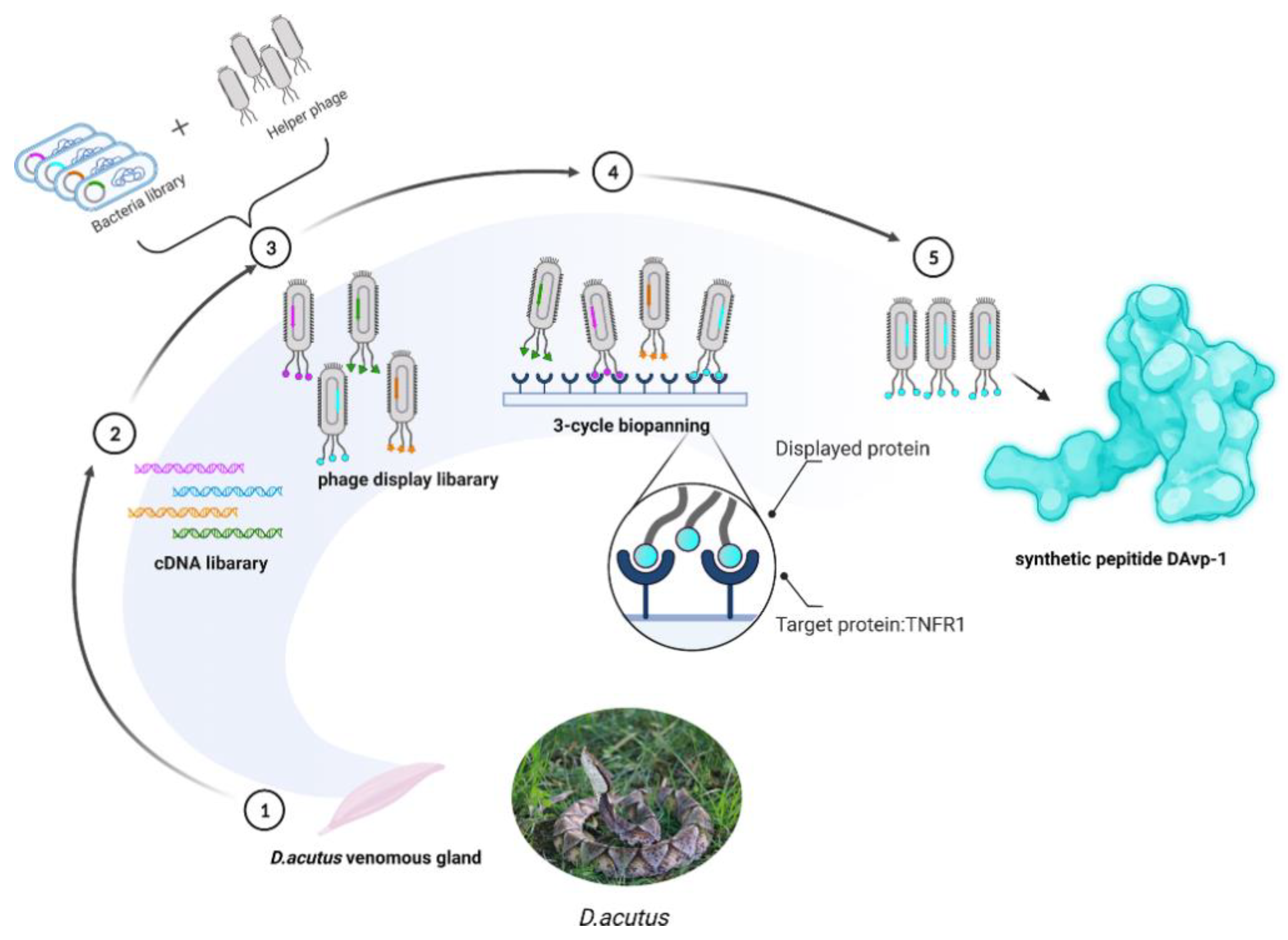
:1. Introduction
2. Results
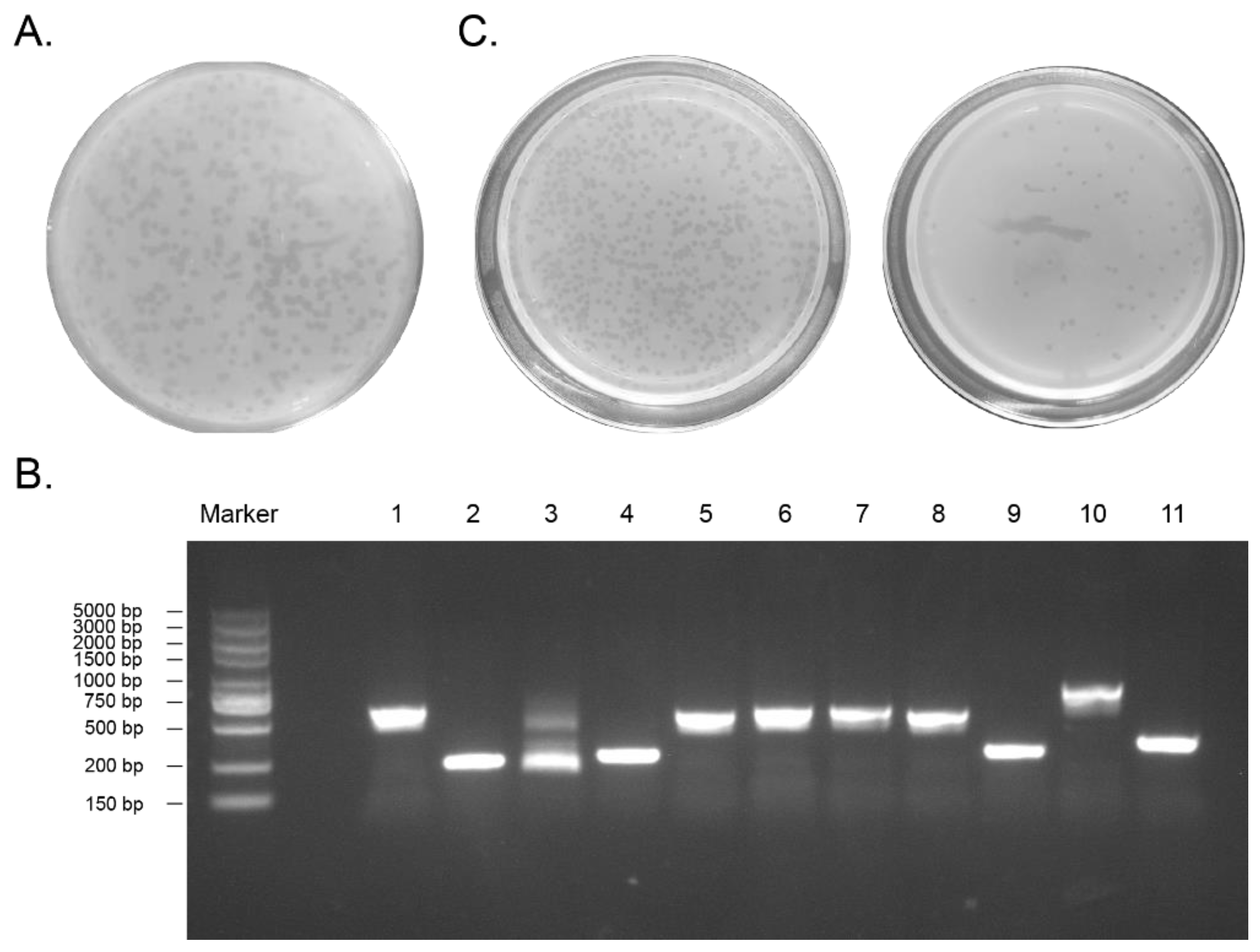
2.1. Construction of the D. acutus Venom Gland T7 Phage Display Library
2.2. Obtaining Peptide DAvp-1
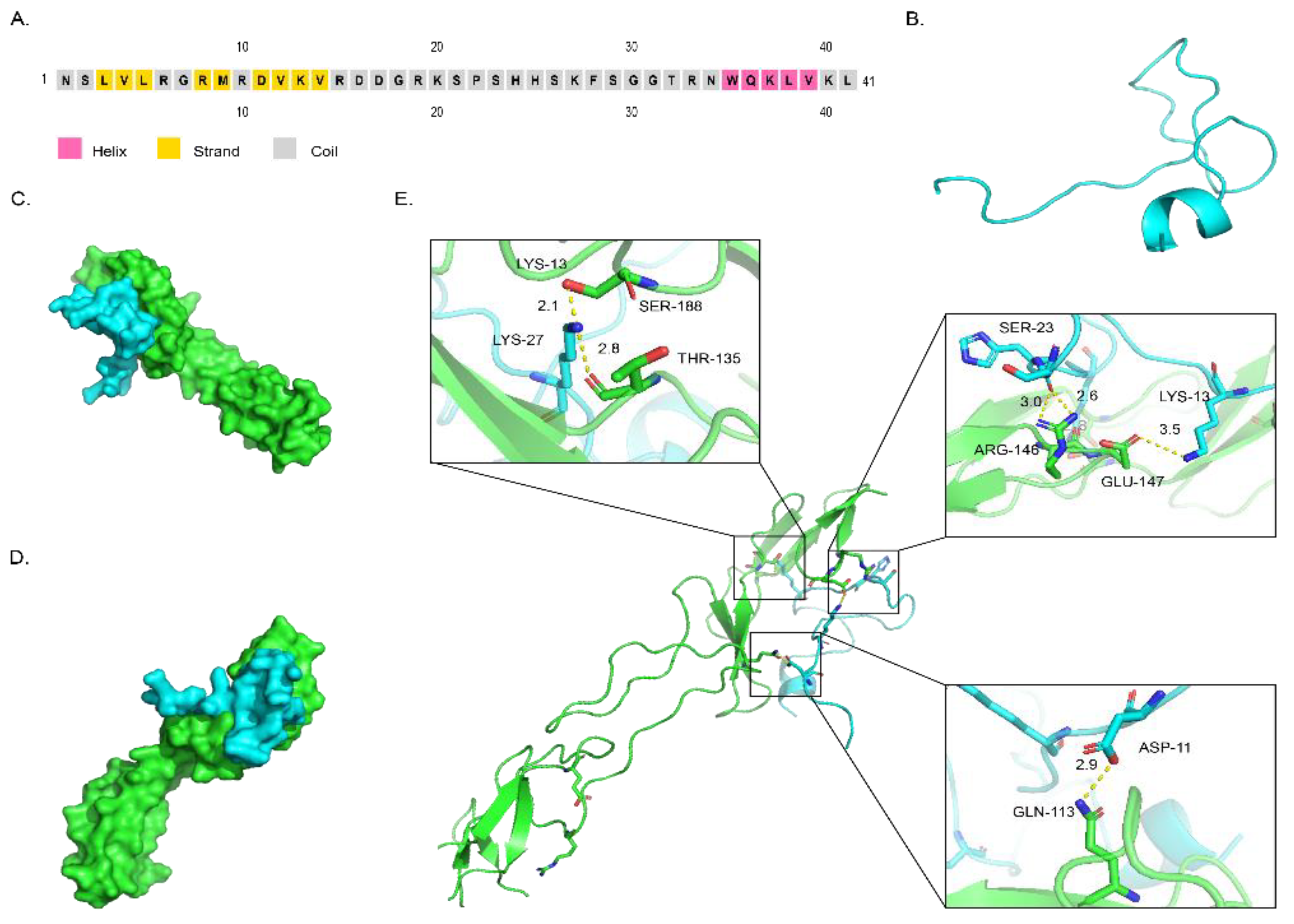
2.3. Structure of DAvp-1 and Docking Model
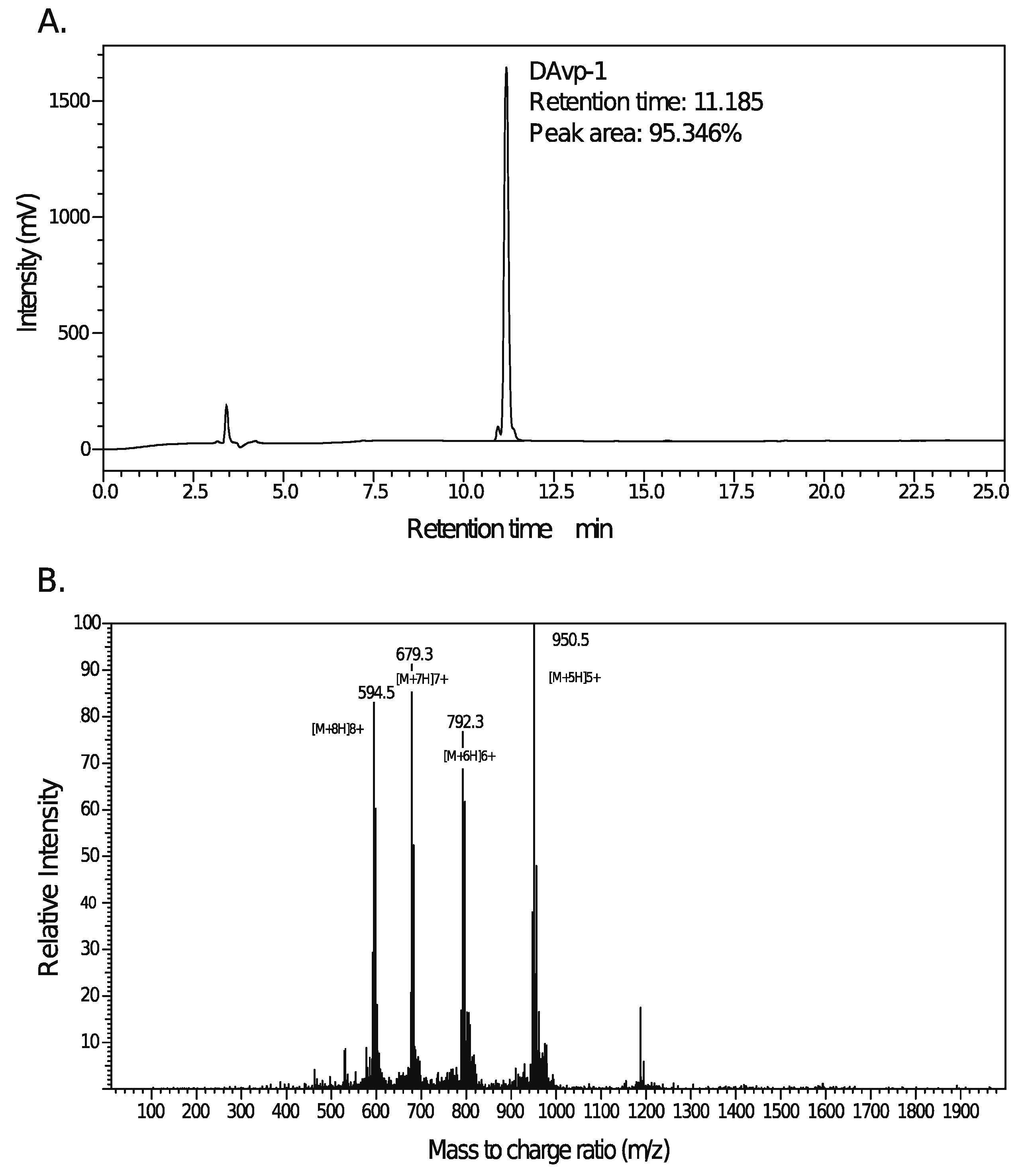
2.4. Peptide Synthesis of DAvp-1
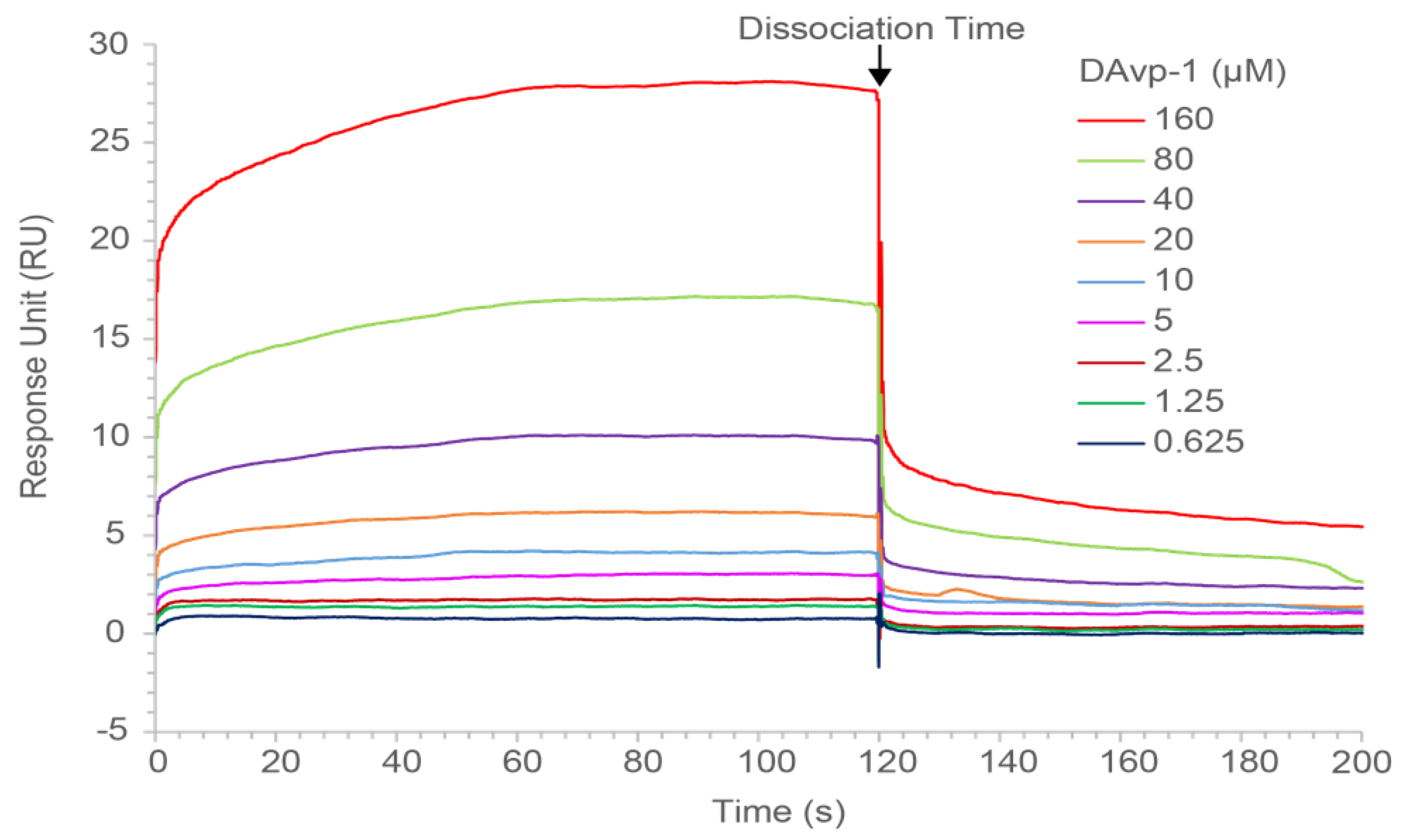
2.5. Surface Plasmon Resonance Measurements
3. Discussion
4. Materials and Methods
4.1. Tissue Sampling and Construction of the cDNA Library
4.2. Construction of the T7 Phage Display Library
4.3. Biopanning
4.4. Structure Modeling and Molecular Docking Study
4.5. Synthetic Peptide
4.6. Characterization of DAvp-1 Binding Properties by SPR
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Tu, A.T. Neurotoxins of animal venoms-snakes. Annu. Rev. Biochem. 1973, 42, 235–258. [Google Scholar] [CrossRef] [PubMed]
- Song, D.G.; Zong, X.; Zhang, H.W.; Wang, T.H.; Yi, H.B.; Luan, C.; Wang, Y.Z. Antimicrobial peptide Cathelicidin-BF prevents intestinal barrier dysfunction in a mouse model of endotoxemia. Int. Immunopharmacol. 2015, 25, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Yu, C.; Zhang, H.; Song, D.; Jiang, D.; Du, H.; Wang, Y. Cathelicidin-BF suppresses intestinal inflammation by inhibiting the nuclear factor-kappa B signaling pathway and enhancing the phagocytosis of immune cells via STAT-1 in weanling piglets. Int. Immunopharmacol. 