Abstract
Toxin–antitoxin systems (TASs) are widely distributed in prokaryotes and encode pairs of genes involved in many bacterial biological processes and mechanisms, including pathogenesis. The TASs have not been extensively studied in Listeria monocytogenes (Lm), a pathogenic bacterium of the Firmicutes phylum causing infections in animals and humans. Using our recently published TASmania database, we focused on the known and new putative TASs in 352 Listeria monocytogenes genomes and identified the putative core gene TASs (cgTASs) with the Pasteur BIGSdb-Lm database and, by complementarity, the putative accessory gene TAS (acTASs). We combined the cgTASs with those of an additional 227 L. monocytogenes isolates from our previous studies containing metadata information. We discovered that the differences in 14 cgTAS alleles are sufficient to separate the four main lineages of Listeria monocytogenes. Analyzing these differences in more details, we uncovered potentially co-evolving residues in some pairs of proteins in cgTASs, probably essential for protein–protein interactions within the TAS complex.
Key Contribution:
Systematic annotation of TASs in a large number of L. monocytogenes assemblies.
1. Introduction
Toxin–antitoxin systems (TASs) were discovered because of their involvement in a biological process called post-segregational killing (PSK), a plasmid maintenance mechanism based on two plasmid-encoded genes: a toxin gene (T) and its antagonistic antitoxin (A) [1,2,3]. In this context, the toxin and antitoxin are equally distributed in the two daughter cells; however, the instability of the antitoxin will lead to an active toxin killing the cell lacking the plasmid because the antitoxin cannot be replaced. This system also could be important under some stress (e.g., antibiotic in the medium), where the toxin is released from its less stable antitoxin partner, leading to a transient metabolic shutdown and growth arrest or cell death, similar to apoptosis in higher organisms. Of particular interest, TASs have been associated with pathogenic bacterial intracellular infection and with quorum sensing [4]. In addition, pandemic bacterial strains have been shown to carry more TASs compared to non-epidemic related species [5].
TAS toxicity relies on various molecular mechanisms targeting diverse biological processes, e.g., cell membrane integrity, assembly of the translational machinery, tRNA and mRNA stability, translation initiation and elongation steps, DNA replication, and ATP synthesis (see [6,7] for reviews). For example, the toxins MazF or PemK target and cut both free mRNA and mRNA bound to the translational machinery, while the toxin ParE targets DNA replication by inhibiting the DNA gyrase [6]. In terms of gene structure, TASs are usually found in operons with either AT or TA orientations. Those TAS operons can be acquired from mobile genetic elements such as plasmids or phages, and are also present in bacterial chromosomes [8], leading to complex heterogeneous genomic landscapes of TASs with both horizontal and vertical transmissions.
Listeria monocytogenes (Lm) is a Gram-positive bacterium and opportunistic food-borne pathogen [9,10,11]. It is the etiological agent of listeriosis in humans and animals causing abortion, septicemia, gastroenteritis, and central nervous system (CNS) infections [12,13]. L. monocytogenes strains are grouped into four distinct phylogenetic lineages called I, II, III, and IV [14,15,16]. Strains belonging to lineages I and II are the most abundant isolates worldwide and particularly Lineage I is frequent in disease [16,17], while Lineage III and IV strains are very rare and predominantly isolated from (asymptomatic) animals [18]. In silico predictions using TADB2 reveal only two TASs in L. monocytogenes EGD-e: lmo0113-0114 and lmo0887-0888 [19]. There are a few studies investigating TAS systems in L. monocytogenes [20,21,22] focusing only on a few L. monocytogenes strains and a few TAS pairs. The following TAS pairs were identified mainly using in silico methods in the strain ATCC19117 (with their corresponding gene names in L. monocytogenes EGD-e): lmo0113-0114, lmo0887-0888, and lmo1301-1302, predicting their 3D structure and potential inhibitory peptides. This report also showed by qPCR that lmo0113 is upregulated upon heat stress [21,22]. Another report investigated the TAS lmo0887-888 of L. monocytogenes EGD-e in more detail, ruling out the classical MazF action, but without demonstrating the exact role of this TAS in L. monocytogenes [20]. They showed that lmo0887-888 does not affect the level of persister formation upon antibiotic treatment, but the expression of σB-dependent genes opuCA and lmo0880 under sub-inhibitory norfloxacin treatment [20].
In this article, we first identified putative core gene TASs in a larger set of 579 L. monocytogenes genomes and discovered how some of these gene pairs have potentially co-evolved. In a second step using putative accessory gene TASs, we correlated our phenotypic metadata to the presence/absence of TAS genes, revealing the potential impact of specific TASs in the pathogenicity of L. monocytogenes strains.
2. Results
2.1. Identification of Core Gene TASs
The TASmania database [23] was queried to identify the putative TASs in the 352 Lm genomes that are currently in the database (list of L. monocytogenes genomes in TASmania, Table S1, list of TASs, Table S2). These 352 genomes were typed using the scheme cgMLST1748 at the Pasteur BIGSdb-Lm web site [24] to obtain the core gene alleles for all strains. By comparing with the TASmania hits we identified n = 14 core gene TASs (cgTASs) (Table 1) and their respective alleles (list of alleles in the 14 cgTASs, Table S3). The current knowledge on TASs in L. monocytogenes is rather scarce and our list included the two cgTASs already identified by TADB2 (lmo0113-0114 and lmo0887-0888). TASmania extended the number of TAS candidates by one order of magnitude in all L. monocytogenes strains, but only a subset of them consists of core genes according to cgMLST1748.

Table 1.
List of 14 putative cgTASs identified in TASmania L.monocytogenes. Gene locus names are taken from the reference EGD-e annotation (NCBI accession number NC_003210.1). The term “Guilt-by-association” refers to potential T or A proteins identified only by their neighborhood to a hit in TASmania [23]. The alternative shading is used to group the pseudo-operon TAS loci.
Out of these 14 cgTASs, two cgTASs appear as orphans (lmo0168 and lmo0887). Their genetic environment was studied in the annotations of the EGD-e strain [25]. In the case of lmo0168, no possible partner was found, as the gene is surrounded by genes located on the other strand, confirming a probable orphan antitoxin. The potential partner of lmo0887 is lmo0888, which likely is a toxin mRNA interferase containing a pemK-like domain and a plasmid_toxin domain (according to TASmania), is a mazEF (according to TADB2); however, it is not identified as a core gene according to the cgMLST1748 scheme and thus must be ignored in our co-evolution analysis below.
