Survey of Toxin–Antitoxin Systems in Erwinia amylovora Reveals Insights into Diversity and Functional Specificity
Abstract
:1. Introduction
2. Results
2.1. Identity and Distribution of Type II and IV TA Systems in Pathogenic and Nonpathogenic Erwinia spp.
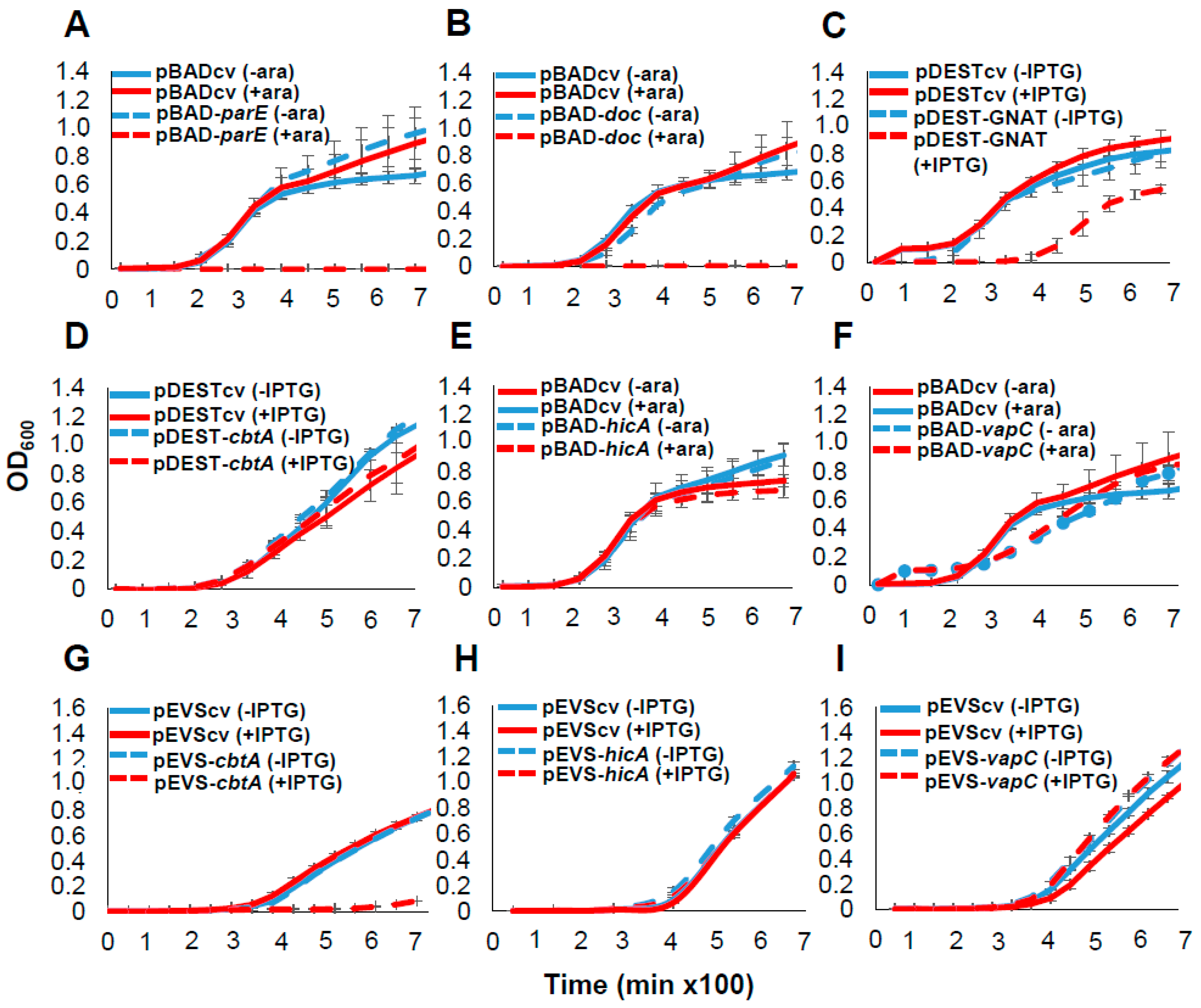
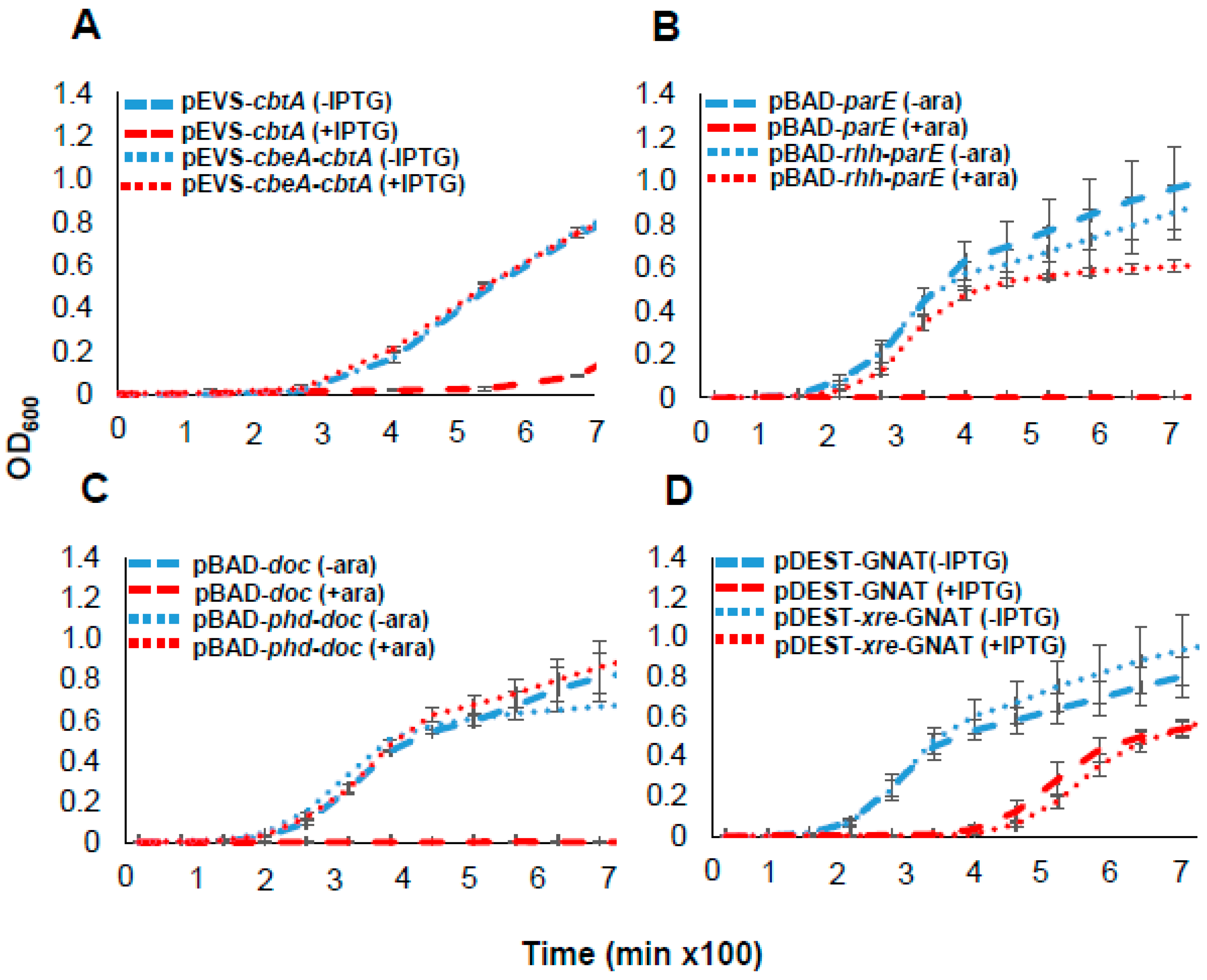
2.2. Functional Validation of Predicted Toxins and TA Systems
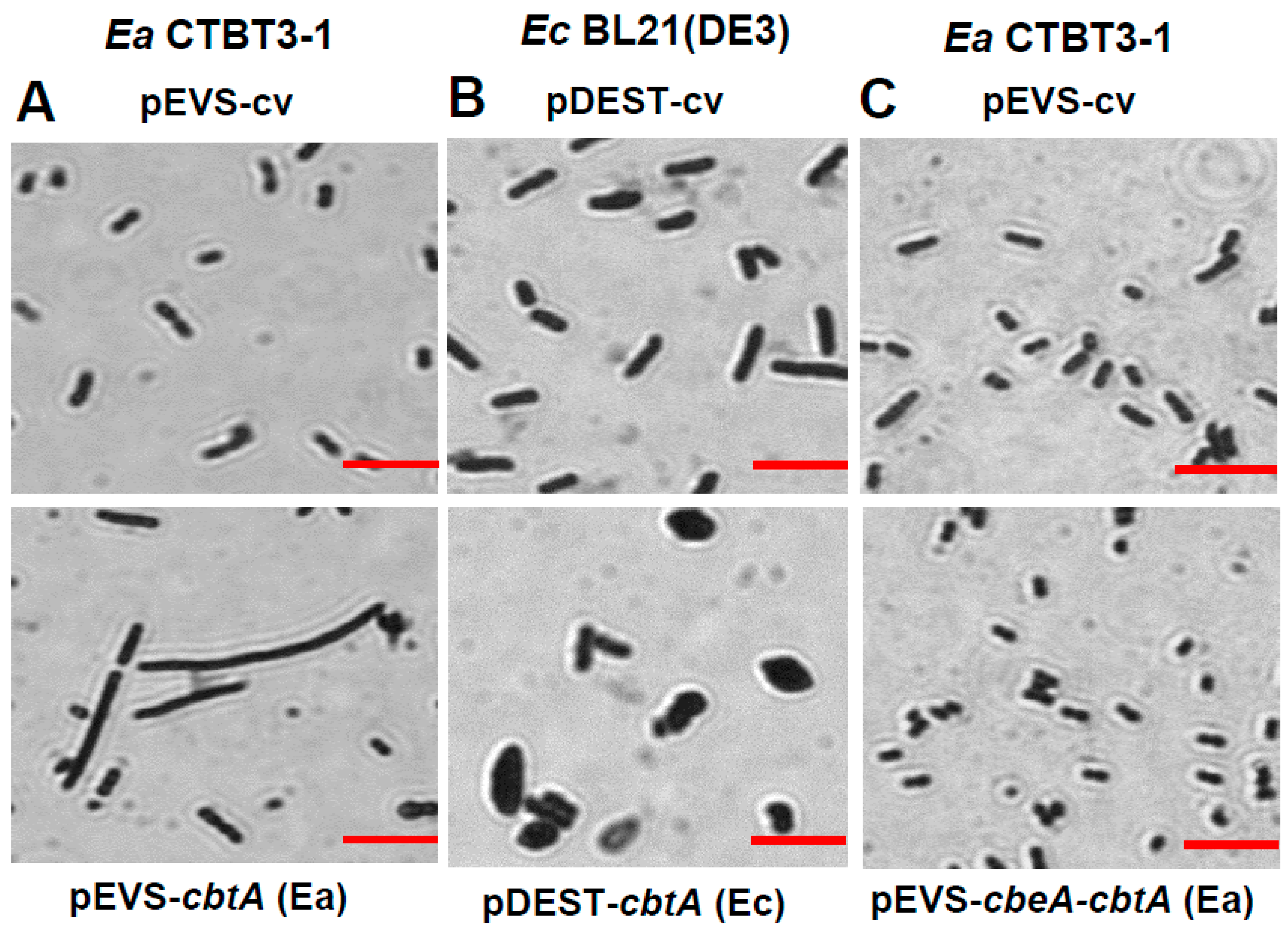
2.3. CbtA Toxin from CTBT3-1 Causes Cell Elongation That Is Suppressed by CbeA
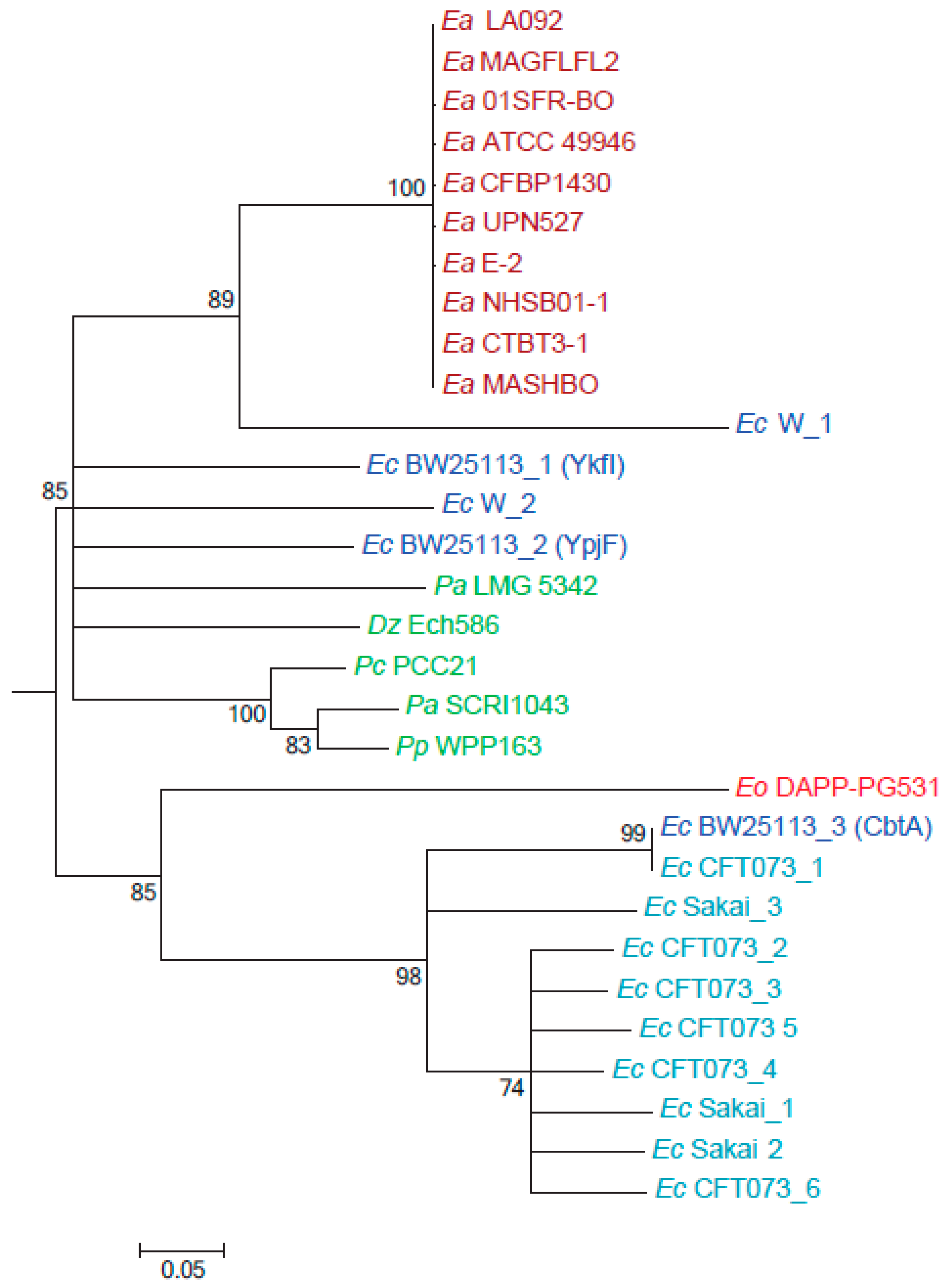
2.4. CbtA (Ea) Is Part of a Separate Clade from CbtA of Pathogenic E. coli
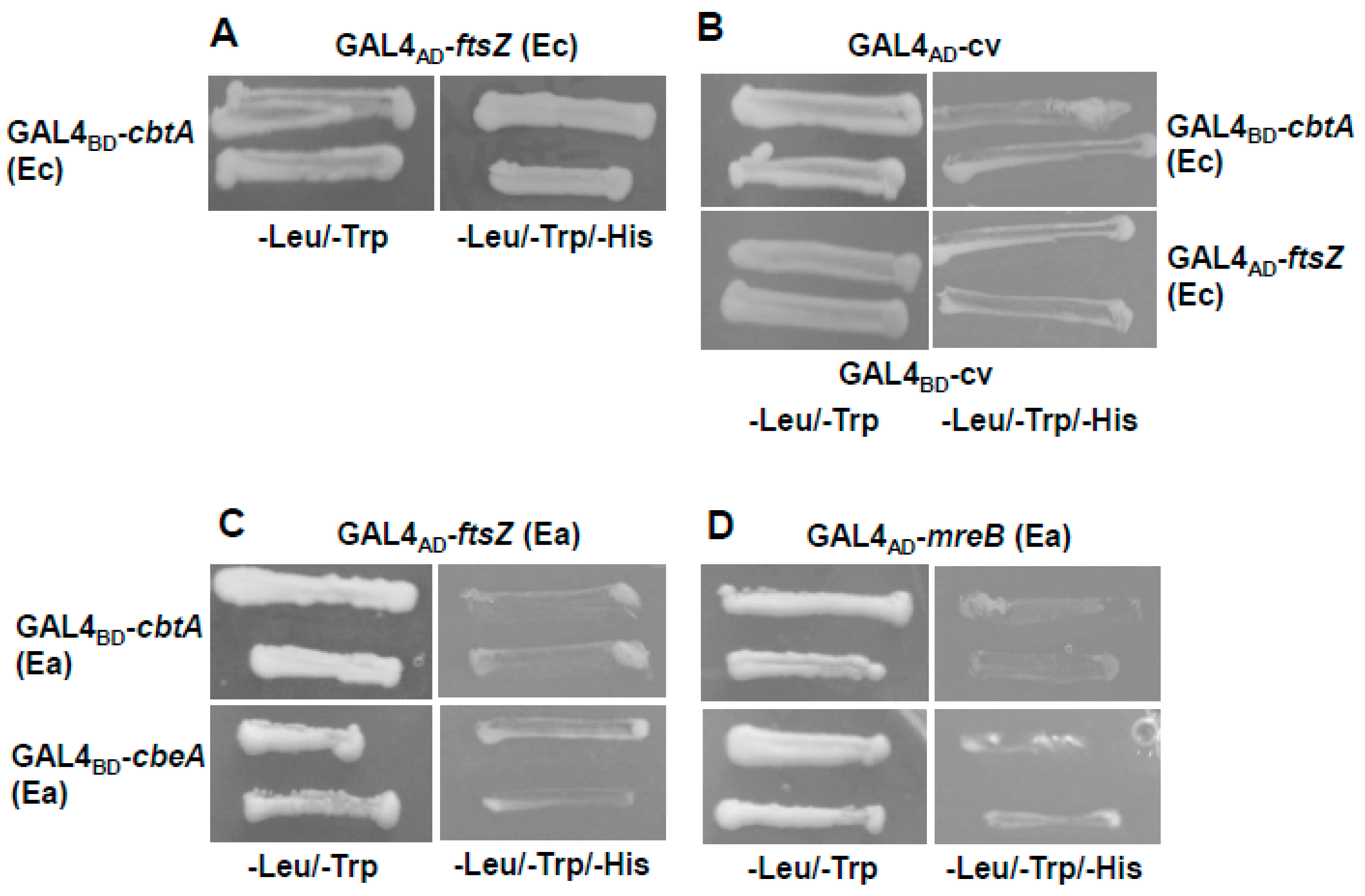
2.5. CbtA (Ea) Did Not Interact with Known Targets of CbtA (Ec) in a Yeast Two-Hybrid Study
3. Discussion
4. Materials and Methods
4.1. Prediction of TA Systems in Erwinia Species
4.2. Phylogenetic Analyses
4.3. Strains, Plasmids and Culture Conditions
4.4. Plasmid Construction
4.5. Monitoring Growth Rates of E. coli and E. amylovora Expressing Toxins or TA Loci
4.6. Microscopic Visualization of Cell Morphology
4.7. Yeast Two-Hybrid Assays