2015, 28, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Gao, J.; Zhang, S.; Wu, S.; Xie, Z.; Ling, G.; Kuang, Y.-Q.; Yang, Y.; Yu, H.; Wang, Y. Identification and Characterization of the First Cathelicidin from Sea Snakes with Potent Antimicrobial and Anti-inflammatory Activity and Special Mechanism. J. Biol. Chem. 2015, 290, 16633–16652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banner, D.W.; Darcy, A.; Janes, W.; Gentz, R.; Schoenfeld, H.J.; Broger, C.; Loetscher, H.; Lesslauer, W. Crystal-structure of the soluble human 55 kd tnf receptor-human tnf-beta complex—Implications for tnf receptor activation. Cell 1993, 73, 431–445. [Google Scholar] [CrossRef]
- Hamley, I.W. Small Bioactive Peptides for Biomaterials Design and Therapeutics. Chem. Rev. 2017, 117, 14015–14041. [Google Scholar] [CrossRef] [Green Version]
- Morimoto, B.H. Therapeutic peptides for CNS indications: Progress and challenges. Bioorganic Med. Chem. 2018, 26, 2859–2862. [Google Scholar] [CrossRef]
- Apostolopoulos, V.; Bojarska, J.; Chai, T.T.; Elnagdy, S.; Kaczmarek, K.; Matsoukas, J.; New, R.; Parang, K.; Lopez, O.P.; Parhiz, H.; et al. A Global Review on Short Peptides: Frontiers and Perspectives. Molecules 2021, 26, 45. [Google Scholar] [CrossRef]
- Liu, P.; Han, L.; Wang, F.; Petrenko, V.A.; Liu, A.H. Gold nanoprobe functionalized with specific fusion protein selection from phage display and its application in rapid, selective and sensitive colorimetric biosensing of Staphylococcus aureus. Biosens. Bioelectron. 2016, 82, 195–203. [Google Scholar] [CrossRef]
