Cholesterol-Lowering Mechanism of Lactobacillus Bile Salt Hydrolase Through Regulation of Bifidobacterium pseudolongum in the Gut Microbiota
Abstract
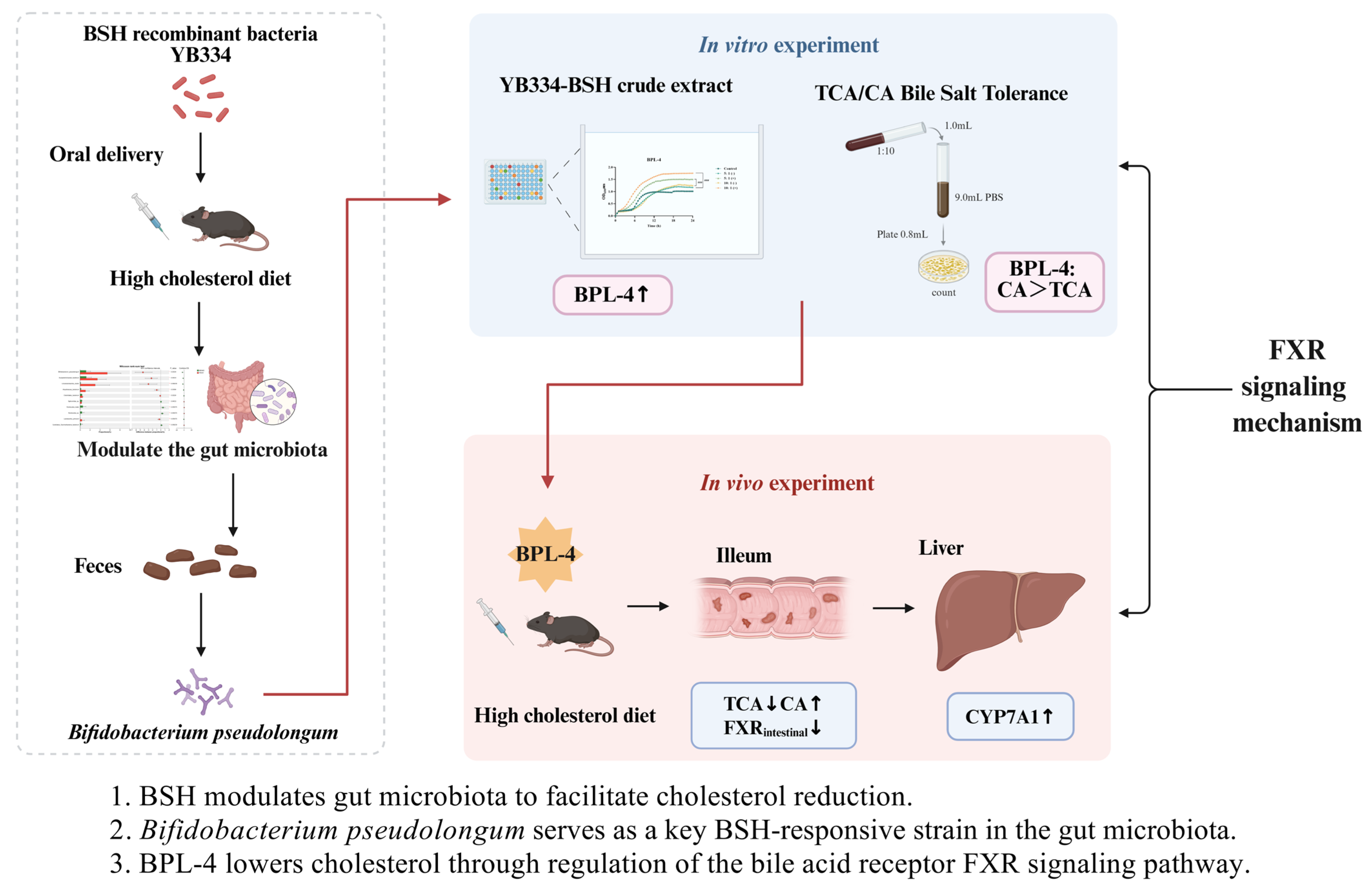
1. Introduction
2. Materials and Methods
2.1. Experimental Strain and Culture
2.2. Animal Experimentation
- Recombinant lactic acid bacteria (NB5462 and YB334): Single colonies were inoculated into liquid medium and cultured overnight for 12 h. The cultures were then diluted in fresh MRS medium to an OD600 nm of approximately 0.1, and incubated at 37 °C until the OD600 nm reached around 0.3. Protein expression was induced by adding the SppIP inducer peptide (amino acid sequence: MAGNSSNFIHKIKQIFTHR; Genscript Biotechnology Co., Ltd., Nanjing, China) at a final concentration of 50 ng/mL. Cultivation continued until an OD600 nm of 2.5 was achieved. The cells were harvested by centrifugation, washed, and resuspended in PBS for storage at 4 °C.