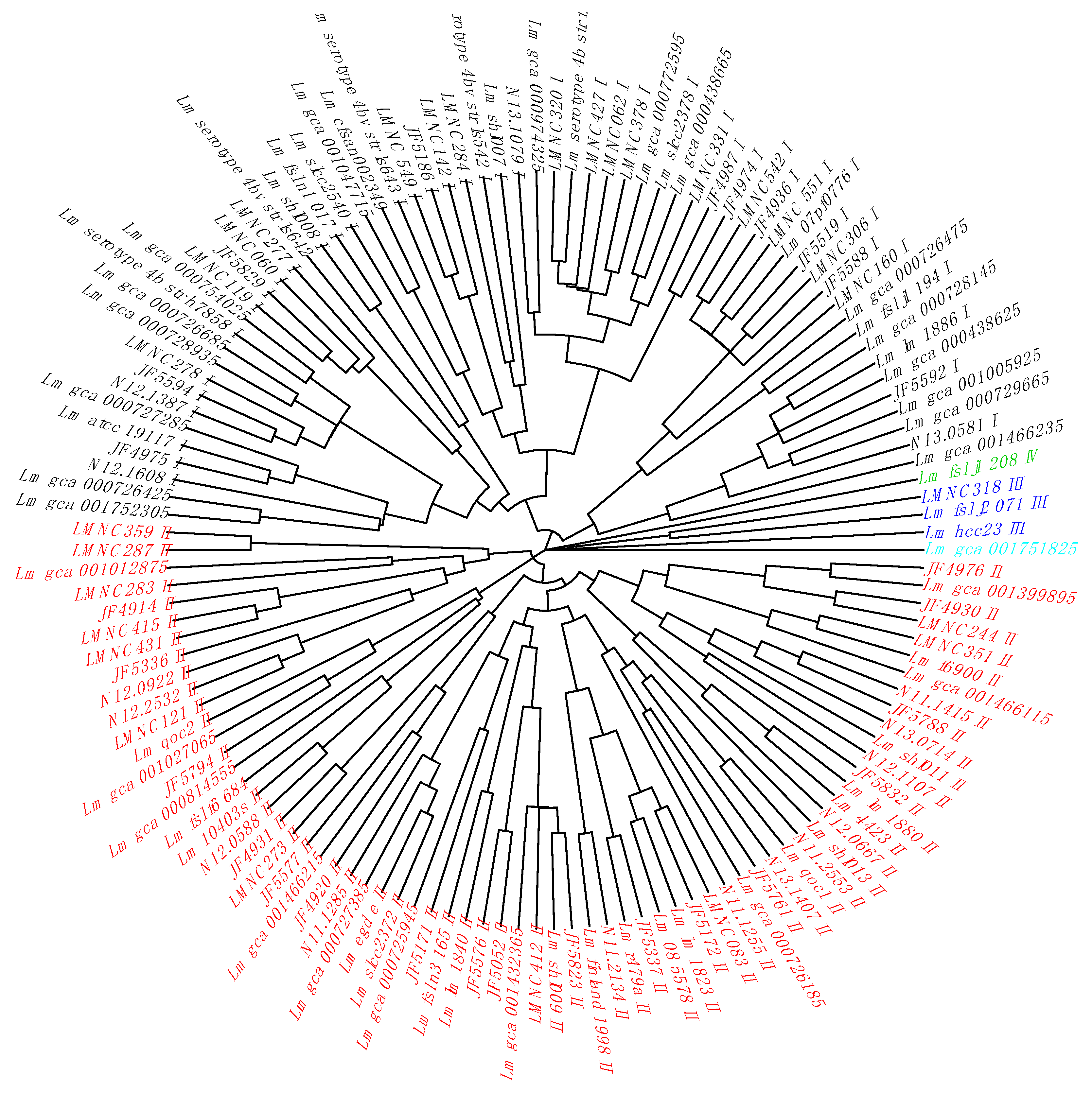
In a second step, we added our own collection of L. monocytogenes strains [26] (n = 227) with their cgMLST alleles (Table S4) and metadata annotation (Table S5). We extracted their non-redundant gene alleles corresponding to the previously identified 14 cgTASs (Table S6). By clustering the non-redundant cgTAS alleles patterns with nominal hierarchical clustering, we obtained the dendrogram (Figure 1) showing that the 14 cgTASs are sufficient to separate the L. monocytogenes lineages (I, II, III, and IV). These results are in agreement with previous MLST analysis using seven housekeeping genes [14]. This clustering allows for characterizing the lineage membership of strains that did not have metadata information available (e.g., Lm_gca_000729665 as Lineage I and Lm_gca_001466115 as Lineage II). However, one cannot infer any causal role for the cgTASs in the lineage separation, as clustering the alleles of some subsets of core genes among the 1748 core genes would potentially show the same lineage split.
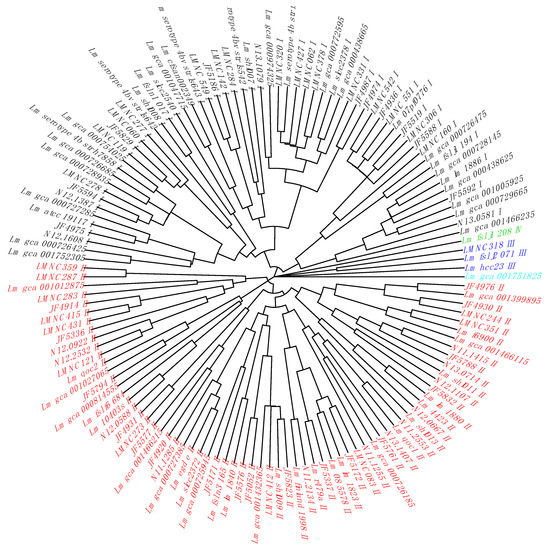
Figure 1.
Dendrogram of the non-redundant L. monocytogenes strains based on the cgTAS alleles. The lineage coloration is based on the known lineages thanks to the metadata. Those without metadata are tentatively colored according to the branch. Black = Lineage I; red = Lineage II; blue = Lineage III; green = Lineage IV; light blue = Listeria innocua as outgroup (wrongly annotated L. monocytogenes in ENA).
2.2. Co-Evolution Analysis
We analyzed the six complete cgTAS pairs for co-evolutionary residues and excluded the orphan cgTASs (lmo0168 and lmo0887). We grouped the sequences by gene, translated them to proteins and fused the toxin (T) alleles with the corresponding antitoxin (A) alleles per genome in a multiple sequence alignment (MSA) using our own Perl script and MAFFT [27]. With this MSA, the BIS2Analyzer server [28] was able to identify potential co-evolutionary residues, i.e. a mutated amino acid in a toxin associated to another mutated residue in the cognate antitoxin of the same TAS (e.g., red arrows in Figure 2 and Table 2). Only two cgTAS pairs (lmo1466-1467 and lmo1309-1310) revealed co-evolving residues between the two partners. For instance, the residue at location 435 (within the first partner lmo1466) co-evolves with the residue at location 828 (within the second partner lmo1467) (Figure 2a and Table 2). More than one residue per partner can be co-evolving as shown with the pair lmo1309-1310 (Figure 2b and Table 2). One cgTAS pair (lmo2793-2794) had co-evolving residues visible on the MSA using Jalview (Figure 2c red arrows), but it failed to reach a significant p-value (<0.05) in BIS2Analyzer.

Figure 2.
The MSA of the fusion pseudo-protein (first partner in orange, second partner in green) with the co-evolving amino acid residues highlighted (inter-protein = red arrows; intra-protein = blue arrows) within Jalview [29]. In that view, all common residues letters are hidden, only varying residues are displayed with their letters. The numbering at the top corresponds to the amino acid positions along the artificially fused protein sequence (same numbers and colors as Table 2). For clarity, only the red arrows (inter-protein) are labeled with residues highlighted in Table 2.

Table 2.
List of residues that are inter-protein co-evolving for each pair of cgTASs according to BIS2Analyzer. Positions numbered for the fusion pseudo-protein; orange = residues in the first partner; green = residues in the second partner. See Materials and Methods for more details. (Intra-protein co-evolving residues are shown in Table A1). Add fusion protein info.