- Smith, G.P.; Petrenko, V.A. Phage display. Chem. Rev. 1997, 97, 391–410. [Google Scholar] [CrossRef]
- Danner, S.; Belasco, J.G. T7 phage display: A novel genetic selection system for cloning RNA-binding proteins from cDNA libraries. Proc. Natl. Acad. Sci. USA 2001, 98, 12954–12959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koivunen, E.; Arap, W.; Rajotte, D.; Lahdenranta, J.; Pasqualini, R. Identification of receptor ligands with phage display peptide libraries. J. Nucl. Med. 1999, 40, 883–888. [Google Scholar]
- Sclavons, C.; Burtea, C.; Boutry, S.; Laurent, S.; Vander Elst, L.; Muller, R.N. Phage Display Screening for Tumor Necrosis Factor-alpha-Binding Peptides: Detection of Inflammation in a Mouse Model of Hepatitis. Int. J. Pept. 2013, 2013, 348409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chirinos-Rojas, C.L.; Steward, M.W.; Partidos, C.D. A peptidomimetic antagonist of TNF-alpha-mediated cytotoxicity identified from a phage-displayed random peptide library. J. Immunol. 1998, 161, 5621–5626. [Google Scholar] [PubMed]
- Zheng, Z.J.; Jiang, H.L.; Huang, Y.; Wang, J.; Qiu, L.; Hu, Z.L.; Ma, X.Y.; Lu, Y.M. Screening of an anti-inflammatory peptide from Hydrophis cyanocinctus and analysis of its activities and mechanism in DSS-induced acute colitis. Sci. Rep. 2016, 6, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.Y.; Huang, Y.; Li, A.; Jiang, H.L.; Wang, J.; Li, J.Z.; Qiu, L.; Li, K.; Lu, Y.M. Hydrostatin-TL1, an Anti-Inflammatory Active Peptide from the Venom Gland of Hydrophis cyanocinctus in the South China Sea. Int. J. Mol. Sci. 2016, 17, 15. [Google Scholar] [CrossRef] [Green Version]
- Kollias, G.; Douni, E.; Kassiotis, G.; Kontoyiannis, D. On the role of tumor necrosis factor and receptors in models of multiorgan failure, rheumatoid arthritis, multiple sclerosis and inflammatory bowel disease. Immunol. Rev. 1999, 169, 175–194. [Google Scholar] [CrossRef]
- Kaminska, B. MAPK signalling pathways as molecular targets for anti-inflammatory therapy—From molecular mechanisms to therapeutic benefits. BBA-Proteins Proteom. 2005, 1754, 253–262. [Google Scholar] [CrossRef]
- Hendrayani, S.F.; Al-Harbi, B.; Al-Ansari, M.M.; Silva, G.; Aboussekhra, A. The inflammatory/cancer-related IL-6/STAT3/NF-kappa B positive feedback loop includes AUF1 and maintains the active state of breast myofibroblasts. Oncotarget 2016, 7, 41974–41985. [Google Scholar] [CrossRef] [Green Version]
- Kyriakis, J.M.; Avruch, J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 2001, 81, 807–869. [Google Scholar] [CrossRef] [Green Version]
- Henriquez-Olguin, C.; Altamirano, F.; Valladares, D.; Lopez, J.R.; Allen, P.D.; Jaimovich, E. Altered ROS production, NF-kappa B activation and interleukin-6 gene expression induced by electrical stimulation in dystrophic mdx skeletal muscle cells. Biochim. Biophys. Acta-Mol. Basis Dis. 2015, 1852, 1410–1419. [Google Scholar] [CrossRef] [Green Version]