- Bifidobacterium pseudolongum BPL-4: A single colony was inoculated into liquid medium and cultured for 16 h overnight. A 1% (v/v) inoculum of this seed culture was transferred to 45 mL of fresh, modified MRS broth supplemented with mupirocin lithium salt and cysteine hydrochloride. After 12 h of incubation, the bacterial cells were collected by centrifugation. The pellet was resuspended in 10% sterile SM and stored at 4 °C.
2.3. Serum Biochemical Parameter Analysis
2.4. Liver Histological Analysis
2.5. Metagenomics Analysis
2.6. Isolation and Identification of B. pseudolongum in Mouse Feces
2.7. BSH Activity Assays
2.8. Effect of Addition of Crude Extract of YB334-BSH Enzyme on the Growth of Different Strains of Bacteria
2.9. TCA/CA Bile Salt Tolerance
2.10. RNA Isolation and RT-qPCR
2.11. Statistical Analysis
3. Results
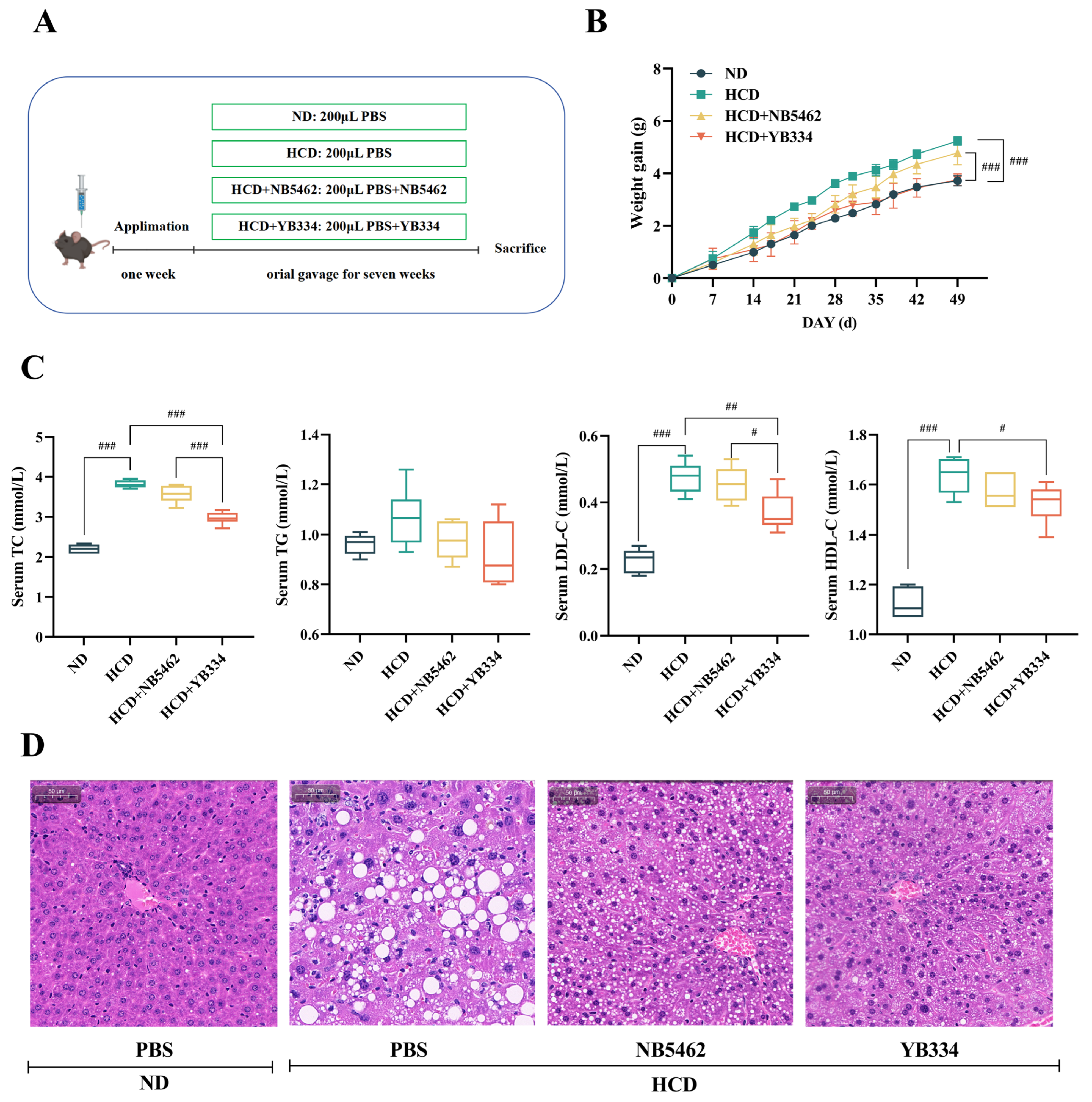
3.1. BSH Recombinant Bacterium YB334 Has Cholesterol-Lowering Properties
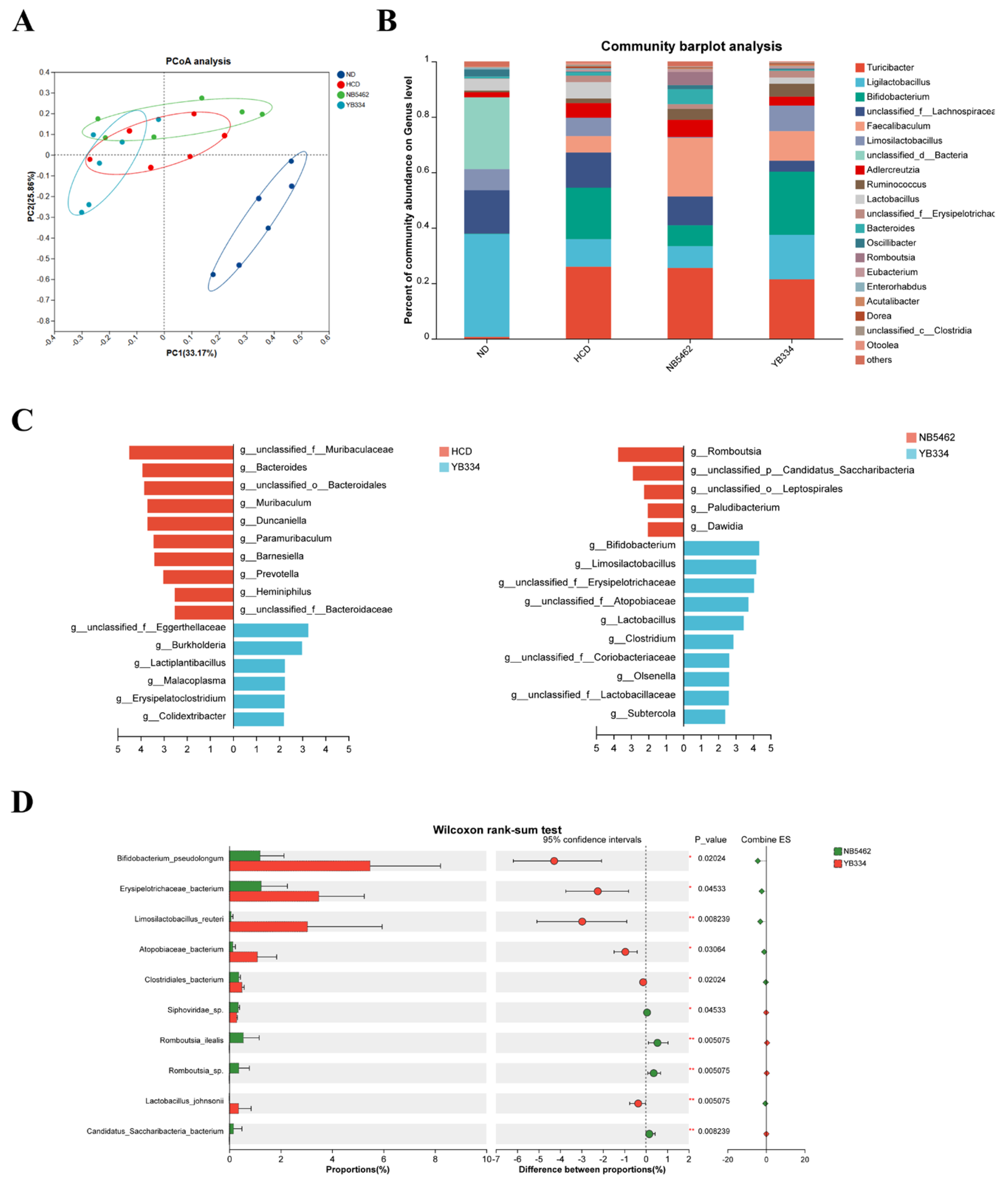
3.2. BSH of Recombinant Bacterium YB334 Regulates Intestinal Flora Distribution
3.3. Screening of B. pseudolongum BPL and Determination of BSH Activity