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shidore, T.; Triplett, L. Toxin-antitoxin systems: Implications for plant disease. Annu. Rev. Phytopathol. 2017, 55, 161–179. [Google Scholar] [CrossRef]
- van der Zwet, T.; Orolaza-Halbrendt, N.; Zeller, W. Front matter. In Fire Blight: History, Biology, and Management; The American Phytopathological Society: Saint Paul, MN, USA, 2016; pp. i–ix. [Google Scholar]
- Zeng, Q.; Cui, Z.; Wang, J.; Childs, K.L.; Sundin, G.W.; Cooley, D.R.; Yang, C.H.; Garofalo, E.; Eaton, A.; Huntley, R.B.; et al. Comparative genomics of Spiraeoideae-infecting Erwinia amylovora strains provides novel insight to genetic diversity and identifies the genetic basis of a low-virulence strain. Mol. Plant Pathol. 2018, 19, 1652–1666. [Google Scholar] [CrossRef] [PubMed]
- Vrancken, K.; Holtappels, M.; Schoofs, H.; Deckers, T.; Valcke, R. Pathogenicity and infection strategies of the fire blight pathogen Erwinia amylovora in Rosaceae: State of the art. Microbiology 2013, 159, 823–832. [Google Scholar] [CrossRef]
- Ordax, M.; Marco-Noales, E.; Lopez, M.M.; Biosca, E.G. Survival strategy of Erwinia amylovora against copper: Induction of the viable-but-nonculturable state. Appl. Environ. Microbiol. 2006, 72, 3482–3488. [Google Scholar] [CrossRef]