2.3. Accessory Gene TAS Analysis
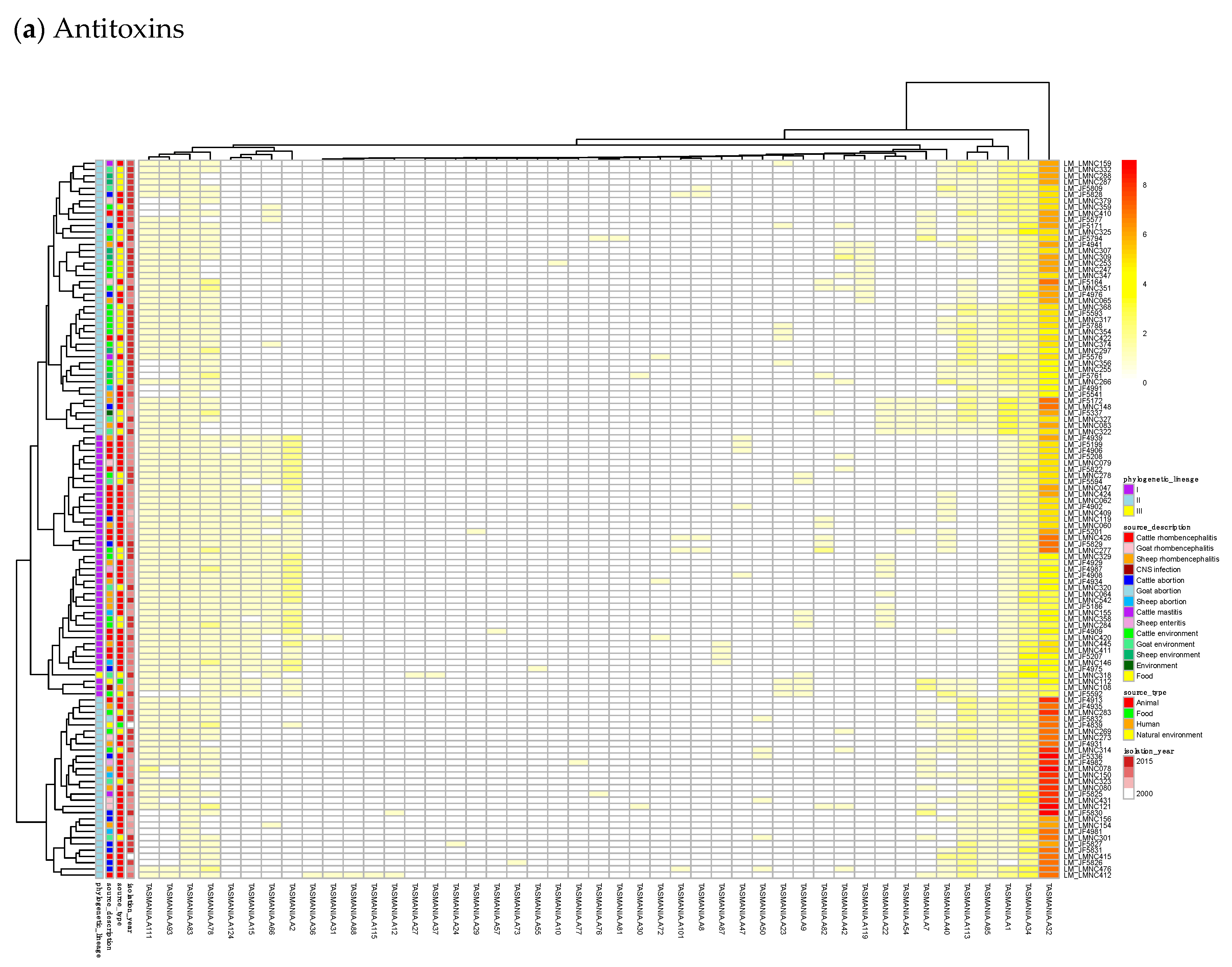
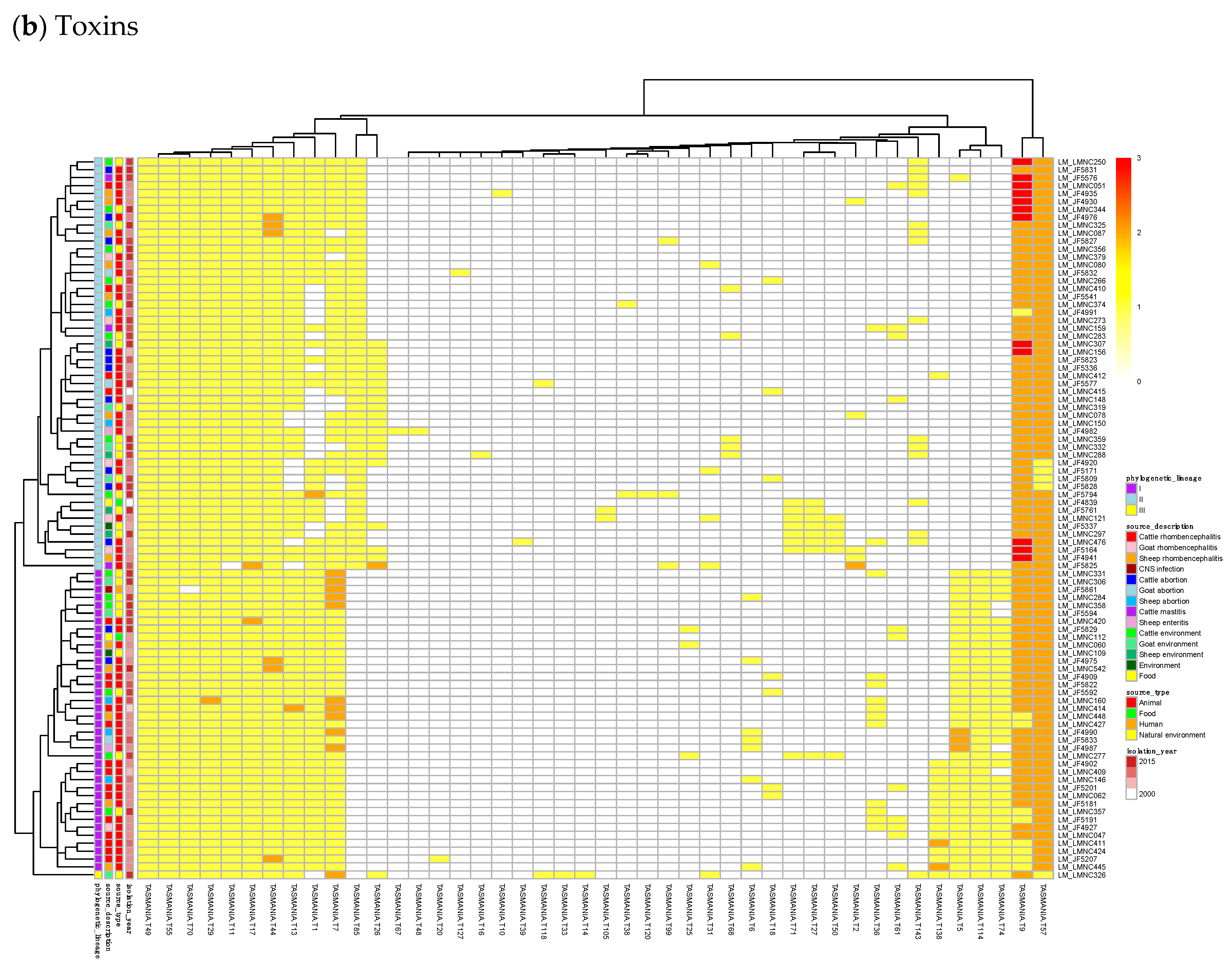
By subtracting the cgTAS hits from the L. monocytogenes strains in TASmania, all other TASs are classified as accessory TASs (acTASs) (Table S7, Figure A1). In order to obtain a better understanding of the role of accessory TASs, we performed a gene analysis with a set of 227 L. monocytogenes strains [26] for which we have the corresponding metadata (Table S5). These isolates are not part of the TASmania database, so we processed them by first predicting protein genes with Prodigal [30] and annotating them with the TASmania HMMs [23]. Finally, we built a heatmap of those acTASs (Table S8) for antitoxins (Figure 3a) and toxins (Figure 3b) after removing the core genes and the redundancy as described above.