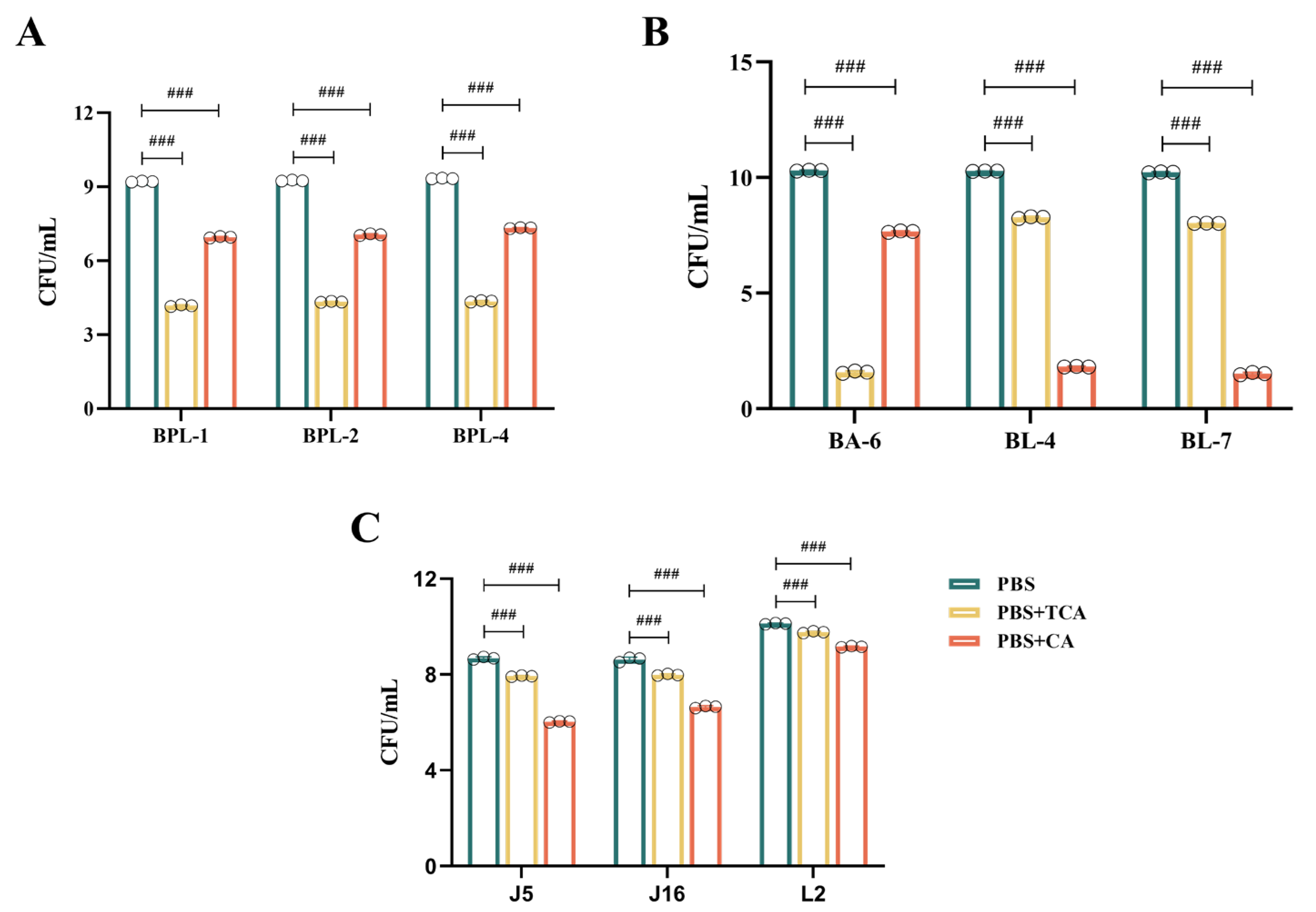
3.4. Effect of Crude Extract of Recombinant Bacterium YB334-BSH on the Growth of Different Strains
- Figure 3A illustrates that, in contrast to the control group, the culture media infused with the YB334-BSH crude extract and inducer markedly enhanced the proliferation of many strains of B. pseudolongum, particularly when administered at a 10:1 supernatant-to-medium ration, demonstrating a significant growth-promoting impact, rising from 106 CFU/mL to 107 CFU/mL. The BPL-4 strain (accession No. PX061997) showed the most significant growth-promoting effects (Figure 3D). Conversely, the uninduced extract did not significantly influence the proliferation of B. pseudolongum;
- AS shown in Figure 3B, crude extracts of YB334-BSH, whether induced or non-induced, did not significantly enhance the development of B. adolescentis BA-6 and B. longum BL-4 and BL-7;
- As shown in Figure 3C, the induced YB334-BSH crude extract showed inhibitory effects on L. johnsonii J5 and J16, as well as L. reuteri L2, decreasing their counts from 107 CFU/mL to 106 CFU/mL.
3.5. Effect of Different Strains on Tolerance to TCA/CA Bile Salts