- Annibaldi, A.; Meier, P. Checkpoints in TNF-Induced Cell Death: Implications in Inflammation and Cancer. Trends Mol. Med. 2018, 24, 49–65. [Google Scholar] [CrossRef] [PubMed]
- Jarosz-Griffiths, H.H.; Holbrook, J.; Lara-Reyna, S.; McDermott, M.F. TNF receptor signalling in autoinflammatory diseases. Int. Immunol. 2019, 31, 639–648. [Google Scholar] [CrossRef]
- Stone, M.B.; Shelby, S.A.; Veatch, S.L. Super-Resolution Microscopy: Shedding Light on the Cellular Plasma Membrane. Chem. Rev. 2017, 117, 7457–7477. [Google Scholar] [CrossRef]
- Netea, M.G.; Balkwill, F.; Chonchol, M.; Cominelli, F.; Donath, M.Y.; Giamarellos-Bourboulis, E.J.; Golenbock, D.; Gresnigt, M.S.; Heneka, M.T.; Hoffman, H.M.; et al. A guiding map for inflammation. Nat. Immunol. 2017, 18, 826–831. [Google Scholar] [CrossRef] [Green Version]
- Weinelt, N.; Karathanasis, C.; Smith, S.; Medler, J.; Malkusch, S.; Fulda, S.; Wajant, H.; Heilemann, M.; van Wijk, S.J.L. Quantitative single-molecule imaging of TNFR1 reveals zafirlukast as antagonist of TNFR1 clustering and TNF alpha-induced NF-& x138;B signaling. J. Leukoc. Biol. 2021, 109, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Efimov, G.A.; Kruglov, A.A.; Tillib, S.V.; Kuprash, D.V.; Nedospasov, S.A. Tumor Necrosis Factor and the consequences of its ablation in vivo. Mol. Immunol. 