Host Transcriptome and Microbial Variation in Relation to Visceral Hyperalgesia †
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Experimental GI Stressor
2.3. Microbiome Profiling
2.4. Whole Genome Gene Expression
2.5. Statistics
2.5.1. Data Analysis of Microbial Abundance, Gene Expression, and IVP
2.5.2. Data Analysis of Microbiome Profiles
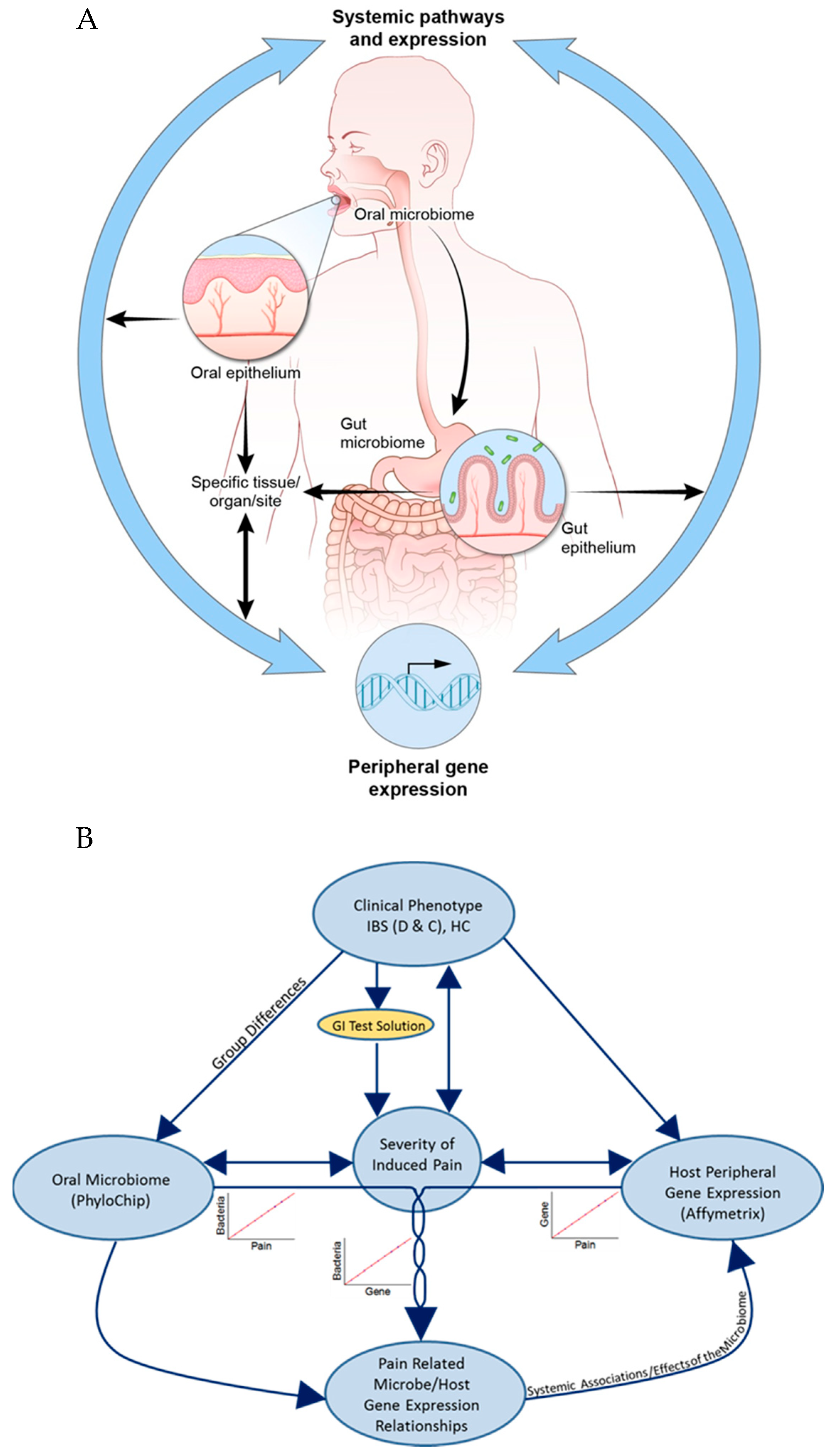
3. Results
3.1. Severity of Induced Visceral Pain (IVP)
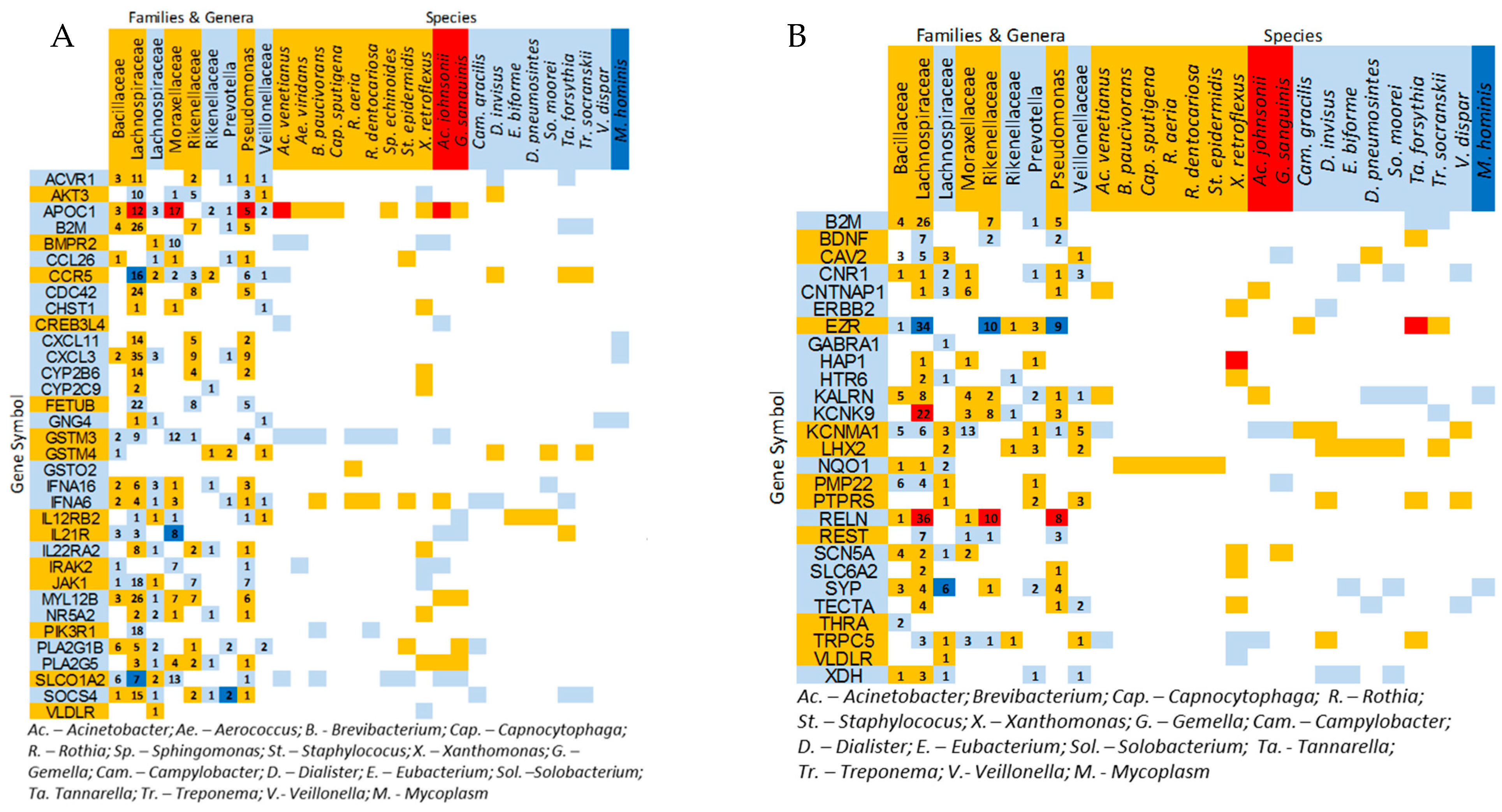
3.2. Microbial and Transcriptome Correlations with Induced Visceral Pain (IVP)
3.3. Correlations Between IVP-Correlated Genes and Microbes
3.4. Microbial Profiles Related to IBS and IBS-Subtypes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Camilleri, M.; Coulie, B.; Tack, J.F. Visceral hypersensitivity: Facts, speculations, and challenges. Gut 2001, 48, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Latremoliere, A.; Woolf, C.J. Central sensitization: A generator of pain hypersensitivity by central neural plasticity. J. Pain 2009, 10, 895–926. [Google Scholar] [CrossRef] [PubMed]
- Marchand, F.; Perretti, M.; McMahon, S.B. Role of the immune system in chronic pain. Nat. Rev. Neurosci. 2005, 6, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Guadagnoli, L.; Hoffert, Y.; Den Hond, S.; Dreesen, E.; van Ryckeghem, D.; Van Damme, S.; Zaman, J.; Van Oudenhove, L. Do we perceive sensations inside and outside of our body differently? Perceptual, emotional, and behavioral differences between visceral and somatic sensation, discomfort, and pain. Neurogastroenterol. Motil 2024, 36, e14787. [Google Scholar] [CrossRef]
- Basbaum, A.I.; Bautista, D.M.; Scherrer, G.; Julius, D. Cellular and molecular mechanisms of pain. Cell 2009, 139, 267–284. [Google Scholar] [CrossRef]
- Gold, M.S.; Gebhart, G.F. Nociceptor sensitization in pain pathogenesis. Nat. Med. 2010, 16, 1248–1257. [Google Scholar] [CrossRef]
- Stein, C.; Clark, J.D.; Oh, U.; Vasko, M.R.; Wilcox, G.L.; Overland, A.C.; Vanderah, T.W.; Spencer, R.H. Peripheral mechanisms of pain and analgesia. Brain Res. Rev. 2009, 60, 90–113. [Google Scholar] [CrossRef]
- Grenham, S.; Clarke, G.; Cryan, J.F.; Dinan, T.G. Brain-gut-microbe communication in health and disease. Front. Physiol. 2011, 2, 16175. [Google Scholar] [CrossRef]
- Mayer, E.A.; Savidge, T.; Shulman, R.J. Brain-gut microbiome interactions and functional bowel disorders. Gastroenterology 2014, 146, 1500–1512. [Google Scholar] [CrossRef]
- Kelly, J.R.; Borre, Y.; O’Brien, C.; Patterson, E.; El Aidy, S.; Deane, J.; Kennedy, P.J.; Beers, S.; Scott, K.; Moloney, G.; et al. Transferring the blues: Depression-associated gut microbiota induces neurobehavioural changes in the rat. J. Psychiatr. Res. 2016, 82, 109–118. [Google Scholar] [CrossRef]
- de Groot, P.F.; Frissen, M.N.; de Clercq, N.C.; Nieuwdorp, M. Fecal microbiota transplantation in metabolic syndrome: History, present and future. Gut Microbes 2017, 8, 253–267. [Google Scholar] [CrossRef] [PubMed]
- Drossman, D.A.; Camilleri, M.; Mayer, E.A.; Whitehead, W.E. AGA technical review on irritable bowel syndrome. Gastroenterology 2002, 123, 2108–2131. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, P.; Mowry, E.M. Gut microbiome and multiple sclerosis. Curr. Neurol. Neurosci. Rep. 2014, 14, 014–0492. [Google Scholar] [CrossRef] [PubMed]