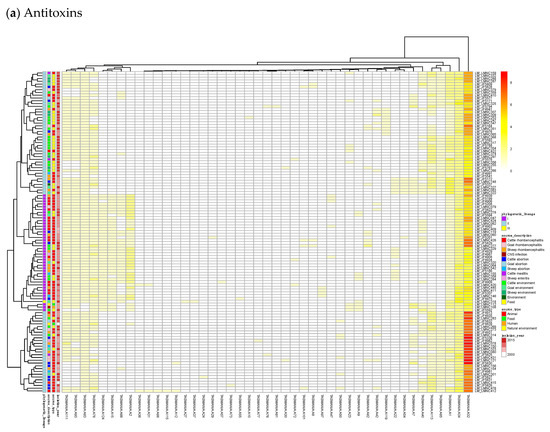
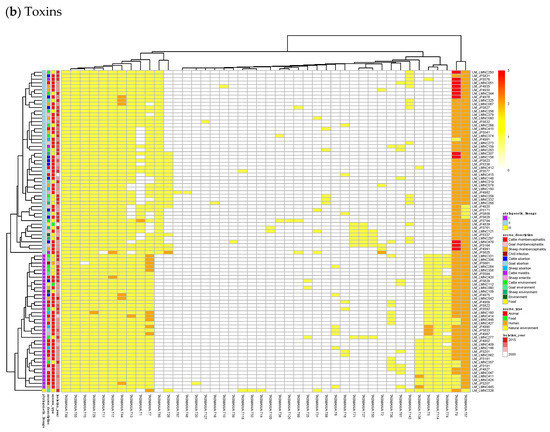
Figure 3.
Heatmaps of the counts of accessory genes for (a) Antitoxins and (b) Toxins in 227 L. monocytogenes isolates. Metadata is given on the left side of the heatmap: lineage classification, source description, source type, and isolation year.
Figure 3a shows on the rightmost column that the antitoxin cluster A32 (HTH_3) is the most abundant and is more frequently found in animal cases of Lineage II. In TASmania, this A32 antitoxin cluster is observed as being paired with at least six different toxin clusters (T2, T13, T14, T38, T48, and T85, whose nearest Pfam identifiers are HipA_C, Zeta_toxin, HipA_C, RelE, Gp49 and Gp49, respectively). All of these pairs were analyzed for their co-occurrence in the L. monocytogenes strains above, but no candidate pair seems to exist in Lineage I, while only A32.T2 (nearest Pfam HTH_3.HipA_C) can be found rarely in Lineage II (Table A2).
In Figure 3b, a small, interesting group of isolates carry one or two genes having a hit to the T138 (nearest Pfam AbiEii) toxin cluster. It is mainly found in Lineage I isolates causing rhombencephalitis, 12 out of 15 (9 in cattle, 2 in goat, and 1 in sheep). The three remaining isolates are not rhombencephalitis, but could be related by their proximity to animals: 1 sheep abortion and 2 environmental isolates. In addition, one case carrying T138 in Lineage II, a cattle rhombencephalitis, is reported. This renders this toxin quite interesting regarding rhombencephalitis. However, in the L. monocytogenes strains of TASmania, no antitoxin partner is known for this toxin that seems to be often encoded in a prophage region.
For the 352 L. monocytogenes strains described in TASmania unfortunately, no metadata is available, but the list of acTASs and their abundance is similar to our annotated dataset, with A32, A1, A2, and A34 (nearest Pfam HTH_3, ParBc, MraZ, and Omega_Repress/Rep_trans, respectively) being the most prevalent antitoxin clusters, and T9, T1, T57, T7, and T44 (nearest Pfam ParE, PhoH, PemK_toxin, Gp49, and HicA_toxin, respectively) being the main toxin clusters, as shown in Figure A1.
Using TASmania, a closer look at the Pfam annotation of the toxin clusters highlights the diversity of the TA systems uncovered in L. monocytogenes. Indeed, ParE targets the DNA gyrase and, along with Gp49, belongs to the Pfam clan called “plasmid_antitox CL0136”, whose members are originally described as plasmid-encoded TASs involved in plasmid maintenance. The PhOH domain is found in cytoplasmic proteins predicted as ATPase and which are induced by phosphate starvation [31]. PemK toxins belong to the Pfam superfamily of CcdB/PemK (CL0624) known as growth inhibitors that can bind to their own promoter and act also as endonucleases. HicA is an mRNA interferase that binds to target mRNA potentially in a translation-independent manner. The Firmicutes phylum has a prevalence of ParE and PemK like toxins in chromosomal TASs, which correspond to the prevalence in L. monocytogenes (Figure A2). More experimental investigation is required to confirm these putative TASs and to understand the conditions that regulate their expression in L. monocytogenes of various pathogenicity.
3. Discussion
Toxins and antitoxins have many roles in the bacterial cells by targeting a broad range of biological processes. Their role in pathogenic bacteria involves plasmid and pathogenicity island maintenance or biofilm formation [4]. We previously stated [23] that the TASs identified in the TASmania database are putative TASs predicted purely in silico by computational means and would benefit from in vivo validation. This is also the case in this report; all TASs that we identified in L. monocytogenes are candidates to be confirmed experimentally.
We obtained those candidates by combining large-scale database searches leveraging on the “guilt-by-association” neighborhood criteria (“guilt-by-association” refers to potential T or A proteins identified only by their neighborhood to a hit in TASmania) with the available metadata.
By looking at L. monocytogenes core genes, we identified 14 putative cgTASs that were analyzed for potential co-evolving residues. In addition, clustering their alleles confirmed the L. monocytogenes lineage separation. However, this lineage split is not necessarily due to the presence of these cgTASs. One of the cgTASs (lmo0168) appears as a putative orphan antitoxin (nearest Pfam “MazE_antitoxin”). Whether this putative antitoxin is expressed and functional remains unclear. If expressed, one could speculate about an interaction in trans with the toxin of another TAS.
Since we unraveled the possibility of co-evolving residues, further work is needed to demonstrate these hypothetical protein–protein contacts between those residues. Interestingly, co-evolution was reported for a Type III toxin between amino acid residues of the toxin CptIN and nucleic acids of its cognate antitoxin ncRNA [32]. An intriguing gene is lmo0888, which is recognized as part of a TAS together with lmo0887, but it is not defined as a core gene contrary to its partner lmo0887. It is not clear to us why this happens, as, intuitively, the whole pair should be part of the core genes. One possible explanation could be that this gene is found in multiple copies in part of the L. monocytogenes genomes and thus was removed from the core genes [24].
Additional knowledge or hypotheses on TASs could be extracted, such as TAS association with diseases. For instance, when looking at the phenotypic link to acTASs, the toxin cluster AbiEii T138 is of particular interest given its frequent association with rhombencephalitis in ruminants. Even though not all strains isolated from rhombencephalitis harbored this toxin, the association of toxin T138 with ruminant rhombencephalitis was significant (p-value = 0.0413, Χ2 test). Looking at the TASs, no obvious partner antitoxin is found for this toxin probably because it is part of a prophage region that is usually less well annotated. Further work remains to be done to demonstrate which pathway or target is activated. The presence of this prophage should be validated for its suitability as a diagnostic tool in the surveillance of animal farms.