2009, 47, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Antoni, C.; Braun, J. Side effects of anti-TNF therapy: Current knowledge. Clin. Exp. Rheumatol. 2002, 20, S152–S157. [Google Scholar]
- Medler, J.; Wajant, H. Tumor necrosis factor receptor-2 (TNFR2): An overview of an emerging drug target. Expert Opin. Ther. Targets 2019, 23, 295–307. [Google Scholar] [CrossRef]
- Medler, J.; Nelke, J.; Weisenberger, D.; Steinfatt, T.; Rothaug, M.; Berr, S.; Hunig, T.; Beilhack, A.; Wajant, H. TNFRSF receptor-specific antibody fusion proteins with targeting controlled Fc gamma R-independent agonistic activity. Cell Death Dis. 2019, 10, 224. [Google Scholar] [CrossRef]
- Lo, C.H.; Schaaf, T.M.; Grant, B.D.; Lim, C.K.W.; Bawaskar, P.; Aldrich, C.C.; Thomas, D.D.; Sachs, J.N. Noncompetitive inhibitors of TNFR1 probe conformational activation states. Sci. Signal. 2019, 12, 1620–1634. [Google Scholar] [CrossRef]
- Lo, C.H.; Vunnam, N.; Lewis, A.K.; Chiu, T.L.; Brummel, B.E.; Schaaf, T.M.; Grant, B.D.; Bawaskar, P.; Thomas, D.D.; Sachs, J.N. An Innovative High-Throughput Screening Approach for Discovery of Small Molecules That Inhibit TNF Receptors. Slas Discov. 2017, 22, 950–961. [Google Scholar] [CrossRef] [Green Version]