- Bessac, A.; Cani, P.D.; Meunier, E.; Dietrich, G.; Knauf, C. Inflammation and Gut-Brain Axis During Type 2 Diabetes: Focus on the Crosstalk Between Intestinal Immune Cells and Enteric Nervous System. Front. Neurosci. 2018, 12, 725. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Garge, N.; Zhang, X.; Sun, W.; O’Connell, T.M.; Bunger, M.K.; Bultman, S.J. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011, 13, 517–526. [Google Scholar] [CrossRef]
- Ericsson, A.C.; Franklin, C.L. Manipulating the Gut Microbiota: Methods and Challenges. ILAR J. 2015, 56, 205–217. [Google Scholar] [CrossRef]
- Farzaei, M.H.; Bahramsoltani, R.; Abdollahi, M.; Rahimi, R. The Role of Visceral Hypersensitivity in Irritable Bowel Syndrome: Pharmacological Targets and Novel Treatments. J. Neurogastroenterol. Motil 2016, 22, 558–574. [Google Scholar] [CrossRef]
- Roediger, W.E. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut 1980, 21, 793–798. [Google Scholar] [CrossRef]
- Karakan, T.; Ozkul, C.; Kupeli Akkol, E.; Bilici, S.; Sobarzo-Sanchez, E.; Capasso, R. Gut-Brain-Microbiota Axis: Antibiotics and Functional Gastrointestinal Disorders. Nutrients 2021, 13, 389. [Google Scholar] [CrossRef]
- Shaikh, S.D.; Sun, N.; Canakis, A.; Park, W.Y.; Weber, H.C. Irritable Bowel Syndrome and the Gut Microbiome: A Comprehensive Review. J. Clin. Med. 2023, 12, 2558. [Google Scholar] [CrossRef]
- Yang, C.Q.; Guo, X.S.; Ji, L.; Wei, Z.B.; Zhao, L.; Zhao, G.T.; Sheng, S.T. Rifaximin Improves Visceral Hyperalgesia via TRPV1 by Modulating Intestinal Flora in the Water Avoidance Stressed Rat. Gastroenterol. Res. Pract. 2020, 2020, 4078681. [Google Scholar] [CrossRef] [PubMed]
- Flemer, B.; Warren, R.D.; Barrett, M.P.; Cisek, K.; Das, A.; Jeffery, I.B.; Hurley, E.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. The oral microbiota in colorectal cancer is distinctive and predictive. Gut 2018, 67, 1454–1463. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Yamashita, R.; Kawashima, J.; Mori, H.; Kurokawa, K.; Fukuda, S.; Gotoh, Y.; Nakamura, K.; Hayashi, T.; Kasahara, Y.; et al. Omics profiles of fecal and oral microbiota change in irritable bowel syndrome patients with diarrhea and symptom exacerbation. J. Gastroenterol. 2022, 57, 748–760. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Hu, Y.; Chen, J.; Su, C.; Zhang, Q.; Huang, C. Oral and fecal microbiota in patients with diarrheal irritable bowel syndrome. Heliyon 2023, 9, e13114. [Google Scholar] [CrossRef]
- Natividad, J.M.; Verdu, E.F. Modulation of intestinal barrier by intestinal microbiota: Pathological and therapeutic implications. Pharmacol. Res. 2013, 69, 42–51. [Google Scholar] [CrossRef]
- Slack, E.; Hapfelmeier, S.; Stecher, B.; Velykoredko, Y.; Stoel, M.; Lawson, M.A.; Geuking, M.B.; Beutler, B.; Tedder, T.F.; Hardt, W.D.; et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science 2009, 325, 617–620. [Google Scholar] [CrossRef]