- Xie, Y.; Wei, Y.; Shen, Y.; Li, X.; Zhou, H.; Tai, C.; Deng, Z.; Ou, H.Y. Tadb 2.0: An updated database of bacterial type II toxin-antitoxin loci. Nucleic Acids Res. 2018, 46, D749–D753. [Google Scholar] [CrossRef]
- Horesh, G.; Harms, A.; Fino, C.; Parts, L.; Gerdes, K.; Heinz, E.; Thomson, N.R. Sling: A tool to search for linked genes in bacterial datasets. Nucleic Acids Res. 2018, 46, e128. [Google Scholar] [CrossRef]
- Sebaihia, M.; Bocsanczy, A.M.; Biehl, B.S.; Quail, M.A.; Perna, N.T.; Glasner, J.D.; DeClerck, G.A.; Cartinhour, S.; Schneider, D.J.; Bentley, S.D.; et al. Complete genome sequence of the plant pathogen Erwinia amylovora strain ATCC 49946. J. Bacteriol. 2010, 192, 2020–2021. [Google Scholar] [CrossRef]
- Heller, D.M.; Tavag, M.; Hochschild, A. CbtA toxin of Escherichia coli inhibits cell division and cell elongation via direct and independent interactions with Ftsz and Mreb. PLoS Genet. 2017, 13, e1007007. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Awano, N.; Inouye, M. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, Ftsz and Mreb. Mol. Microbiol. 2011, 79, 109–118. [Google Scholar] [CrossRef]