- Paul, A.T.; Gohil, V.M.; Bhutani, K.K. Modulating TNF-alpha signaling with natural products. Drug Discov. Today 2006, 11, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Bindu, S.; Mazumder, S.; Bandyopadhyay, U. Non-steroidal anti-inflammatory drugs (NSAIDs) and organ damage: A current perspective. Biochem. Pharmacol. 2020, 180, 21. [Google Scholar] [CrossRef] [PubMed]
- Kay, B.K.; Castagnoli, L. Mapping protein-protein interactions with phage-displayed combinatorial peptide libraries. Curr. Protoc. Cell Biol. 2003, 17, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Ooms, F. Molecular modeling and computer aided drug design. Examples of their applications in medicinal chemistry. Curr. Med. Chem. 2000, 7, 141–158. [Google Scholar] [CrossRef]
- Barradas-Bautista, D.; Rosell, M.; Pallara, C.; Fernandez-Recio, J. Structural prediction of protein-protein interactions by docking: Application to biomedical problems. In Protein-Protein Interactions in Human Disease, Pt A; Donev, R., Ed.; Advances in Protein Chemistry and Structural Biology; Elsevier: Amsterdam, The Netherlands, 2018; Volume 110, pp. 203–249. [Google Scholar]
- Ahmad, N.M. Solubility-driven lead optimisation: Recent examples and personal perspectives. Bioorg. Med. Chem. Lett. 2016, 26, 2975–2979. [Google Scholar] [CrossRef]
- Green, R.J.; Frazier, R.A.; Shakesheff, K.M.; Davies, M.C.; Roberts, C.J.; Tendler, S.J.B. Surface plasmon resonance analysis of dynamic biological interactions with biomaterials. Biomaterials 2000, 21, 1823–1835. [Google Scholar] [CrossRef]