- Docktor, M.J.; Paster, B.J.; Abramowicz, S.; Ingram, J.; Wang, Y.E.; Correll, M.; Jiang, H.; Cotton, S.L.; Kokaras, A.S.; Bousvaros, A. Alterations in diversity of the oral microbiome in pediatric inflammatory bowel disease. Inflamm. Bowel Dis. 2012, 18, 935–942. [Google Scholar] [CrossRef]
- Han, Y.W.; Wang, X. Mobile microbiome: Oral bacteria in extra-oral infections and inflammation. J. Dent. Res. 2013, 92, 485–491. [Google Scholar] [CrossRef]
- Rautava, J.; Pinnell, L.J.; Vong, L.; Akseer, N.; Assa, A.; Sherman, P.M. Oral Microbiome Composition Changes in Mouse Models of Colitis. J. Gastroenterol. Hepatol. 2014, 2, 12713. [Google Scholar] [CrossRef]
- Said, H.S.; Suda, W.; Nakagome, S.; Chinen, H.; Oshima, K.; Kim, S.; Kimura, R.; Iraha, A.; Ishida, H.; Fujita, J.; et al. Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res. 2014, 21, 15–25. [Google Scholar] [CrossRef]
- Wang, Y.; Xue, J.; Zhou, X.; You, M.; Du, Q.; Yang, X.; He, J.; Zou, J.; Cheng, L.; Li, M.; et al. Oral microbiota distinguishes acute lymphoblastic leukemia pediatric hosts from healthy populations. PLoS ONE 2014, 9, e102116. [Google Scholar] [CrossRef] [PubMed]
- Yeoh, N.; Burton, J.P.; Suppiah, P.; Reid, G.; Stebbings, S. The role of the microbiome in rheumatic diseases. Curr. Rheumatol. Rep. 2013, 15, 012–0314. [Google Scholar] [CrossRef]
- L’Heureux, J.E.; Corbett, A.; Ballard, C.; Vauzour, D.; Creese, B.; Winyard, P.G.; Jones, A.M.; Vanhatalo, A. Oral microbiome and nitric oxide biomarkers in older people with mild cognitive impairment and APOE4 genotype. PNAS Nexus 2025, 4, pgae543. [Google Scholar] [CrossRef] [PubMed]
- Arevalo-Caro, C.; Lopez, D.; Sanchez Milan, J.A.; Lorca, C.; Mulet, M.; Arboleda, H.; Losada Amaya, S.; Serra, A.; Gallart-Palau, X. Periodontal Indices as Predictors of Cognitive Decline: Insights from the PerioMind Colombia Cohort. Biomedicines 2025, 13, 205. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Schloss, P.D. Dynamics and associations of microbial community types across the human body. Nature 2014, 509, 357–360. [Google Scholar] [CrossRef]
- Del Valle-Pinero, A.Y.; Van Deventer, H.E.; Fourie, N.H.; Martino, A.C.; Patel, N.S.; Remaley, A.T.; Henderson, W.A. Gastrointestinal permeability in patients with irritable bowel syndrome assessed using a four probe permeability solution. Clin. Chim. Acta 2013, 418, 97–101. [Google Scholar] [CrossRef]
- Henderson, W.A.; Shankar, R.; Taylor, T.J.; Del Valle-Pinero, A.Y.; Kleiner, D.E.; Kim, K.H.; Youssef, N.N. Inverse relationship of interleukin-6 and mast cells in children with inflammatory and non-inflammatory abdominal pain phenotypes. World J. Gastrointest. Pathophysiol. 2012, 3, 102–108. [Google Scholar] [CrossRef]
- Ford, A.C.; Bercik, P.; Morgan, D.G.; Bolino, C.; Pintos-Sanchez, M.I.; Moayyedi, P. Validation of the Rome III criteria for the diagnosis of irritable bowel syndrome in secondary care. Gastroenterology 2013, 145, 1262–1270.e1. [Google Scholar] [CrossRef]
- Bertin, L.; Zanconato, M.; Crepaldi, M.; Marasco, G.; Cremon, C.; Barbara, G.; Barberio, B.; Zingone, F.; Savarino, E.V. The Role of the FODMAP Diet in IBS. Nutrients 2024, 16, 370. [Google Scholar] [CrossRef]