By using large-scale in silico analysis of toxin–antitoxin systems in Listeria monocytogenes, we demonstrated that knowledge could be extracted from combined genome sequences and associated metadata. This includes potential co-evolutionary residues, the detection of putative new toxin or antitoxin partners, as well as the suspected role for a specific prophage TAS in rhombencephalitis in ruminants.
4. Materials and Methods
4.1. Data Used
Sequences were obtained from ENSEMBL.Bacteria [33] (352 L. monocytogenes strains chosen because they are included in our TASmania database) and from our previous work PRJEB15123, PRJEB15195 [34,35], and PRJEB22706 [26] (227 L. monocytogenes strains collected by our group with their associated metadata).
4.2. Identification of Putative TAS Genes using the TASmania Database
TAS candidates were extracted from our TASmania database https://bugfri.unibe.ch/tasmania [23] in all 352 L. monocytogenes strains included in the TASmania database with an R script directly querying the database. This list of candidates (Table S2) allows for the discovery of new TAS pairs.
4.3. Identification of Core Gene TASs
In a first step, typing of all the 352 L. monocytogenes strains was performed at the BIGSdb-Lm https://bigsdb.pasteur.fr/listeria/database [24] with a full genome sequence query on the cgMLST1748 scheme, in order to identify the alleles of each of the 1748 core genes. In the second step, the candidate TAS genes of TASmania were crossed with the core genes, leading to a list of 14 core gene TASs (cgTASs) (Table 1). Core genes (cgMLST1748) are 1748 genes that were identified among all L. monocytogenes comparing thousands of L. monocytogenes genomes [24]. Accessory genes are only found in a subset of L. monocytogenes strains.
Another set, including 227 L. monocytogenes field isolates not belonging to TASmania, was added to the study. For these isolates, we knew the cgMLST profiles from a preceding study (Aguilar-Bultet, manuscript in preparation). The cgTAS genes identified above were identified in these 227 genomes by BLASTn and combined with the alleles of the 352 L. monocytogenes strains. From this, a matrix with allele information of all isolates, but containing only the 14 cgTAS loci previously identified from the combination TASmania-BIGSdb, was obtained.
4.4. Removing Redundancy and Clustering
First, the strains containing more than 50% of alleles with no hits in the 14 core gene set were removed. Second, only one genome was kept as representative for all the strains with 100% identical cgTAS allele patterns (in the 14 cgTASs).
The representative cgTAS patterns (Table S6) were clustered according to the cgMSLT allele identifiers using the Nominal Clustering nomclust R package [36]. The Nominal Clustering performs hierarchical cluster analysis (HCA) with objects characterized by nominal (categorical) variables with a distance measured by the Goodall 1 dissimilarity measure. The dendrogram obtained with hclust was then plotted with plot as a fan with labels colored according to the lineages (Figure 1).
4.5. Identification of Accessory Gene TASs
The accessory gene TASs (acTASs) were deduced by subtracting the cgTASs from the list of TASs extracted from TASmania. Redundancy among the 100% identical acTAS pattern was removed by keeping a single representative genome. The heatmaps were calculated with R package pheatmap, allowing for the addition of metadata (Tables S5 and S8).
4.6. Identification of Potential Co-Evolving Residues
The BIS2Analyzer web site [28] was used to identify potential co-evolving residues. The input MSA for each of the 6 cgTAS pairs was built with an in-house Perl script and computed with MAFFT (File F1). The Dimension parameter was set to 2, allowing up to 2 exceptions on a column, and all other parameters were kept as defaults (Table 2). The residue positions and p-values (Fischer test) were evaluated by the CLAG score [37] and the server, respectively.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6651/12/1/29/s1, Table S1: genomes, Table S2: all TAShits, Table S3, 14cg TAS Allele, Table S4: All Sample Allele, Table S5: metadata, Table S6: NonRedundant 14cgTAS TASmania BIGSdb Allele, Table S7: acTAS TASMANIA, Table S8: acTAS_additionalstrains.
Author Contributions
Methodology, software, validation, formal analysis, funding acquisition, writing—review and editing: J.A.A.; resources, writing—review and editing: A.O.; formal analysis, data curation, writing—review and editing: H.A.; formal analysis, data curation, and writing—review and editing: L.A.-B.; formal analysis, supervision, project administration, funding acquisition, and writing—original draft preparation: L.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Swiss National Science Foundation (CRSII3_160703 & CRSII3_147692 http://www.snf.ch), and J.A.A. was funded by the Swiss Excellence Fellowships for Foreign Scholars (ESKAS).
Acknowledgments
We thank Sylvain Brisse, Marc Lecuit, and Alexandra Moura for helping us with the strain typing via their cgMLST1748 platform at Pasteur. We are grateful to Joachim Frey for the initial funding of the Listeria monocytogenes project.
Conflicts of Interest
The authors declare that there is no conflict of interest.
Appendix A
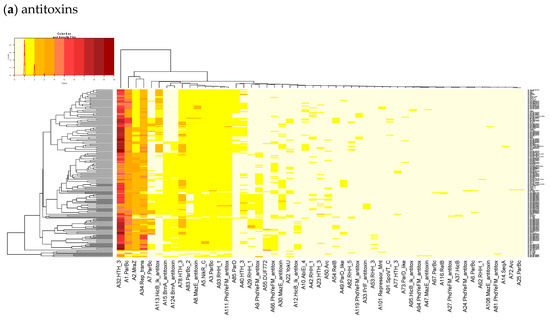
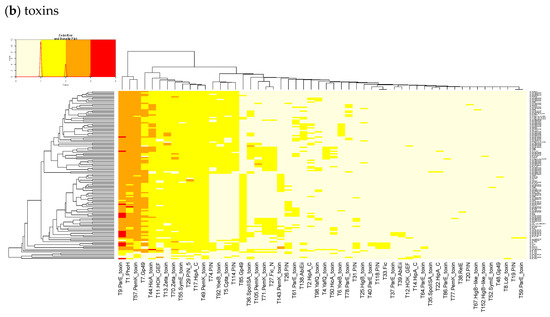
Figure A1.
Heatmaps of the counts of accessory TAS genes (non-redundant for the strains) for (a) Antitoxins and (b) Toxins in 322 L. monocytogenes of TASMANIA. The closest Pfam name is given to the TASMANIA cluster.
Figure A1.
Heatmaps of the counts of accessory TAS genes (non-redundant for the strains) for (a) Antitoxins and (b) Toxins in 322 L. monocytogenes of TASMANIA. The closest Pfam name is given to the TASMANIA cluster.
