Gene–Lifestyle Interactions in Renal Dysfunction: Polygenic Risk Modulation via Plant-Based Diets, Coffee Intake, and Bioactive Compound Interactions
Abstract
1. Introduction
2. Methods
2.1. Participants
2.2. Demographic, Anthropometric, and Biochemical Measurements
2.3. Assessment of Kidney Function
2.4. Usual Food Intake Assessment Using a Semi-Quantitative Food Frequency Questionnaire (SQFFQ)
2.5. Dietary Pattern Classification and Inflammatory Index
2.6. Genotyping and Quality Control
2.7. Selection and Characterization of the Genetic Variants Associated with Renal Dysfunction Risk
2.8. PRS Development and Gene–Lifestyle Interactions
2.9. Molecular Docking Analysis of Missense Mutation with Food Compounds
2.10. Statistical Analysis
3. Results
3.1. Demographic and Lifestyle Characteristics
3.2. Biochemical Parameters Related to Metabolic Syndrome
3.3. Lifestyles and Nutrient Intake
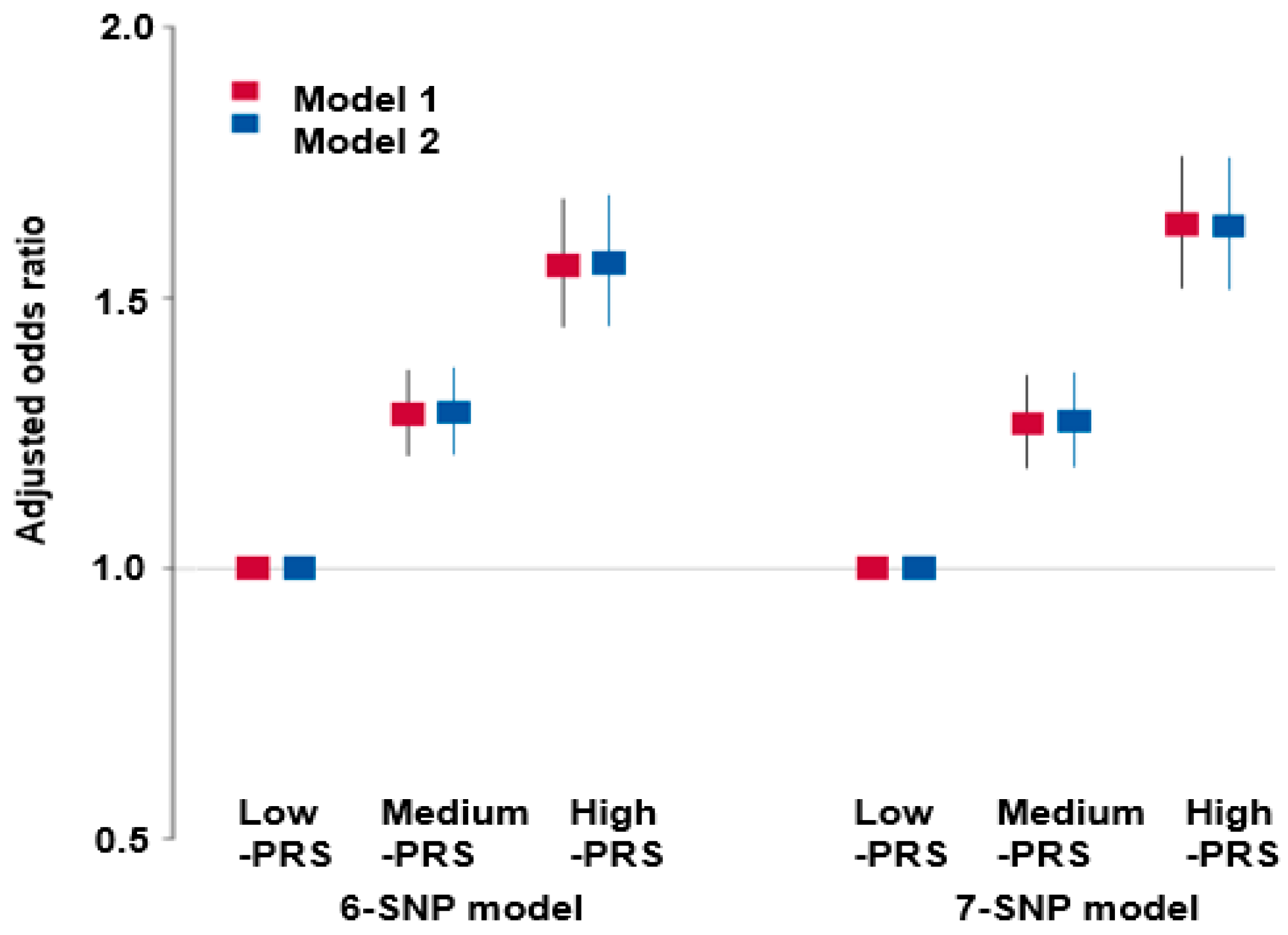
3.4. Polygenic Variants and Their Interaction Related to Renal Dysfunction Risk
3.5. Metabolic Functions Related to Genes Involved in the Hypo-eGFR
3.6. Genetic Variants-Lifestyle Interaction with Hypo-eGFR
3.7. Bioactive Compound Interaction with CPS1 rs1047891 Missense Mutation (Thr1406) and Molecular Docking
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kovesdy, C.P. Epidemiology of chronic kidney disease: An update 2022. Kidney Int. Suppl. 2022, 12, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Teo, B.W.; Chan, G.C.; Leo, C.C.H.; Tay, J.C.; Chia, Y.-C.; Siddique, S.; Turana, Y.; Chen, C.-H.; Cheng, H.-M.; Hoshide, S.; et al. Hypertension and chronic kidney disease in Asian populations. J. Clin. Hypertens. 2021, 23, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Bragadottir, G.; Redfors, B.; Ricksten, S.-E. Assessing glomerular filtration rate (GFR) in critically ill patients with acute kidney injury—True GFR versus urinary creatinine clearance and estimating equations. Crit. Care 2013, 17, R108. [Google Scholar] [CrossRef] [PubMed]
- Shahbaz, H.; Gupta, M. Creatinine Clearance; StatPearls: Treasure Island, FL, USA, 2021. [Google Scholar]
- Teo, B.W.; Zhang, L.; Guh, J.Y.; Tang, S.C.W.; Jha, V.; Kang, D.H.; Tanchanco, R.; Hooi, L.S.; Praditpornsilpa, K.; Kong, X.; et al. Glomerular Filtration Rates in Asians. Adv. Chronic Kidney Dis. 2018, 25, 41–48. [Google Scholar] [CrossRef]
- Yanai, H.; Adachi, H.; Hakoshima, M.; Katsuyama, H. Molecular, Biological, and Clinical Understanding of the Pathophysiology and Treatments of Hyperuricemia and Its Association with Metabolic Syndrome, Cardiovascular Diseases and Chronic Kidney Disease. Int. J. Mol. Sci. 2021, 22, 9221. [Google Scholar] [CrossRef]