- Fourie, N.H.; Wang, D.; Abey, S.K.; Sherwin, L.B.; Joseph, P.V.; Rahim-Williams, B.; Ferguson, E.G.; Henderson, W.A. The microbiome of the oral mucosa in irritable bowel syndrome. Gut Microbes 2016, 7, 286–301. [Google Scholar] [CrossRef]
- Vacca, M.; Celano, G.; Calabrese, F.M.; Portincasa, P.; Gobbetti, M.; De Angelis, M. The Controversial Role of Human Gut Lachnospiraceae. Microorganisms 2020, 8, 573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Song, L.; Wang, Y.; Liu, C.; Zhang, L.; Zhu, S.; Liu, S.; Duan, L. Beneficial effect of butyrate-producing Lachnospiraceae on stress-induced visceral hypersensitivity in rats. J. Gastroenterol. Hepatol. 2019, 34, 1368–1376. [Google Scholar] [CrossRef]
- Gosalbes, M.J.; Durban, A.; Pignatelli, M.; Abellan, J.J.; Jimenez-Hernandez, N.; Perez-Cobas, A.E.; Latorre, A.; Moya, A. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS ONE 2011, 6, 0017447. [Google Scholar] [CrossRef] [PubMed]
- Kittelmann, S.; Seedorf, H.; Walters, W.A.; Clemente, J.C.; Knight, R.; Gordon, J.I.; Janssen, P.H. Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PLoS ONE 2013, 8, e47879. [Google Scholar] [CrossRef] [PubMed]
- Charrier, C.; Duncan, G.J.; Reid, M.D.; Rucklidge, G.J.; Henderson, D.; Young, P.; Russell, V.J.; Aminov, R.I.; Flint, H.J.; Louis, P. A novel class of CoA-transferase involved in short-chain fatty acid metabolism in butyrate-producing human colonic bacteria. Microbiology 2006, 152 Pt 1, 179–185. [Google Scholar] [CrossRef]
- Duncan, S.H.; Barcenilla, A.; Stewart, C.S.; Pryde, S.E.; Flint, H.J. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl. Environ. Microbiol. 2002, 68, 5186–5190. [Google Scholar] [CrossRef]
- Louis, P.; Duncan, S.H.; McCrae, S.I.; Millar, J.; Jackson, M.S.; Flint, H.J. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J. Bacteriol. 2004, 186, 2099–2106. [Google Scholar] [CrossRef]
- McIntyre, A.; Gibson, P.R.; Young, G.P. Butyrate production from dietary fibre and protection against large bowel cancer in a rat model. Gut 1993, 34, 386–391. [Google Scholar] [CrossRef]
- Pryde, S.E.; Duncan, S.H.; Hold, G.L.; Stewart, C.S.; Flint, H.J. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 2002, 217, 133–139. [Google Scholar] [CrossRef]
- Hijova, E.; Chmelarova, A. Short chain fatty acids and colonic health. Bratisl. Lek. Listy. 2007, 108, 354–358. [Google Scholar]
- Sun, C.Q.; O’Connor, C.J.; Turner, S.J.; Lewis, G.D.; Stanley, R.A.; Roberton, A.M. The effect of pH on the inhibition of bacterial growth by physiological concentrations of butyric acid: Implications for neonates fed on suckled milk. Chem. Biol. Interact. 1998, 113, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Zeng, A.P.; Ross, A.; Biebl, H.; Tag, C.; Gunzel, B.; Deckwer, W.D. Multiple product inhibition and growth modeling of clostridium butyricum and klebsiella pneumoniae in glycerol fermentation. Biotechnol. Bioeng. 1994, 44, 902–911. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Carroll, I.M.; Ringel-Kulka, T.; Keku, T.O.; Chang, Y.H.; Packey, C.D.; Sartor, R.B.; Ringel, Y. Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 301, G799–G807. [Google Scholar] [CrossRef]