- Figure 4A illustrates that the B. pseudolongum BPL-1, BPL-2, and BPL-4 displayed a notable tolerance to TCA, with a decline from 109 CFU/mL to 104 CFU/mL; however, they exhibited superior tolerance to CA, with a decrease from 109 CFU/mL to 107 CFU/mL;
- As shown in Figure 4B, B. adolescentis BA-6 was highly sensitive to TCA, declining from 1010 CFU/mL to 101 CFU/mL, while it exhibited relatively good tolerance to CA, decreasing from 1010 CFU/mL to 107 CFU/mL. Conversely, B. longum BL-4 and BL-7 demonstrated poor tolerance to CA, diminishing from 1010 CFU/mL to 101 CFU/mL, but moderate tolerance to TCA, decreasing from 1010 CFU/mL to 108 CFU/mL;
- Figure 4C illustrates that L. johnsonii J5 and J16, along with L. reuteri L2, demonstrate a notable resistance to both TCA and CA. TCA concentrations diminished from 108 CFU/mL to 107 CFU/mL, from 108 CFU/mL to 107 CFU/mL, and from 1010 CFU/mL to 109 CFU/mL, respectively. CA diminished from 108 CFU/mL to 106 CFU/mL, from 108 CFU/mL to 106 CFU/mL, and from 1010 CFU/mL to 109 CFU/mL, respectively. They also demonstrated the highest tolerance to TCA.