- Schrauben, S.J.; Hsu, J.Y.; Amaral, S.; Anderson, A.H.; Feldman, H.I.; Dember, L.M. Effect of Kidney Function on Relationships between Lifestyle Behaviors and Mortality or Cardiovascular Outcomes: A Pooled Cohort Analysis. J. Am. Soc. Nephrol. 2021, 32, 663–675. [Google Scholar] [CrossRef]
- Kelly, J.T.; Su, G.; Zhang, L.; Qin, X.; Marshall, S.; González-Ortiz, A.; Clase, C.M.; Campbell, K.L.; Xu, H.; Carrero, J.-J. Modifiable Lifestyle Factors for Primary Prevention of CKD: A Systematic Review and Meta-Analysis. J. Am. Soc. Nephrol. 2021, 32, 239–253. [Google Scholar] [CrossRef]
- Kim, Y.; Han, B.G. Cohort Profile: The Korean Genome and Epidemiology Study (KoGES) Consortium. Int. J. Epidemiol. 2017, 46, e20. [Google Scholar] [CrossRef]
- Zhang, T.; Park, S. Energy Intake-Dependent Genetic Associations with Obesity Risk: BDNF Val66Met Polymorphism and Interactions with Dietary Bioactive Compounds. Antioxidants 2025, 14, 170. [Google Scholar] [CrossRef]
- Böger, C.A. It’s not only the kidneys—Genetic determinants of glomerular filtration marker levels. Nephrol. Dial. Transplant. 2013, 28, 2397–2398. [Google Scholar] [CrossRef][Green Version]
- Ryu, J.; Lee, C. Association of glycosylated hemoglobin with the gene encoding CDKAL1 in the Korean Association Resource (KARE) study. Hum. Mutat. 2012, 33, 655–659. [Google Scholar] [CrossRef] [PubMed]
- Montasser, M.E.; Shimmin, L.C.; Gu, D.; Chen, J.; Gu, C.; Kelly, T.N.; Jaquish, C.E.; Rice, T.K.; Rao, D.C.; Cao, J.; et al. Variations in genes that regulate blood pressure are associated with glomerular filtration rate in Chinese. PLoS ONE 2014, 9, e92468. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Kato, K.; Fujimaki, T.; Yokoi, K.; Oguri, M.; Watanabe, S.; Metoki, N.; Yoshida, H.; Satoh, K.; Aoyagi, Y.; et al. Association of genetic variants with chronic kidney disease in Japanese individuals. Clin. J. Am. Soc. Nephrol. 2009, 4, 883–890. [Google Scholar] [CrossRef]
- Daily, J.W.; Park, S. Interaction of BDNF rs6265 variants and energy and protein intake in the risk for glucose intolerance and type 2 diabetes in middle-aged adults. Nutrition 2017, 33, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Daily, J.W.; Zhang, X.; Jin, H.S.; Lee, H.J.; Lee, Y.H. Interactions with the MC4R rs17782313 variant, mental stress and energy intake and the risk of obesity in Genome Epidemiology Study. Nutr. Metab. 2016, 13, 38. [Google Scholar] [CrossRef]
- Park, S.; Ahn, J.; Lee, B.K. Self-rated Subjective Health Status Is Strongly Associated with Sociodemographic Factors, Lifestyle, Nutrient Intakes, and Biochemical Indices, but Not Smoking Status: KNHANES 2007-2012. J. Korean Med. Sci. 2015, 30, 1279–1287. [Google Scholar] [CrossRef]