- Taverniti, V.; Guglielmetti, S. Methodological issues in the study of intestinal microbiota in irritable bowel syndrome. World J. Gastroenterol. 2014, 20, 8821–8836. [Google Scholar]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef]
- Hofer, U. Microbiome: Pro-inflammatory Prevotella? Nat. Rev. Microbiol. 2014, 12, 25. [Google Scholar]
- Yakob, M.; Soder, B.; Meurman, J.H.; Jogestrand, T.; Nowak, J.; Soder, P.O. Prevotella nigrescens and Porphyromonas gingivalis are associated with signs of carotid atherosclerosis in subjects with and without periodontitis. J. Periodontal. Res. 2011, 46, 749–755. [Google Scholar] [CrossRef]
- Scher, J.U.; Sczesnak, A.; Longman, R.S.; Segata, N.; Ubeda, C.; Bielski, C.; Rostron, T.; Cerundolo, V.; Pamer, E.G.; Abramson, S.B.; et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013, 5, 01202. [Google Scholar] [CrossRef]
- Rigsbee, L.; Agans, R.; Shankar, V.; Kenche, H.; Khamis, H.J.; Michail, S.; Paliy, O. Quantitative profiling of gut microbiota of children with diarrhea-predominant irritable bowel syndrome. Am. J. Gastroenterol. 2012, 107, 1740–1751. [Google Scholar] [CrossRef]
- Francavilla, R.; Ercolini, D.; Piccolo, M.; Vannini, L.; Siragusa, S.; De Filippis, F.; De Pasquale, I.; Di Cagno, R.; Di Toma, M.; Gozzi, G.; et al. Salivary microbiota and metabolome associated with celiac disease. Appl. Environ. Microbiol. 2014, 80, 3416–3425. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, I.; Tap, J.; Roudot-Thoraval, F.; Roperch, J.P.; Letulle, S.; Langella, P.; Corthier, G.; Tran Van Nhieu, J.; Furet, J.P. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE 2011, 6, 0016393. [Google Scholar] [CrossRef] [PubMed]
- Yue, T.; Tan, H.; Wang, C.; Liu, Z.; Yang, D.; Ding, Y.; Xu, W.; Yan, J.; Zheng, X.; Weng, J.; et al. High-risk genotypes for type 1 diabetes are associated with the imbalance of gut microbiome and serum metabolites. Front. Immunol. 2022, 13, 1033393. [Google Scholar] [CrossRef]
- Lee, K.J.; Tack, J. Altered intestinal microbiota in irritable bowel syndrome. Neurogastroenterol. Motil 2010, 22, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Jumas-Bilak, E.; Jean-Pierre, H.; Carlier, J.P.; Teyssier, C.; Bernard, K.; Gay, B.; Campos, J.; Morio, F.; Marchandin, H. Dialister micraerophilus sp. nov. and Dialister propionicifaciens sp. nov., isolated from human clinical samples. Int. J. Syst. Evol. Microbiol. 2005, 55 Pt 6, 2471–2478. [Google Scholar] [CrossRef]
- Chavez, L.; Bais, A.S.; Vingron, M.; Lehrach, H.; Adjaye, J.; Herwig, R. In silico identification of a core regulatory network of OCT4 in human embryonic stem cells using an integrated approach. BMC Genom. 2009, 10, 1471–2164. [Google Scholar] [CrossRef]
- Schafer, K.H.; Van Ginneken, C.; Copray, S. Plasticity and neural stem cells in the enteric nervous system. Anat. Rec. 2009, 292, 1940–1952. [Google Scholar] [CrossRef]
- Obata, Y.; Pachnis, V. The Effect of Microbiota and the Immune System on the Development and Organization of the Enteric Nervous System. Gastroenterology 2016, 151, 836–844. [Google Scholar] [CrossRef]
- Posada-Quintero, H.F.; Kong, Y.; Chon, K.H. Objective pain stimulation intensity and pain sensation assessment using machine learning classification and regression based on electrodermal activity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2021, 321, R186–R196. [Google Scholar] [CrossRef]