- Ellert-Miklaszewska, A.; Poleszak, K.; Kaminska, B. Short peptides interfering with signaling pathways as new therapeutic tools for cancer treatment. Future Med. Chem. 2017, 9, 199–221. [Google Scholar] [CrossRef]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editorand analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, P.; Jin, B.W.; Li, H.; Huang, S.Y. HPEPDOCK: A web server for blind peptide-protein docking based on a hierarchical algorithm. Nucleic Acids Res. 2018, 46, W443–W450. [Google Scholar] [CrossRef] [PubMed]
- Delano, W.L. The PyMOL Molecular Graphics System. Available online: http://www.pymol.org (accessed on 20 October 2021).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, K.; Liu, Y.; Tang, Y. Screening of TNFR1 Binding Peptides from Deinagkistrodon acutus Venom through Phage Display. Toxins 2022, 14, 155. https://doi.org/10.3390/toxins14020155
Zhang K, Liu Y, Tang Y. Screening of TNFR1 Binding Peptides from Deinagkistrodon acutus Venom through Phage Display. Toxins. 2022; 14(2):155. https://doi.org/10.3390/toxins14020155
Chicago/Turabian StyleZhang, Kangran, Yang Liu, and Yezhong Tang. 2022. "Screening of TNFR1 Binding Peptides from Deinagkistrodon acutus Venom through Phage Display" Toxins 14, no. 2: 155. https://doi.org/10.3390/toxins14020155
APA StyleZhang, K., Liu, Y., & Tang, Y. (2022). Screening of TNFR1 Binding Peptides from Deinagkistrodon acutus Venom through Phage Display. Toxins, 14(2), 155. https://doi.org/10.3390/toxins14020155