- Michels, W.M.; Grootendorst, D.C.; Verduijn, M.; Elliott, E.G.; Dekker, F.W.; Krediet, R.T. Performance of the Cockcroft-Gault, MDRD, and New CKD-EPI Formulas in Relation to GFR, Age, and Body Size. Clin. J. Am. Soc. Nephrol. 2010, 5, 1003–1009. [Google Scholar] [CrossRef]
- Ma, Y.; Zhan, J.; Xu, G. Reference values of glomerular filtration rate for healthy adults in southern China: A cross-sectional survey. Ther. Adv. Chronic Dis. 2021, 12, 20406223211035287. [Google Scholar] [CrossRef]
- Park, S.; Zhang, X.; Lee, N.R.; Jin, H.S. TRPV1 Gene Polymorphisms Are Associated with Type 2 Diabetes by Their Interaction with Fat Consumption in the Korean Genome Epidemiology Study. J. Nutrigenet. Nutrigenom. 2016, 9, 47–61. [Google Scholar] [CrossRef]
- Ahn, Y.; Kwon, E.; Shim, J.E.; Park, M.K.; Joo, Y.; Kimm, K.; Park, C.; Kim, D.H. Validation and reproducibility of food frequency questionnaire for Korean genome epidemiologic study. Eur. J. Clin. Nutr. 2007, 61, 1435–1441. [Google Scholar] [CrossRef]
- Steel, R.G.; Torrie, J.H.; Dickey, D.A. Principles and Procedures of Statistics: A Biological Approach; McGraw-Hill: New York, NY, USA, 1997. [Google Scholar]
- Hébert, J.R.; Shivappa, N.; Wirth, M.D.; Hussey, J.R.; Hurley, T.G. Perspective: The Dietary Inflammatory Index (DII)-Lessons Learned, Improvements Made, and Future Directions. Adv. Nutr. 2019, 10, 185–195. [Google Scholar] [CrossRef]
- van Woudenbergh, G.J.; Theofylaktopoulou, D.; Kuijsten, A.; Ferreira, I.; van Greevenbroek, M.M.; van der Kallen, C.J.; Schalkwijk, C.G.; Stehouwer, C.D.; Ocké, M.C.; Nijpels, G.; et al. Adapted dietary inflammatory index and its association with a summary score for low-grade inflammation and markers of glucose metabolism: The Cohort study on Diabetes and Atherosclerosis Maastricht (CODAM) and the Hoorn study. Am. J. Clin. Nutr. 2013, 98, 1533–1542. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Kim, M.K. Relationship of sodium intake with obesity among Korean children and adolescents: Korea National Health and Nutrition Examination Survey. Br. J. Nutr. 2016, 115, 834–841. [Google Scholar] [CrossRef] [PubMed]
- Rabbee, N.; Speed, T.P. A genotype calling algorithm for Affymetrix SNP arrays. Bioinformatics 2006, 22, 7–12. [Google Scholar] [CrossRef]
- Uma Jyothi, K.; Reddy, B.M. Gene-gene and gene-environment interactions in the etiology of type 2 diabetes mellitus in the population of Hyderabad, India. Meta Gene 2015, 5, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Liu, L.; Zhou, J.; Zhang, T.; Daily, J.W.; Park, S. Bioactive Components of Houttuynia cordata Thunb and Their Potential Mechanisms Against COVID-19 Using Network Pharmacology and Molecular Docking Approaches. J. Med. Food 2022, 25, 355–366. [Google Scholar] [CrossRef]