| Chronic Visceral Hypersensitivity | Healthy Controls | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | Age | Race | Condition Subtype | IVP | Sex | Age | Race | Condition Subtype | IVP | ||
| n = 18 | Male | 44 | Caucasian | Diarrhea | 81 | n = 18 | Female | 30 | Caucasian | n/a | 10 |
| Male | 15 | Caucasian | Mixed | 77 | Female | 23 | Mixed/Other | n/a | 6.2 | ||
| Female | 30 | African-American | Diarrhea | 65 | Female | 24 | African-American | n/a | 0 | ||
| Male | 45 | African-American | Constipation | 64 | Male | 23 | Caucasian | n/a | 0 | ||
| Male | 26 | African-American | Diarrhea | 54 | Male | 40 | Caucasian | n/a | 0 | ||
| Female | 32 | African-American | Constipation | 48 | Male | 29 | Asian | n/a | 0 | ||
| Female | 26 | African-American | Constipation | 42 | Female | 33 | African-American | n/a | 0 | ||
| Female | 23 | African-American | Diarrhea | 29 | Female | 24 | Caucasian | n/a | 0 | ||
| Female | 27 | Caucasian | Diarrhea | 28 | Female | 24 | Caucasian | n/a | 0 | ||
| Female | 29 | African-American | Constipation | 27 | Male | 24 | African-American | n/a | 0 | ||
| Female | 24 | Caucasian | Diarrhea | 25 | Male | 31 | Caucasian | n/a | 0 | ||
| Male | 30 | Caucasian | Constipation | 16 | Female | 37 | African-American | n/a | 0 | ||
| Female | 26 | Asian | Diarrhea | 11 | Female | 21 | African-American | n/a | 0 | ||
| Female | 24 | Caucasian | Diarrhea | 11 | Female | 21 | Asian | n/a | 0 | ||
| Female | 24 | Caucasian | Constipation | 5 | Female | 16 | Mixed/Other | n/a | 0 | ||
| Male | 26 | Caucasian | Constipation | 4 | Female | 43 | Mixed/Other | n/a | 0 | ||
| Male | 31 | Asian | Mixed | 0 | Male | 22 | Caucasian | n/a | 0 | ||
| Female | 24 | Mixed/Other | Constipation | 0 | Female | 28 | African-American | n/a | 0 | ||
| Mean | 44% male | 28.11 | 44% Caucasian | 32.61 | Mean | 33% male | 27.39 | 38% Caucasian | 0.90 | ||
| SD | 7.09 | 38% African-American | 26.76 | SD | 7.17 | 33% African-American | 2.70 | ||||
| Canonical Pathway | p |
|---|---|
| Role of Oct4 in Mammalian Embryonic Stem Cell Pluripotency | 0.0004 |
| Pathogeneses of Multiple Sclerosis | 0.0007 |
| Human Embryonic Stem Cell Pluripotency | 0.002 |
| Role of NANOG in Mammalian Embryonic Stem Cell Pluripotency | 0.002 |
| Axonal Guidance Signaling | 0.005 |
| Aspartate Degradation II | 0.009 |
| IL-17A Signaling in Airway Cells | 0.01 |
| Virus Entry via Entry via Endocytic Pathways | 0.01 |
| Gα12/13 Signaling | 0.01 |
| OTUs Common in IBS but Uncommon in HC | |||||||
|---|---|---|---|---|---|---|---|
| Phylum | Family | Genus | Species | IBS (n = 18) | IBS-D (n = 7) | IBS-C (n = 9) | HC (n = 18) |
| Bacteroidetes | Flavobacteriaceae | unclassified | unclassified | 13 | 3 | 7 | 8 |
| Bacteroidetes | Prevotellaceae | Prevotella | unclassified | 6 | 2 | 2 | 1 |
| Bacteroidetes | Prevotellaceae | Prevotella | unclassified | 14 | 4 | 7 | 8 |
| Bacteroidetes | Prevotellaceae | Prevotella | unclassified | 15 | 5 | 7 | 9 |
| Bacteroidetes | Rikenellaceae | unclassified | unclassified | 6 | 2 | 4 | 1 |