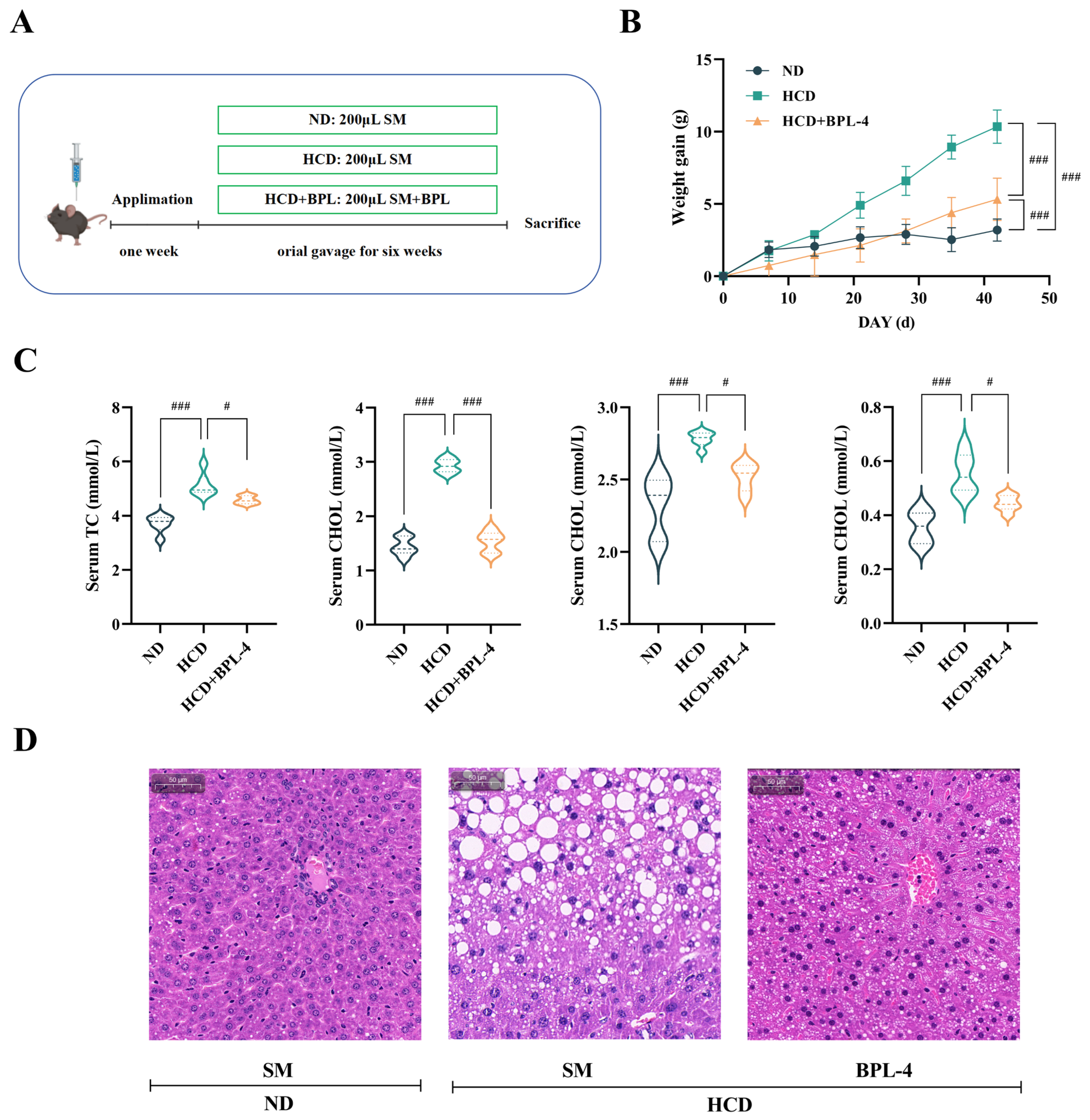
3.6. B. pseudolongum BPL-4 Has Cholesterol-Lowering Effects
3.7. BSH Indirectly Regulates Intestinal FXR in B. pseudolongum BPL-4
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BSH | Bile salt hydrolase |
| FXR | Farnesoid X receptor |
| CYP7A1 | Cholesterol 7α-hydroxylase |
| GF | Germ-free |
| SPF | Specific pathogen-free |
| MRS | de Man, Rogosa and Sharpe |
| ND | Normal diet |
| HCD | High-cholesterol diet |
| PBS | Phosphate-buffered saline |
| SM | Skim milk |
| HDL-C | High-density lipoprotein cholesterol |
| LDL-C | Low-density lipoprotein cholesterol |
| TC | Total cholesterol |
| TG | Triglycerides |
| H&E | Hematoxylin and eosin |
| PCR | Polymerase chain reaction |
| BLAST | Basic local alignment search tool |
| GCA | Glycocholic acid |
| TCA | Taurocholic acid |
| LC-MS/MS | Liquid chromatography-tandem mass spectrometry |
| CA | Cholic acid |
| RT-qPCR | Quantitative reverse transcription polymerase chain reaction |
| PCoA | Principal coordinates analysis |
| LefSe | Linear discriminant analysis effect size |
| LDA | Linear discriminant analysis |
| FDR | False discovery rate |
| NCBI | National center for biotechnology information |
| SHP | Short heterodimer partner |
| FGF15 | Fibroblast growth factor 15 |
| IBABP | Intestinal bile acid binding protein |
| CYP27A1 | Sterol 27-hydroxylase |
| CYP8B1 | Sterol 12α-hydroxylase |
| CAT | Carvacrol and thymol |
| cGMP-PKG | Cyclic guanosine monophosphate-protein kinase G pathway |
| DSS-induced | Dextran sulfate sodium-induced |
| T-β-MCA | Tauro-β-muricholic acid |
References
- Martin, S.S.; Aday, A.W.; Almarzooq, Z.I.; Anderson, C.A.M.; Arora, P.; Avery, C.L.; Baker-Smith, C.M.; Barone Gibbs, B.; Beaton, A.Z.; Boehme, A.K.; et al. 2024 Heart Disease and Stroke Statistics: A Report of US and Global Data From the American Heart Association. Circulation 2024, 149, e347–e913. [Google Scholar] [CrossRef]
- Sharma, K.; Mohan, S.; Hossain, S.A.; Shah, S.; Konat, A.; Shah, K.; Mehta, S.; Tavethia, J.J.; Sarvaiya, J.N.; Joshi, S.; et al. Prevalence of traditional cardiovascular risk factors in high-risk Kyrgyzstan population as compared to Indians—An Indo-Kyrgyz cardiometabolic study. J. Fam. Med. Prim. Care 2024, 13, 5621–5625. [Google Scholar] [CrossRef]
- Baigent, C.; Landray, M.J.; Reith, C.; Emberson, J.; Wheeler, D.C.; Tomson, C.; Wanner, C.; Krane, V.; Cass, A.; Craig, J.; et al. The effects of lowering LDL cholesterol with simvastatin plus ezetimibe in patients with chronic kidney disease (Study of Heart and Renal Protection): A randomised placebo-controlled trial. Lancet 2011, 377, 2181–2192. [Google Scholar] [CrossRef]
- Xu, Z.; Arnold, M.; Stevens, D.; Kaptoge, S.; Pennells, L.; Sweeting, M.J.; Barrett, J.; Di Angelantonio, E.; Wood, A.M. Prediction of Cardiovascular Disease Risk Accounting for Future Initiation of Statin Treatment. Am. J. Epidemiol. 2021, 190, 2000–2014. [Google Scholar] [CrossRef]
- Li, Y.; Chen, M.; Ma, Y.; Yang, Y.; Cheng, Y.; Ma, H.; Ren, D.; Chen, P. Regulation of viable/inactivated/lysed probiotic Lactobacillus plantarum H6 on intestinal microbiota and metabolites in hypercholesterolemic mice. NPJ Sci. Food 2022, 6, 50. [Google Scholar] [CrossRef]