- Yang, Y.; Shi, C.-Y.; Xie, J.; Dai, J.-H.; He, S.-L.; Tian, Y. Identification of potential dipeptidyl peptidase (DPP)-IV inhibitors among Moringa oleifera phytochemicals by virtual screening, molecular docking analysis, ADME/T-based prediction, and in vitro analyses. Molecules 2020, 25, 189. [Google Scholar] [CrossRef]
- SAS Inc. SAS/STAT® 14.2 User’s Guide; SAS Inc.: Cary, NC, USA, 2016. [Google Scholar]
- Inaguma, D.; Kitagawa, A.; Yanagiya, R.; Koseki, A.; Iwamori, T.; Kudo, M.; Yuzawa, Y. Increasing tendency of urine protein is a risk factor for rapid eGFR decline in patients with CKD: A machine learning-based prediction model by using a big database. PLoS ONE 2020, 15, e0239262. [Google Scholar] [CrossRef]
- Köttgen, A.; Pattaro, C.; Böger, C.A.; Fuchsberger, C.; Olden, M.; Glazer, N.L.; Parsa, A.; Gao, X.; Yang, Q.; Smith, A.V.; et al. New loci associated with kidney function and chronic kidney disease. Nat. Genet. 2010, 42, 376–384. [Google Scholar] [CrossRef]
- Kaesler, N.; Babler, A.; Floege, J.; Kramann, R. Cardiac Remodeling in Chronic Kidney Disease. Toxins 2020, 12, 161. [Google Scholar] [CrossRef]
- Silveira Rossi, J.L.; Barbalho, S.M.; Reverete de Araujo, R.; Bechara, M.D.; Sloan, K.P.; Sloan, L.A. Metabolic syndrome and cardiovascular diseases: Going beyond traditional risk factors. Diabetes/Metab. Res. Rev. 2022, 38, e3502. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, S.; Kim, Y.; Lee, Y.; Kang, M.W.; Kim, K.; Kim, Y.C.; Han, S.S.; Lee, H.; Lee, J.P.; et al. Causal effects of relative fat, protein, and carbohydrate intake on chronic kidney disease: A Mendelian randomization study. Am. J. Clin. Nutr. 2021, 113, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Cirillo, M.; Lombardi, C.; Chiricone, D.; De Santo, N.G.; Zanchetti, A.; Bilancio, G. Protein intake and kidney function in the middle-age population: The contrast between cross-sectional and longitudinal data. Nephrol. Dial. Transplant. 2014, 29, 1733–1740. [Google Scholar] [CrossRef] [PubMed]
- Oosterwijk, M.M.; Navis, G.; Bakker, S.J.L.; Laverman, G.D. Personalized Nutrition in Patients with Type 2 Diabetes and Chronic Kidney Disease: The Two-Edged Sword of Dietary Protein Intake. J. Pers. Med. 2022, 12, 300. [Google Scholar] [CrossRef]
- Jung, H.W.; Kim, S.W.; Kim, I.Y.; Lim, J.Y.; Park, H.S.; Song, W.; Yoo, H.J.; Jang, H.C.; Kim, K.; Park, Y.; et al. Protein Intake Recommendation for Korean Older Adults to Prevent Sarcopenia: Expert Consensus by the Korean Geriatric Society and the Korean Nutrition Society. Ann. Geriatr. Med. Res. 2018, 22, 167–175. [Google Scholar] [CrossRef]
- He, J.; Mills, K.T.; Appel, L.J.; Yang, W.; Chen, J.; Lee, B.T.; Rosas, S.E.; Porter, A.; Makos, G.; Weir, M.R.; et al. Urinary Sodium and Potassium Excretion and CKD Progression. J. Am. Soc. Nephrol. 2016, 27, 1202–1212. [Google Scholar] [CrossRef]