| Bacteroidetes | Rikenellaceae | unclassified | unclassified | 15 | 4 | 9 * | 9 |
| Bacteroidetes | unclassified | unclassified | unclassified | 10 | 2 | 6 | 5 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 7 | 3 | 1 | 2 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 9 | 3 | 5 | 2 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 9 | 3 | 5 | 3 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 10 | 2 | 6 | 2 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 14 | 4 | 9 | 9 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 14 | 5 | 7 | 9 |
| Firmicutes | Ruminococcaceae | unclassified | unclassified | 9 | 2 | 6 | 3 |
| Firmicutes | Streptococcaceae | Streptococcus | lutetiensis | 11 | 2 | 8 | 4 |
| Firmicutes | unclassified | unclassified | unclassified | 8 | 2 | 4 | 2 |
| Firmicutes | unclassified | unclassified | unclassified | 8 | 3 | 4 | 2 |
| Fusobacteria | Fusobacteriaceae | Fusobacterium | unclassified | 6 | 2 | 3 | 0 * |
| Fusobacteria | Fusobacteriaceae | Fusobacterium | unclassified | 6 | 0 * | 6 | 1 |
| Proteobacteria | Alcaligenaceae | unclassified | unclassified | 6 | 3 | 2 | 1 |
| Proteobacteria | Chromatiaceae | unclassified | unclassified | 9 | 3 | 6 | 4 |
| Proteobacteria | Desulfobacteraceae | unclassified | unclassified | 15 | 5 | 9 * | 8 |
| Proteobacteria | Pseudomonadaceae | Pseudomonas | unclassified | 6 | 2 | 3 | 1 |
| Proteobacteria | unclassified | unclassified | unclassified | 6 | 1 | 3 | 1 |
| Proteobacteria | unclassified | unclassified | unclassified | 9 | 3 | 5 | 2 |
| Proteobacteria | unclassified | unclassified | unclassified | 9 | 4 | 3 | 3 |
| Spirochaetes | Spirochaetaceae | Treponema | unclassified | 8 | 1 | 4 | 3 |
| OTUs Common in HC but Uncommon in IBS | |||||||
|---|---|---|---|---|---|---|---|
| Phylum | Family | Genus | Species | IBS (n = 18) | IBS-D (n = 7) | IBS-C (n = 9) | HC (n = 18) |
| Actinobacteria | Coriobacteriaceae | unclassified | unclassified | 1 | 1 | 0 * | 6 |
| Actinobacteria | Micrococcaceae | Arthrobacter | unclassified | 3 | 1 | 2 | 8 |
| Actinobacteria | Streptomycetaceae | Streptomyces | unclassified | 8 | 1 | 6 | 13 |
| Bacteroidetes | Flavobacteriaceae | Capnocytophaga | ochracea | 6 | 3 | 3 | 13 |
| Bacteroidetes | Porphyromonadaceae | Porphyromonas | unclassified | 3 | 1 | 2 | 9 |
| Bacteroidetes | Rikenellaceae | unclassified | unclassified | 7 | 3 | 2 | 15 |
| Bacteroidetes | unclassified | unclassified | unclassified | 5 | 1 | 4 | 10 |
| Firmicutes | Bacillaceae | Bacillus | unclassified | 4 | 1 | 3 | 9 |
| Firmicutes | Bacillaceae | Bacillus | unclassified | 6 | 2 | 4 | 11 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 0 * | 0 * | 0 * | 5 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 0 * | 0 * | 0 * | 5 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 0 * | 0 * | 0 * | 6 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 1 | 0 * | 1 | 6 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 1 | 1 | 0 | 6 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 1 | 0 * | 1 | 6 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 2 | 2 | 0 * | 7 |