- Wen, Z.; Wang, P.; Sun, C.; Guo, Y.; Wang, X. Interaction of type IV toxin/antitoxin systems in cryptic prophages of Escherichia coli K-12. Toxins 2017, 9, 77. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.M.; Shaw, K.J. A novel family of Escherichia coli toxin-antitoxin gene pairs. J. Bacteriol. 2003, 185, 6600–6608. [Google Scholar] [CrossRef]
- Brynildsrud, O.; Gulla, S.; Feil, E.J.; Norstebo, S.F.; Rhodes, L.D. Identifying copy number variation of the dominant virulence factors msa and p22 within genomes of the fish pathogen Renibacterium salmoninarum. Microb. Genom. 2016, 2, e000055. [Google Scholar] [CrossRef]
- Elliott, K.T.; Cuff, L.E.; Neidle, E.L. Copy number change: Evolving views on gene amplification. Future Microbiol. 2013, 8, 887–899. [Google Scholar] [CrossRef]
- Masuda, H.; Tan, Q.; Awano, N.; Wu, K.P.; Inouye, M. YeeU enhances the bundling of cytoskeletal polymers of Mreb and Ftsz, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol. Microbiol. 2012, 84, 979–989. [Google Scholar] [CrossRef]
- Chan, W.T.; Yeo, C.C.; Sadowy, E.; Espinosa, M. Functional validation of putative toxin-antitoxin genes from the Gram-positive pathogen Streptococcus pneumoniae: Phd-doc is the fourth bona-fide operon. Front. Microbiol. 2014, 5, 677. [Google Scholar] [CrossRef]