- Cirillo, M.; Bilancio, G.; Cavallo, P.; Palladino, R.; Terradura-Vagnarelli, O.; Laurenzi, M. Sodium intake and kidney function in the general population: An observational, population-based study. Clin. Kidney J. 2021, 14, 647–655. [Google Scholar] [CrossRef]
- McMahon, E.J.; Campbell, K.L.; Bauer, J.D.; Mudge, D.W. Altered dietary salt intake for people with chronic kidney disease. Cochrane Database Syst. Rev. 2015, 18, CD010070. [Google Scholar] [CrossRef]
- Okada, Y.; Sim, X.; Go, M.J.; Wu, J.-Y.; Gu, D.; Takeuchi, F.; Takahashi, A.; Maeda, S.; Tsunoda, T.; Chen, P.; et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in East Asian populations. Nat. Genet. 2012, 44, 904–909. [Google Scholar] [CrossRef]
- Kim, H.; Caulfield, L.E.; Garcia-Larsen, V.; Steffen, L.M.; Grams, M.E.; Coresh, J.; Rebholz, C.M. Plant-Based Diets and Incident CKD and Kidney Function. Clin. J. Am. Soc. Nephrol. 2019, 14, 682–691. [Google Scholar] [CrossRef]
- Xu, K.; Cui, X.; Wang, B.; Tang, Q.; Cai, J.; Shen, X. Healthy adult vegetarians have better renal function than matched omnivores: A cross-sectional study in China. BMC Nephrol. 2020, 21, 268. [Google Scholar] [CrossRef] [PubMed]
- Ashkar, F.; Bhullar, K.S.; Wu, J. The Effect of Polyphenols on Kidney Disease: Targeting Mitochondria. Nutrients 2022, 14, 3115. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, O.J.; Pirastu, N.; Poole, R.; Fallowfield, J.A.; Hayes, P.C.; Grzeszkowiak, E.J.; Taal, M.W.; Wilson, J.F.; Parkes, J.; Roderick, P.J. Coffee Consumption and Kidney Function: A Mendelian Randomization Study. Am. J. Kidney Dis. 2020, 75, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Kanbay, M.; Siriopol, D.; Copur, S.; Tapoi, L.; Benchea, L.; Kuwabara, M.; Rossignol, P.; Ortiz, A.; Covic, A.; Afsar, B. Effect of Coffee Consumption on Renal Outcome: A Systematic Review and Meta-Analysis of Clinical Studies. J. Ren. Nutr. 2021, 31, 5–20. [Google Scholar] [CrossRef]
- Vervloet, M.G.; Hsu, S.; de Boer, I.H. Vitamin D supplementation in people with chronic kidney disease. Kidney Int. 2023, 104, 698–706. [Google Scholar] [CrossRef]
- Yeung, W.-C.G.; Toussaint, N.D.; Badve, S.V. Vitamin D therapy in chronic kidney disease: A critical appraisal of clinical trial evidence. Clin. Kidney J. 2024, 17, sfae227. [Google Scholar] [CrossRef]
- Nitzahn, M.; Lipshutz, G.S. CPS1: Looking at an ancient enzyme in a modern light. Mol. Genet. Metab. 2020, 131, 289–298. [Google Scholar] [CrossRef]

| High-GFR (n = 51,084) | Low-GFR (n = 7617) | Adjusted OR and 95% CI | |
|---|---|---|---|
| Age (years) 2 | 53.3 ± 0.03 | 56.5 ± 0.08 *** | 2.156 (1.975–2.354) |
| Gender (male, N %) | 16,105 (33.6) | 3332 (43.7) +++ | 0.701 (0.649–0.756) |
| Education <High school | 7268 (19.6) | 1223 (21.2) | 1 |
| High school | 27,133 (73.2) | 4172 (72.2) | 1.130 (0.978–1.306) |