| Firmicutes | Lachnospiraceae | Coprococcus | unclassified | 2 | 1 | 1 | 8 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 3 | 1 | 2 | 9 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 3 | 0 * | 3 | 9 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 3 | 2 | 1 | 10 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 4 | 1 | 3 | 10 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 6 | 4 | 1 | 11 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 4 | 0 * | 4 | 12 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 8 | 3 | 5 | 13 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 9 | 3 | 5 | 14 |
| Firmicutes | Lachnospiraceae | unclassified | unclassified | 13 | 4 | 8 | 18 * |
| Firmicutes | Streptococcaceae | Streptococcus | unclassified | 3 | 1 | 2 | 8 |
| Firmicutes | Streptococcaceae | Streptococcus | gordonii | 10 | 4 | 6 | 15 |
| Firmicutes | unclassified | unclassified | unclassified | 3 | 1 | 2 | 8 |
| Firmicutes | Veillonellaceae | Dialister | invisus | 11 | 4 | 5 | 16 |
| Proteobacteria | Burkholderiaceae | unclassified | unclassified | 3 | 2 | 1 | 8 |
| Proteobacteria | Campylobacteraceae | Campylobacter | unclassified | 5 | 1 | 4 | 11 |
| Proteobacteria | Neisseriaceae | Neisseria | unclassified | 6 | 1 | 5 | 11 |
| Proteobacteria | Pasteurellaceae | Haemophilus | parainfluenzae | 3 | 1 | 2 | 9 |
| Proteobacteria | Pseudomonadaceae | Pseudomonas | unclassified | 2 | 1 | 1 | 7 |
| Proteobacteria | Pseudomonadaceae | Pseudomonas | unclassified | 5 | 2 | 3 | 10 |
| Proteobacteria | Pseudomonadaceae | Pseudomonas | unclassified | 7 | 2 | 4 | 12 |
| Proteobacteria | Pseudomonadaceae | Pseudomonas | unclassified | 13 | 3 | 9 * | 18 * |
| Proteobacteria | unclassified | unclassified | unclassified | 1 | 0 * | 1 | 7 |
| Verrucomicrobia | unclassified | unclassified | unclassified | 0 * | 0 * | 0 * | 5 |
| WS3 | PRR-10 | unclassified | unclassified | 9 | 2 | 7 | 16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, C.J.; Prescott, S.; Fourie, N.H.; Abey, S.K.; Sherwin, L.B.; Rahim-Williams, B.; Joseph, P.V.; Posada-Quintero, H.; Hoffman, R.K.; Henderson, W.A. Host Transcriptome and Microbial Variation in Relation to Visceral Hyperalgesia. Nutrients 2025, 17, 921. https://doi.org/10.3390/nu17050921
Costa CJ, Prescott S, Fourie NH, Abey SK, Sherwin LB, Rahim-Williams B, Joseph PV, Posada-Quintero H, Hoffman RK, Henderson WA. Host Transcriptome and Microbial Variation in Relation to Visceral Hyperalgesia. Nutrients. 2025; 17(5):921. https://doi.org/10.3390/nu17050921
Chicago/Turabian StyleCosta, Christopher J., Stephanie Prescott, Nicolaas H. Fourie, Sarah K. Abey, LeeAnne B. Sherwin, Bridgett Rahim-Williams, Paule V. Joseph, Hugo Posada-Quintero, Rebecca K. Hoffman, and Wendy A. Henderson. 2025. "Host Transcriptome and Microbial Variation in Relation to Visceral Hyperalgesia" Nutrients 17, no. 5: 921. https://doi.org/10.3390/nu17050921
APA StyleCosta, C. J., Prescott, S., Fourie, N. H., Abey, S. K., Sherwin, L. B., Rahim-Williams, B., Joseph, P. V., Posada-Quintero, H., Hoffman, R. K., & Henderson, W. A. (2025). Host Transcriptome and Microbial Variation in Relation to Visceral Hyperalgesia. Nutrients, 17(5), 921. https://doi.org/10.3390/nu17050921