- Jiang, Y.; Pogliano, J.; Helinski, D.R.; Konieczny, I. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 2002, 44, 971–979. [Google Scholar] [CrossRef]
- Yuan, J.; Yamaichi, Y.; Waldor, M.K. The three Vibrio cholerae chromosome II-encoded ParE toxins degrade chromosome I following loss of chromosome II. J. Bacteriol. 2011, 193, 611–619. [Google Scholar] [CrossRef]
- Unterholzner, S.J.; Hailer, B.; Poppenberger, B.; Rozhon, W. Characterisation of the StbD/E toxin-antitoxin system of pep36, a plasmid of the plant pathogen Erwinia pyrifoliae. Plasmid 2013, 70, 216–225. [Google Scholar] [CrossRef]
- Helaine, S.; Cheverton, A.M.; Watson, K.G.; Faure, L.M.; Matthews, S.A.; Holden, D.W. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 2014, 343, 204–208. [Google Scholar] [CrossRef]
- Lee, M.D. GToTree: User-friendly workflow for phylogenomics. bioRxiv 2019. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (itol) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Guzman, L.M.; Berlin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef]
- Dunn, A.K.; Millikan, D.S.; Adin, D.M.; Bose, J.L.; Stabb, E.V. New rfp- and pES21derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 2006, 72, 802–810. [Google Scholar] [CrossRef]