| >High school | 2687 (7.24) | 412 (7.09) | 1.194 (0.912–1.562) |
| MetS (N, %) 3 | 6998 (13.7) | 2148 (28.2) +++ | 1.991 (1.781–2.225) |
| CVD (N, %) 4 | 1890 (3.72) | 799 (10.5) +++ | 1.920 (1.625–2.268) |
| BMI (kg/m2) 5 | 23.9 ± 0.03 | 24.3 ± 0.05 *** | 1.374 (1.244–1.518) |
| Waist Cir. (cm) 6 | 80.6 ± 0.03 | 81.7 ± 0.18 *** | 1.304 (1.168–1.456) |
| Serum glucose (mg/dL) 7 | 95.1 ± 0.09 | 97.6 ± 0.48 ** | 1.853 (1.632–2.105) |
| Blood HbA1c (%) 8 | 5.71 ± 0.004 | 5.96 ± 0.026 *** | 1.598 (1.339–1.908) |
| Serum HDL (mg/dL) 9 | 53.9 ± 0.06 | 51.1 ± 0.31 *** | 0.662 (0.598–0.733) |
| Serum LDL (mg/dL) 10 | 118 ± 0.15 | 120 ± 0.39 *** | 1.277 (1.143–1.427) |
| Serum TG (mg/dL) 11 | 124 ± 0.39 | 130 ± 1.00 *** | 1.340 (1.210–1.483) |
| SBP (mmHg) 12 | 122 ± 0.07 | 122 ± 0.17 | 1.142 (1.032–1.263) |
| DBP (mmHg) 13 | 75.7 ± 0.04 | 75.8 ± 0.11 | 1.103 (0.943–1.289) |
| Hypertension (N, %) | 2814 (5.51) | 1150 (15.1) +++ | 2.089 (1.887–2.312) |
| Insulin resistance (N, %) | 3918 (7.67) | 975 (12.8)*** | 1.473 (1.271–1.707) |
| Serum CRP (mg/dL) 14 | 0.137 ± 0.002 | 0.154 ± 0.005 ** | 1.714 (1.254–2.344) |
| Serum creatinine (mg/dL) | 0.79 ± 0.001 | 1.24 ± 0.003 | |
| Serum uric acid (mg/dL) 15 | 4.60 ± 0.005 | 5.24 ± 0.01 *** | 4.531 (4.036–5.087) |
| Blood urinary nitrogen (mg/dL) 16 | 14.4 ± 0.02 | 18.2 ± 0.05 | 3.247 (2.935–3.591) |
| Albumin (mg/dL) 17 | 4.62 ± 0.001 | 4.59 ± 0.006 *** | 0.753 (0.683–0.831) |
| AST (U/L) 18 | 23.6 ± 0.11 | 24.7 ± 0.30 *** | 1.189 (0.957–1.477) |
| ALT (U/L) 19 | 22.3 ± 0.11 | 23.0 ± 0.29 * | 1.170 (1.010–1.355) |
| Total bilirubin 20 | 0.73 ± 0.002 | 0.74 ± 0.005 * | 2.432 (2.306–2.565) |
| Urinary protein 21 | 1.09 ± 0.002 | 1.33 ± 0.01 *** | 4.653 (3.919–5.524) |
| CHR 1 | SNP 2 | Position | Mi 3 | Ma 4 | OR 5 | SE 6 | Adjusted p-Value 7 | Adjusted p-Value 8 | MAF 9 | p-Value for HWE 10 | Gene Names | Location |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | rs1047891 (Thr1406Asn) | 211540507 | A | C | 1.172 | 0.0241 | 4.61 × 10−11 | 2.98 × 10−5 | 0.179 | 0.1156 | CPS1 | Missense |
| 2 | rs3770636 | 170202833 | G | T | 0.881 | 0.0245 | 2.3 × 10−7 | 0.00692 | 0.200 | 0.0692 | LRP2 | Intron |
| 4 | rs5020545 | 77414988 | T | C | 1.149 | 0.0247 | 1.83 × 10−8 | 0.00724 | 0.171 | 0.6204 | SHROOM3 | Intron |
| 5 | rs3812036 | 176813404 | T | C | 1.122 | 0.0227 | 3.97 × 10−7 | 0.0003224 | 0.219 | 0.7723 | SLC34A1 | Intron |
| 6 | rs4715517 | 54973761 | A | C | 1.283 | 0.0396 | 2.88 × 10−10 | 0.0002845 | 0.054 | 0.3498 | HCRTR2 | 5′-UTR |
| 6 | rs140652052 | 33547810 | C | A | 1.259 | 0.0464 | 6.95 × 10−7 | 0.009824 | 0.038 | 0.8193 | BAK1 | Intron |
| 7 | rs139132767 | 116726021 | G | A | 1.579 | 0.0616 | 1.22 × 10−13 | 0.008934 | 0.018 | 0.2941 | ST7 | Intron |