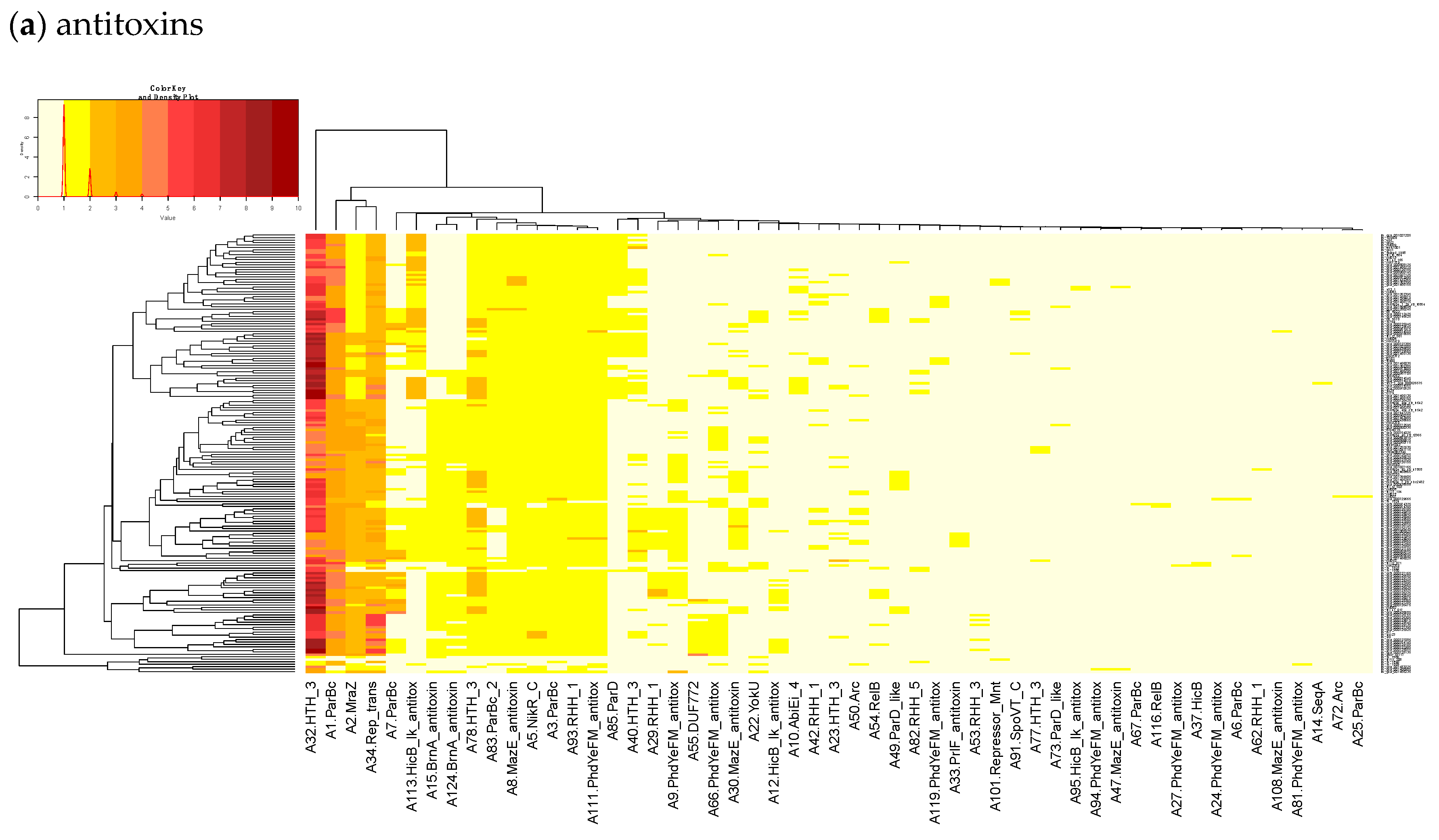
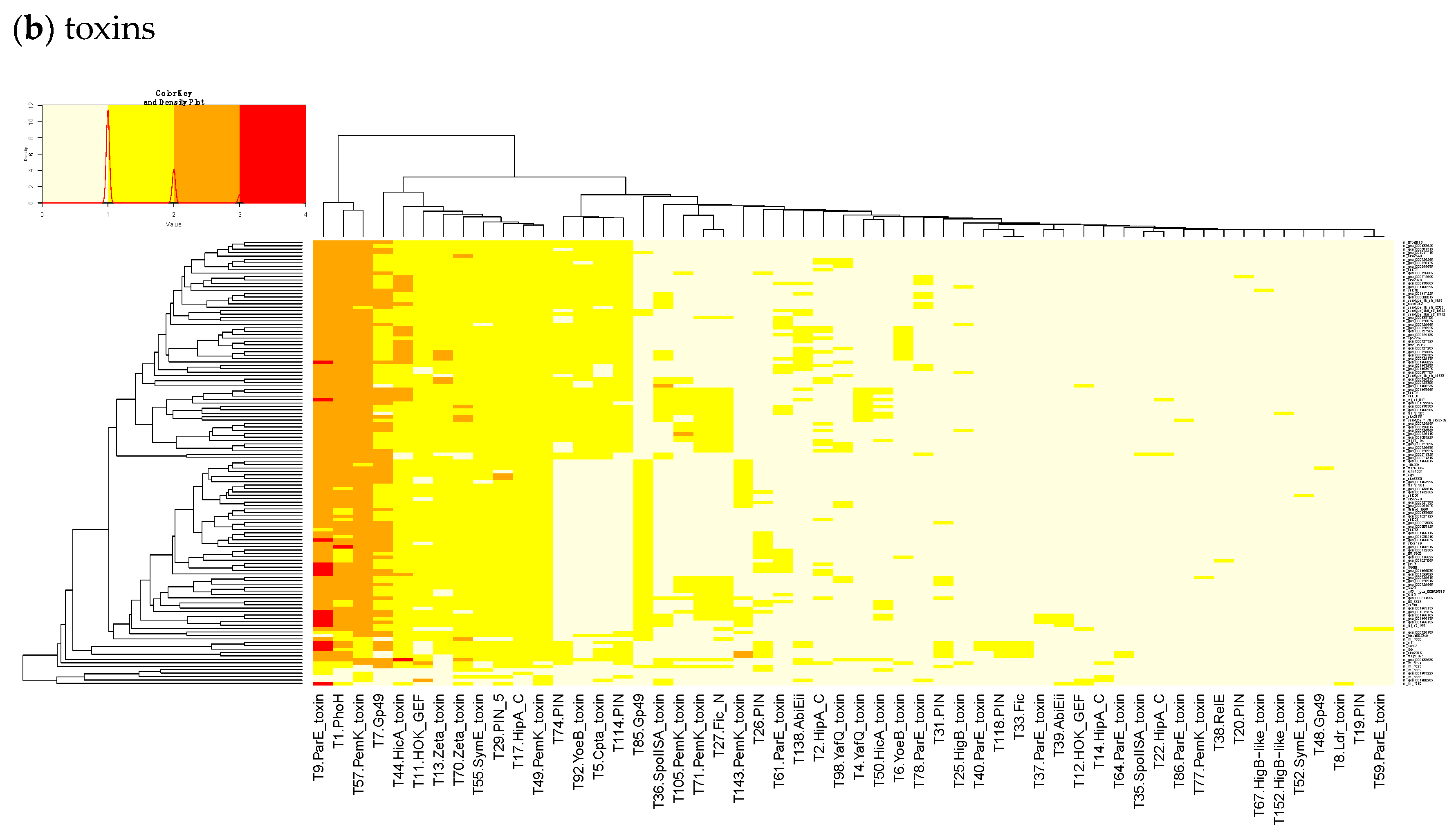
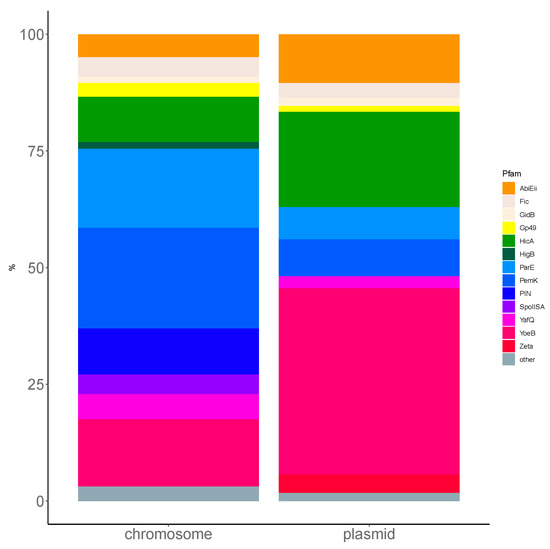
Figure A2.