- Puttarat, N.; Kasorn, A.; Vitheejongjaroen, P.; Chantarangkul, C.; Tangwattanachuleeporn, M.; Taweechotipatr, M. Beneficial Effects of Indigenous Probiotics in High-Cholesterol Diet-Induced Hypercholesterolemic Rats. Nutrients 2023, 15, 2710. [Google Scholar] [CrossRef]
- Reis, S.A.; Conceição, L.L.; Rosa, D.D.; Siqueira, N.P.; Peluzio, M.C.G. Mechanisms responsible for the hypocholesterolaemic effect of regular consumption of probiotics. Nutr. Res. Rev. 2017, 30, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.B.; Lew, L.C.; Yeo, S.K.; Nair Parvathy, S.; Liong, M.T. Probiotics and the BSH-related cholesterol lowering mechanism: A Jekyll and Hyde scenario. Crit. Rev. Biotechnol. 2015, 35, 392–401. [Google Scholar] [CrossRef]
- Joyce, S.A.; Gahan, C.G. Bile Acid Modifications at the Microbe-Host Interface: Potential for Nutraceutical and Pharmaceutical Interventions in Host Health. Annu. Rev. Food Sci. Technol. 2016, 7, 313–333. [Google Scholar] [CrossRef] [PubMed]
- Horáčková, Š.; Plocková, M.; Demnerová, K. Importance of microbial defence systems to bile salts and mechanisms of serum cholesterol reduction. Biotechnol. Adv. 2018, 36, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Foley, M.H.; O’Flaherty, S.; Barrangou, R.; Theriot, C.M. Bile salt hydrolases: Gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract. PLoS Pathog. 2019, 15, e1007581. [Google Scholar] [CrossRef]
- Dong, Z.; Lee, B.H. Bile salt hydrolases: Structure and function, substrate preference, and inhibitor development. Protein Sci. 2018, 27, 1742–1754. [Google Scholar] [CrossRef]
- Joyce, S.A.; Shanahan, F.; Hill, C.; Gahan, C.G. Bacterial bile salt hydrolase in host metabolism: Potential for influencing gastrointestinal microbe-host crosstalk. Gut Microb. 2014, 5, 669–674. [Google Scholar] [CrossRef]
- Huang, F.; Zheng, X.; Ma, X.; Jiang, R.; Zhou, W.; Zhou, S.; Zhang, Y.; Lei, S.; Wang, S.; Kuang, J.; et al. Theabrownin from Pu-erh tea attenuates hypercholesterolemia via modulation of gut microbiota and bile acid metabolism. Nat. Commun. 2019, 10, 4971. [Google Scholar] [CrossRef]
- Wang, G.; Huang, W.; Xia, Y.; Xiong, Z.; Ai, L. Cholesterol-lowering potentials of Lactobacillus strain overexpression of bile salt hydrolase on high cholesterol diet-induced hypercholesterolemic mice. Food Funct. 2019, 10, 1684–1695. [Google Scholar] [CrossRef]
- Miremadi, F.; Sherkat, F.; Stojanovska, L. Hypocholesterolaemic effect and anti-hypertensive properties of probiotics and prebiotics: A review. J. Funct. Foods 2016, 25, 497–510. [Google Scholar] [CrossRef]
- Zhu, H.; Zhao, F.; Zhang, W.; Xia, W.; Chen, Y.; Liu, Y.; Fan, Z.; Zhang, Y.; Yang, Y. Cholesterol-lowering effect of bile salt hydrolase from a Lactobacillus johnsonii strain mediated by FXR pathway regulation. Food Funct. 2022, 13, 725–736. [Google Scholar] [CrossRef]
- Joyce, S.A.; MacSharry, J.; Casey, P.G.; Kinsella, M.; Murphy, E.F.; Shanahan, F.; Hill, C.; Gahan, C.G. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci. USA 2014, 111, 7421–7426. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Seaton, S.C.; Ndousse-Fetter, S.; Adhikari, A.A.; DiBenedetto, N.; Mina, A.I.; Banks, A.S.; Bry, L.; Devlin, A.S. A selective gut bacterial bile salt hydrolase alters host metabolism. Elife 2018, 7, e37182. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Kuang, W.; Yang, J.; Liu, Y.; Yang, M.; Chen, Y.; Zhu, H.; Yang, Y. Cholesterol lowering in diet-induced hypercholesterolemic mice using Lactobacillus bile salt hydrolases with different substrate specificities. Food Funct. 2024, 15, 1340–1354. [Google Scholar] [CrossRef] [PubMed]