| 12 | rs141574969 | 111319366 | G | A | 1.142 | 0.0257 | 2.15 × 10−7 | 0.007747 | 0.159 | 0.1384 | CCDC63 | Intron |
| 15 | rs1031755 | 53951435 | C | A | 0.8967 | 0.0198 | 3.49 × 10−8 | 0.00043 | 0.403 | 0.4817 | WDR72 | Intron |
| 22 | rs6001939 | 40892794 | T | C | 0.9098 | 0.0213 | 9.21 × 10−7 | 0.001771 | 0.293 | 0.2773 | MRTFA | Intron |
| Gene Set | No. Genes | Beta | Beta SD | SE | p-Value |
|---|---|---|---|---|---|
| GO BP: Protein autoprocessing | 28 | 0.59988 | 0.02312 | 0.15852 | 7.73 × 10−5 |
| GO BP: Regulation of receptor recycling | 22 | 0.66108 | 0.02259 | 0.17705 | 9.46 × 10−5 |
| Acevedo liver cancer dn | 516 | 0.1403 | 0.02291 | 0.03774 | 0.0001007 |
| GO BERT: Core oligodendrocyte differentiation | 40 | 0.48444 | 0.02231 | 0.14096 | 0.0002951 |
| GO CC: Neuronal ribonucleoprotein granule | 3 | 1.6359 | 0.02065 | 0.48121 | 0.0003383 |
| Biocarta LDL pathway | 6 | 1.2646 | 0.02257 | 0.37738 | 0.0004037 |
| Bhat esr1 targets via akt1 up | 273 | 0.17603 | 0.02104 | 0.05297 | 0.0004456 |
| GO CC: Proton transporting atp synthase complex catalytic core f1 | 6 | 1.0336 | 0.01845 | 0.31139 | 0.0004519 |
| GO BP: Negative regulation of receptor recycling | 5 | 1.0453 | 0.01703 | 0.31814 | 0.0005096 |
| GO BP: Negative regulation of myosin light chain phosphatase activity | 5 | 1.4803 | 0.02412 | 0.4553 | 0.0005756 |
| PRS 6 | Low-PRS (n = 17,680) | Medium-PRS (n = 26,672) | High-PRS (n = 9476) | PRS Interaction p-Value |
|---|---|---|---|---|
| Low-ABD 1 High-ABD | 1 1 | 1.247 (1.151–1.351) 1.396 (1.250–1.558) | 1.534 (1.390–1.693) 1.665 (1.452–1.910) | 0.2522 |
| Low-PBD 1 High-PBD | 1 1 | 1.371 (1.266–1.484) 1.162 (1.039–1.299) | 1.628 (1.475–1.797) 1.457 (1.275–1.686) | 0.0215 |
| Low-WSD 1 High-WSD | 1 1 | 1.305 (1.199–1.420) 1.285 (1.162–1.421) | 1.555 (1.400–1.728) 1.611 (1.424–1.822) | 0.9430 |
| Low-RMD 1 High-RMD | 1 1 | 1.331 (1.230–1.440) 1.232 (1.100–1.379) | 1.592 (1.444–1.755) 1.558 (1.354–1.792) | 0.0756 |
| Low-Sodium 2 High-Sodium | 1 1 | 1.301 (1.179–1.435) 1.298 (1.191–1.414) | 1.501 (1.328–1.697) 1.649 (1.484–1.833) | 0.8257 |
| Low-Alcohol 3 High-Alcohol | 1 1 | 1.226 (1.124–1.336) 1.391 (1.261–1.534) | 1.526 (1.376–1.694) 1.639 (1.445–1.859) | 0.2312 |
| Non-smoker Former + Current smokers | 1 1 | 1.284 (1.187–1.388) 1.338 (1.192–1.502) | 1.620 (1.472–1.784) 1.522 (1.318–1.757) | 0.4421 |
| Low Exercise Regular Exercise 4 | 1 1 | 1.140 (1.033–1.259) 1.419 (1.304–1.545) | 1.431 (1.267–1.615) 1.709 (1.538–1.898) | 0.0596 |
| Low-Coffee 5 High-Coffee | 1 1 | 1.311 (1.220–1.409) 1.248 (1.078–1.445) | 1.655 (1.513–1.809) 1.344 (1.121–1.611) | 0.0092 |
| Low-Tea 6 High-Tea | 1 1 | 1.307 (1.212–1.409) 1.272 (1.119–1.446) | 1.609 (1.466–1.765) 1.496 (1.275–1.754) | 0.2290 |