Comparison of the Firmicutes toxin closest to the Pfam family distribution between plasmid and chromosome. For simplicity, only TASs from the non-ambiguous two-gene AT/TA pseudo-operons were analyzed. The xT/Tx or Ax/xA pairs were ignored. L. monocytogenes strains, as Firmicutes in general, present a high prevalence of PemK and ParE-like chromosomal toxins, while YoeB and HicA toxins are more prevalent in plasmids.
Figure A2.
Comparison of the Firmicutes toxin closest to the Pfam family distribution between plasmid and chromosome. For simplicity, only TASs from the non-ambiguous two-gene AT/TA pseudo-operons were analyzed. The xT/Tx or Ax/xA pairs were ignored. L. monocytogenes strains, as Firmicutes in general, present a high prevalence of PemK and ParE-like chromosomal toxins, while YoeB and HicA toxins are more prevalent in plasmids.
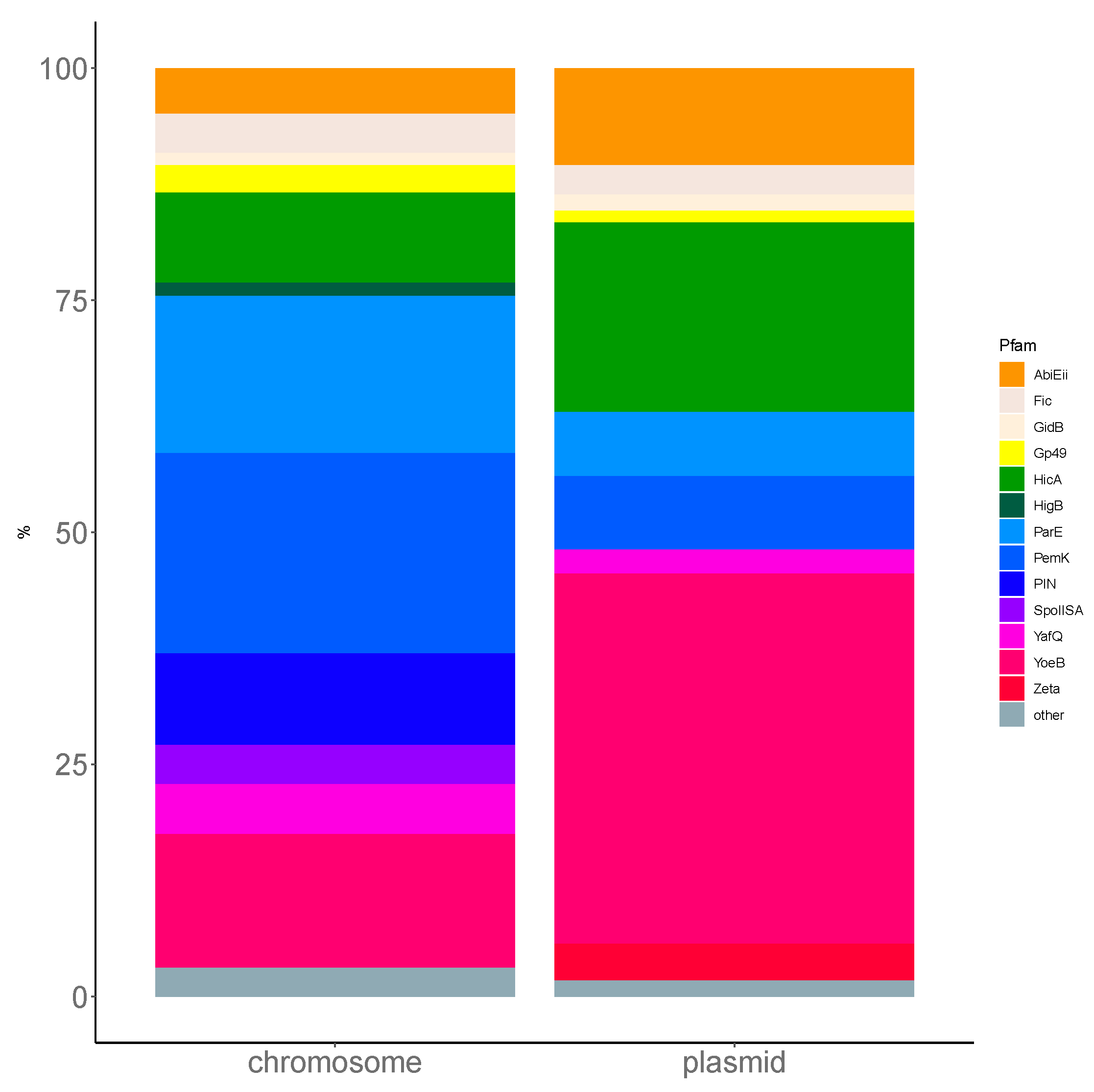

Table A1.
List of residues that are significantly co-evolving intra-protein for each pair of cgTASs.
Table A1.
List of residues that are significantly co-evolving intra-protein for each pair of cgTASs.
| Lmo1466-1467 | 4.024153e-16 |
| intra-protein Lmo1466 | |||||||||||||||||||||
| 4.719742e-07 |
| intra-protein Lmo1466 | ||||||||||||||||||||||
| 4.342162e-06 |
| intra-protein Lmo1466 | ||||||||||||||||||||||
| Lmo1309-1310 | 3.977592e-11 |
| intra-protein Lmo1310 |

Table A2.
List of possible A32 TAS pairs in TASMANIA (all bacteria) vs. Lm strains.
Table A2.
List of possible A32 TAS pairs in TASMANIA (all bacteria) vs. Lm strains.