- Simpson, P.J.; Fitzgerald, G.F.; Stanton, C.; Ross, R.P. The evaluation of a mupirocin-based selective medium for the enumeration of bifidobacteria from probiotic animal feed. J. Microbiol. Methods 2004, 57, 9–16. [Google Scholar] [CrossRef]
- Nair, P.P.; Annapure, U.S. Fermentation dynamics of bile salt hydrolase production in Heyndrickxia coagulans ATCC 7050 and Lactiplantibacillus plantarum ATCC 10012: Addressing ninhydrin assay limitations with a novel HPTLC-MS method. J. Microbiol. Methods 2024, 226, 107050. [Google Scholar] [CrossRef] [PubMed]
- Frappier, M.; Auclair, J.; Bouasker, S.; Gunaratnam, S.; Diarra, C.; Millette, M. Screening and Characterization of Some Lactobacillaceae for Detection of Cholesterol-Lowering Activities. Probiotics Antimicrob. Proteins 2022, 14, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Zhang, F.; Yuan, W.; Wei, Y.; Wei, M.; Zhou, Y.; Yang, Y.; Chang, Y.; Wu, X. Hepatic cholesterol accumulation ascribed to the activation of ileum Fxr-Fgf15 pathway inhibiting hepatic Cyp7a1 in high-fat diet-induced obesity rats. Life Sci. 2019, 232, 116638. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Huang, D.; Dong, Z.; Wang, X.; Ning, M.; Xia, J.; Shen, S.; Wu, S.; Shi, Y.; Wang, J.; et al. FXR Signaling-Mediated Bile Acid Metabolism Is Critical for Alleviation of Cholesterol Gallstones by Lactobacillus Strains. Microbiol. Spectr. 2022, 10, e0051822. [Google Scholar] [CrossRef]
- Sayin, S.I.; Wahlström, A.; Felin, J.; Jäntti, S.; Marschall, H.U.; Bamberg, K.; Angelin, B.; Hyötyläinen, T.; Orešič, M.; Bäckhed, F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013, 17, 225–235. [Google Scholar] [CrossRef]
- De Gottardi, A.; Touri, F.; Maurer, C.A.; Perez, A.; Maurhofer, O.; Ventre, G.; Bentzen, C.L.; Niesor, E.J.; Dufour, J.F. The bile acid nuclear receptor FXR and the bile acid binding protein IBABP are differently expressed in colon cancer. Dig. Dis. Sci. 2004, 49, 982–989. [Google Scholar] [CrossRef]
- Jin, L.; Dang, H.; Wu, J.; Yuan, L.; Chen, X.; Yao, J. Supplementation of Weizmannia coagulans BC2000 and Ellagic Acid Inhibits High-Fat-Induced Hypercholesterolemia by Promoting Liver Primary Bile Acid Biosynthesis and Intestinal Cholesterol Excretion in Mice. Microorganisms 2023, 11, 264. [Google Scholar] [CrossRef]
- Jayakrishnan, T.T.; Sangwan, N.; Barot, S.V.; Farha, N.; Mariam, A.; Xiang, S.; Aucejo, F.; Conces, M.; Nair, K.G.; Krishnamurthi, S.S.; et al. Multi-omics machine learning to study host-microbiome interactions in early-onset colorectal cancer. NPJ Precis. Oncol. 2024, 8, 146. [Google Scholar] [CrossRef]
- Jia, B.; Zou, Y.; Han, X.; Bae, J.W.; Jeon, C.O. Gut microbiome-mediated mechanisms for reducing cholesterol levels: Implications for ameliorating cardiovascular disease. Trends Microbiol. 2023, 31, 76–91. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, R.; Song, J.; Sohaib, M.; Wang, S.; Mao, J.; Qi, J.; Xiong, X.; Zhou, W.; Guo, L. Limosilactobacillus reuteri consumption significantly reduces the total cholesterol concentration without affecting other cardiovascular disease risk factors in adults: A systematic review and meta-analysis. Nutr. Res. 2023, 117, 1–14. [Google Scholar] [CrossRef]