- Nakayama, M.; Kikuno, R.; Ohara, O. Protein-protein interactions between large proteins: Two-hybrid screening using a functionally classified library composed of long cDNAs. Genome Res. 2002, 12, 1773–1784. [Google Scholar] [CrossRef]

| No | Locus Tag (CFBP1430) | Length(aa) | Strand | Family/Domain | TAfinder | SLING |
|---|---|---|---|---|---|---|
| Chromosome | ||||||
| 1 | EAMY_0399 | 107 | + | YeeU/CbeA (A) | P | P |
| EAMY_0400 | 106 | + | YeeV/CbtA (T) | |||
| 2 | EAMY_0402 | 82 | + | RHH (A) | P | P |
| EAMY_0403 | 91 | + | RelE/ParE (T) | |||
| 3 | EAMY_0675 | 222 | - | DUF4258 | NP | P |
| EAMY_0676 | 100 | - | - | |||
| 4 | EAMY_1249 | 135 | + | Xre (A) | P | NP |
| EAMY_1250 | 137 | + | GNAT (T) | |||
| 5 * | EAMY_1411 | 240 | + | Pfam 00196 (A) | P (QS system) | NP |
| EAMY_1412 | 207 | + | COG03916 (T) | |||
| 6 | EAMY_1772 | 73 | + | PhD (A) | P | P |
| EAMY_1773 | 123 | + | Fic/Doc(T) | |||
| 7 | EAMY_1828 | 59 | + | HicB (A) | P | P |
| EAMY_1829 | 136 | + | HicA (T) | |||
| 8 | EAMY_2645 | 144 | - | RatA | NP | P |
| EAMY_2644 | 95 | - | - | |||
| 9 | EAMY_2779 | 46 | - | ParE (T) | P | NP |