| Low-Vitamin D 7 High-Vitamin D | 1 1 | 1.327 (1.169–1.505) 1.280 (1.191–1.376) | 1.574 (1.440–1.721) 1.521 (1.300–1.780) | 0.2066 |
| CPS1 rs1047891 | Major | Heterozygote | Minor | |
| Low-Vitamin D 7 High-Vitamin D | 1 1 | 1.235 (1.099–1.388) 1.176 (1.099–1.257) | 1.393 (1.190–1.632) 1.124 (0.833–1.517) | 0.0436 |
| Food Components | Foods | WT | MT |
|---|---|---|---|
| Docking Energy, ΔG (kcal mol−1) | |||
| Cichoriin | Chicory | −10.1 | −8.6 |
| Malvidin 3-alpha-L-galactoside | Blue berry | −10.1 | −8.6 |
| Glyceollin II | Soybeans | −10.6 | −8.7 |
| Stigmasteryl glucoside | Soybean oil | −10.2 | −8.5 |
| alpha-Carotene | Carrot | −10.9 | −8.6 |
| (5R,5’R,6S,8′R)-Luteochrome | Sweet potato | −10.2 | −7.3 |
| Tuberoside B | Allium tuberosum | −10.6 | −8.7 |
| Cycloartanyl ferulate | Rice bran oil | −10.3 | −8.6 |
| delta-Carotene | Carrot tomatoes | −10.5 | −7.7 |
| 19’-Hexanoyloxymytiloxanthin | Mussel | −10.2 | −8 |
| (R)-Hispaglabridin A | Licorice | −10.2 | −8.6 |
| 5,6,7,8-Tetrahydroxy-3’,4’-dimethoxyflavone | Seville orange | −10.6 | −8.7 |
| (3S,3’S,all-E)-Zeaxanthin | Shrimp | −10.9 | −8.3 |
| (E)-4-(3,7-Dimethyl-2,6-octadienyl)-1,3,5-trihydroxyxanthone | Garcinia livingstonei | −10.1 | −8.5 |
| 28-Hydroxymangiferonic acid | Mango | −10.5 | −8.5 |
| Soyasaponin ag | Soybeans, pulses | −11.3 | −8.2 |
| (S)-Nerolidol 3-O-[a-L-Rhamnopyranosyl-(1->4)-a-L-rhamnopyranosyl-(1->2)-[4-(4-hydroxy-3-methoxycinnamoyl)-(E)-a-L-rhamnopyranosyl-(1->6)]-b-D-glucopyranoside] | Eriobotrya japonica | −10.5 | −8.6 |
| 3alpha-12-Ursene-3,24-diol | Boswellia serrata | −10.6 | −8.5 |
| Hovenidulcioside B2 | Hovenia dulcis | −10.5 | −8.6 |
| Vitamin D3 | Egg yolk, liver, tuna | −7.6 | −9.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.; Kim, D.-S.; Park, S. Gene–Lifestyle Interactions in Renal Dysfunction: Polygenic Risk Modulation via Plant-Based Diets, Coffee Intake, and Bioactive Compound Interactions. Nutrients 2025, 17, 916. https://doi.org/10.3390/nu17050916
Liu M, Kim D-S, Park S. Gene–Lifestyle Interactions in Renal Dysfunction: Polygenic Risk Modulation via Plant-Based Diets, Coffee Intake, and Bioactive Compound Interactions. Nutrients. 2025; 17(5):916. https://doi.org/10.3390/nu17050916
Chicago/Turabian StyleLiu, Meiling, Da-Sol Kim, and Sunmin Park. 2025. "Gene–Lifestyle Interactions in Renal Dysfunction: Polygenic Risk Modulation via Plant-Based Diets, Coffee Intake, and Bioactive Compound Interactions" Nutrients 17, no. 5: 916. https://doi.org/10.3390/nu17050916
APA StyleLiu, M., Kim, D.-S., & Park, S. (2025). Gene–Lifestyle Interactions in Renal Dysfunction: Polygenic Risk Modulation via Plant-Based Diets, Coffee Intake, and Bioactive Compound Interactions. Nutrients, 17(5), 916. https://doi.org/10.3390/nu17050916