| First in Pair | Second in Pair | Pair Present in Lm | Remark |
|---|---|---|---|
| A32 | T13 (Zeta toxin) | No | |
| A32 | T14 (HipA_C) | No | |
| A32 | T2 (HipA_C) | Yes | Only found rarely in Lineage II, in animals with mastitis (cattle) or rhomboencephalitis (goat and sheep) |
| T85 (Gp49) | A32 | No | |
| T48 (Gp49) | A32 | No | |
| T38 (relE) | A32 | No |
References
- Jaffé, A.; Ogura, T.; Hiraga, S. Effects of the ccd function of the F plasmid on bacterial growth. J. Bacteriol. 1985, 163, 841–849. [Google Scholar] [PubMed]
- Gerdes, K.; Rasmussen, P.B.; Molin, S. Unique type of plasmid maintenance function: Postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 1986, 83, 3116–3120. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Larsen, J.E.; Molin, S. Stable inheritance of plasmid R1 requires two different loci. J. Bacteriol. 1985, 161, 292–298. [Google Scholar] [PubMed]
- Lobato-Márquez, D.; Díaz-Orejas, R.; García-del Portillo, F. Toxin-antitoxins and bacterial virulence. FEMS Microbiol. Rev. 2016, 40, 592–609. [Google Scholar] [CrossRef] [PubMed]
- Georgiades, K.; Raoult, D. Genomes of the Most Dangerous Epidemic Bacteria Have a Virulence Repertoire Characterized by Fewer Genes but More Toxin-Antitoxin Modules. PLoS ONE 2011, 6, e17962. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Brodersen, D.E.; Mitarai, N.; Gerdes, K. Toxins, Targets, and Triggers: An Overview of Toxin-Antitoxin Biology. Mol. Cell 2018, 70, 768–784. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Park, J.-H.; Inouye, M. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 2011, 45, 61–79. [Google Scholar] [CrossRef]
- Hayes, F.; Van Melderen, L. Toxins-antitoxins: Diversity, evolution and function. Crit. Rev. Biochem. Mol. Biol. 2011, 46, 386–408. [Google Scholar] [CrossRef]
- Conly, J.; Johnston, B. Listeria: A persistent food-borne pathogen. Can. J. Infect. Dis Med. Microbiol. 2008, 19, 327–328. [Google Scholar] [CrossRef]
- Vázquez-Boland, J.A.; Kuhn, M.; Berche, P.; Chakraborty, T.; Domínguez-Bernal, G.; Goebel, W.; González-Zorn, B.; Wehland, J.; Kreft, J. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 2001, 14, 584–640. [Google Scholar] [CrossRef]
- Farber, J.M.; Peterkin, P.I. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 1991, 55, 476–511. [Google Scholar] [PubMed]
- Low, J.C.; Donachie, W. A review of Listeria monocytogenes and listeriosis. Vet. J. 1997, 153, 9–29. [Google Scholar] [CrossRef]
- Oevermann, A.; Zurbriggen, A.; Vandevelde, M. Rhombencephalitis Caused by Listeria monocytogenes in Humans and Ruminants: A Zoonosis on the Rise? Interdiscip. Perspect. Infect. Dis. 2010, 2010, 632513. [Google Scholar] [CrossRef] [PubMed]
- Ward, T.J.; Ducey, T.F.; Usgaard, T.; Dunn, K.A.; Bielawski, J.P. Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl. Environ. Microbiol. 2008, 74, 7629–7642. [Google Scholar] [CrossRef] [PubMed]
- Den Bakker, H.C.; Bundrant, B.N.; Fortes, E.D.; Orsi, R.H.; Wiedmann, M. A population genetics-based and phylogenetic approach to understanding the evolution of virulence in the genus Listeria. Appl. Environ. Microbiol. 2010, 76, 6085–6100. [Google Scholar] [CrossRef]
- Orsi, R.H.; den Bakker, H.C.; Wiedmann, M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 2011, 301, 79–96. [Google Scholar] [CrossRef]
- Chenal-Francisque, V.; Lopez, J.; Cantinelli, T.; Caro, V.; Tran, C.; Leclercq, A.; Lecuit, M.; Brisse, S. Worldwide distribution of major clones of Listeria monocytogenes. Emerg. Infect. Dis. 2011, 17, 1110–1112. [Google Scholar] [CrossRef]
- Kuenne, C.; Billion, A.; Mraheil, M.A.; Strittmatter, A.; Daniel, R.; Goesmann, A.; Barbuddhe, S.; Hain, T.; Chakraborty, T. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics 2013, 14, 47. [Google Scholar] [CrossRef]
- Xie, Y.; Wei, Y.; Shen, Y.; Li, X.; Zhou, H.; Tai, C.; Deng, Z.; Ou, H.-Y. TADB 2.0: An updated database of bacterial type II toxin-antitoxin loci. Nucleic Acids Res. 2018, 46, D749–D753. [Google Scholar] [CrossRef]
- Curtis, T.; Takeuchi, I.; Gram, L.; Knudsen, G. The Influence of the Toxin/Antitoxin mazEF on Growth and Survival of Listeria monocytogenes under Stress. Toxins 2017, 9, 31. [Google Scholar] [CrossRef]
- Kalani, B.S.; Irajian, G.; Lotfollahi, L.; Abdollahzadeh, E.; Razavi, S. Putative type II toxin-antitoxin systems in Listeria monocytogenes isolated from clinical, food, and animal samples in Iran. Microb. Pathog. 2018, 122, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Mohammadzadeh, R.; Shivaee, A.; Ohadi, E.; Kalani, B.S. In Silico Insight into the Dominant Type II Toxin–Antitoxin Systems and Clp Proteases in Listeria monocytogenes and Designation of Derived Peptides as a Novel Approach to Interfere with this System. Int. J. Pept. Res. Ther. 2019, 1–11. [Google Scholar] [CrossRef]
- Akarsu, H.; Bordes, P.; Mansour, M.; Bigot, D.-J.; Genevaux, P.; Falquet, L. TASmania: A bacterial Toxin-Antitoxin Systems database. PLoS Comput. Biol. 2019, 15, e1006946. [Google Scholar] [CrossRef] [PubMed]
- Moura, A.; Criscuolo, A.; Pouseele, H.; Maury, M.M.; Leclercq, A.; Tarr, C.; Björkman, J.T.; Dallman, T.; Reimer, A.; Enouf, V.; et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2016, 2, 16185. [Google Scholar] [CrossRef] [PubMed]
- Bierne, H.; Sabet, C.; Personnic, N.; Cossart, P. Internalins: A complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect. 2007, 9, 1156–1166. [Google Scholar] [CrossRef]
- Aguilar-Bultet, L.; Nicholson, P.; Rychener, L.; Dreyer, M.; Gözel, B.; Origgi, F.C.; Oevermann, A.; Frey, J.; Falquet, L. Genetic Separation of Listeria monocytogenes Causing Central Nervous System Infections in Animals. Front. Cell. Infect. Microbiol. 2018, 8, 20. [Google Scholar] [CrossRef]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef]
- Oteri, F.; Nadalin, F.; Champeimont, R.; Carbone, A. BIS2Analyzer: A server for co-evolution analysis of conserved protein families. Nucleic Acids Res. 2017, 45, W307–W314. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Kim, S.K.; Makino, K.; Amemura, M.; Shinagawa, H.; Nakata, A. Molecular analysis of the phoH gene, belonging to the phosphate regulon in Escherichia coli. J. Bacteriol. 1993, 175, 1316–1324. [Google Scholar] [CrossRef] [PubMed]
- Rao, F.; Short, F.L.; Voss, J.E.; Blower, T.R.; Orme, A.L.; Whittaker, T.E.; Luisi, B.F.; Salmond, G.P.C. Co-evolution of quaternary organization and novel RNA tertiary interactions revealed in the crystal structure of a bacterial protein-RNA toxin-antitoxin system. Nucleic Acids Res. 2015, 43, 9529–9540. [Google Scholar] [CrossRef] [PubMed]
- Kersey, P.J.; Allen, J.E.; Allot, A.; Barba, M.; Boddu, S.; Bolt, B.J.; Carvalho-Silva, D.; Christensen, M.; Davis, P.; Grabmueller, C.; et al. Ensembl Genomes 2018: An integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2018, 46, D802–D808. [Google Scholar] [CrossRef] [PubMed]
- Balandyté, L.; Brodard, I.; Frey, J.; Oevermann, A.; Abril, C. Ruminant rhombencephalitis-associated Listeria monocytogenes alleles linked to a multilocus variable-number tandem-repeat analysis complex. Appl. Environ. Microbiol. 2011, 77, 8325–8335. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dreyer, M.; Aguilar-Bultet, L.; Rupp, S.; Guldimann, C.; Stephan, R.; Schock, A.; Otter, A.; Schüpbach, G.; Brisse, S.; Lecuit, M.; et al. Listeria monocytogenes sequence type 1 is predominant in ruminant rhombencephalitis. Sci. Rep. 2016, 6, 36419. [Google Scholar] [CrossRef] [PubMed]
- Šulc, Z.; Řezanková, H. Evaluation of Recent Similarity Measures for Categorical Data. In Proceedings of the 17th International Science Conference Applications of Mathematics and Statistics in Economics, Jerzmanowice, Poland, 27–31 August 2014; Wydawnictwo Uniwersytetu Ekonomicznego we Wrocławiu: Wrocław, Poland; pp. 249–258. [Google Scholar]
- Dib, L.; Carbone, A. CLAG: An unsupervised non hierarchical clustering algorithm handling biological data. BMC Bioinform. 2012, 13, 194. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).