| EAMY_2780 | 75 | - | RHH (A) | |||
| Plasmid | ||||||
| 1 | EAMY_3728 | 81 | + | PhD (A) | P | P |
| EAMY_3729 | 130 | + | VapC/PIN (T) | |||
| 2 | EAMY_3729 | 130 | + | VapC (T) | P | NP |
| EAMY_3730 | 57 | + | Pfam 01527 (A) | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shidore, T.; Zeng, Q.; Triplett, L.R. Survey of Toxin–Antitoxin Systems in Erwinia amylovora Reveals Insights into Diversity and Functional Specificity. Toxins 2019, 11, 206. https://doi.org/10.3390/toxins11040206
Shidore T, Zeng Q, Triplett LR. Survey of Toxin–Antitoxin Systems in Erwinia amylovora Reveals Insights into Diversity and Functional Specificity. Toxins. 2019; 11(4):206. https://doi.org/10.3390/toxins11040206
Chicago/Turabian StyleShidore, Teja, Quan Zeng, and Lindsay R. Triplett. 2019. "Survey of Toxin–Antitoxin Systems in Erwinia amylovora Reveals Insights into Diversity and Functional Specificity" Toxins 11, no. 4: 206. https://doi.org/10.3390/toxins11040206
APA StyleShidore, T., Zeng, Q., & Triplett, L. R. (2019). Survey of Toxin–Antitoxin Systems in Erwinia amylovora Reveals Insights into Diversity and Functional Specificity. Toxins, 11(4), 206. https://doi.org/10.3390/toxins11040206