- Morinaga, K.; Kusada, H.; Tamaki, H. Bile Salt Hydrolases with Extended Substrate Specificity Confer a High Level of Resistance to Bile Toxicity on Atopobiaceae Bacteria. Int. J. Mol. Sci. 2022, 23, 10980. [Google Scholar] [CrossRef]
- Çetin, B.; Aktaş, H. Monitoring probiotic properties and safety evaluation of antilisterial Enterococcus faecium strains with cholesterol-lowering potential from raw Cow’s milk. Food Biosci. 2024, 16, 104532. [Google Scholar] [CrossRef]
- Wang, G.; Yu, H.; Feng, X.; Tang, H.; Xiong, Z.; Xia, Y.; Ai, L.; Song, X. Specific bile salt hydrolase genes in Lactobacillus plantarum AR113 and relationship with bile salt resistance. LWT Food Sci. Technol. 2021, 145, 111208. [Google Scholar] [CrossRef]
- Li, F.; Jiang, C.; Krausz, K.W.; Li, Y.; Albert, I.; Hao, H.; Fabre, K.M.; Mitchell, J.B.; Patterson, A.D.; Gonzalez, F.J. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat. Commun. 2013, 4, 2384. [Google Scholar] [CrossRef] [PubMed]
- Miyata, M.; Tanaka, T.; Takahashi, K.; Funaki, A.; Sugiura, Y. Cholesterol-lowering effects of taurine through the reduction of ileal FXR signaling due to the alteration of ileal bile acid composition. Amino Acids 2021, 10, 1523–1532. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Wang, H.; Bukhari, I.; Zhao, Y.; Huang, H.; Yu, Y.; Sun, X.; Mi, Y.; Mei, L.; Zheng, P. Effects of cholesterol-lowering probiotics on non-alcoholic fatty liver disease in FXR gene knockout mice. Front. Nutr. 2023, 10, 1121203. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Zhang, X.; Liu, W.; Wei, H.; Liang, W.; Zhou, Y.; Ding, Y.; Ji, F.; Cheung, A.H.K.; Wong, N.; et al. Bifidobacterium pseudolongum-generated acetate suppresses non-alcoholic fatty liver disease-associated hepatocellular carcinoma. J. Hepatol. 2023, 79, 1352–1365. [Google Scholar] [CrossRef]
- Zhang, K.; Xu, Y.; Zheng, Y.; Zhang, T.; Wu, Y.; Yan, Y.; Lei, Y.; Cao, X.; Wang, X.; Yan, F.; et al. Bifidobacterium pseudolongum-Derived Bile Acid from Dietary Carvacrol and Thymol Supplementation Attenuates Colitis via cGMP-PKG-mTORC1 Pathway. Adv. Sci 2024, 11, e2406917. [Google Scholar] [CrossRef]
- Kusada, H.; Arita, M.; Tohno, M.; Tamaki, H. Isolation of a Highly Thermostable Bile Salt Hydrolase With Broad Substrate Specificity From Lactobacillus paragasseri. Front. Microbiol. 2022, 13, 810872. [Google Scholar] [CrossRef]


| Strains | Descriptions a | Source or Reference |
|---|---|---|
| Lactobacillus johnsonii | ||
| YH334 | Wide type | Laboratory screening [17] |
| J5 | Wide type | This study |
| J16 | Wide type | This study |
| Lactiplantibacillus plantarum | ||
| YB334 | Emr, the pSIP334 plasmid was introduced into strain WCFS1Δbsh | Laboratory construction [17] |
| NB5462 | Emr, the pSIP334 plasmid was introduced into strain WCFS1Δbsh | Laboratory construction [17] |
| Bifidobacterium pseudolongum | ||
| BPL | Wide type | This study |
| Bifidobacterium adolescentis | ||
| BA-6 | Wide type | This study |
| Bifidobacterium longum | ||
| BL-4 | Wide type | This study |
| BL-7 | Wide type | This study |
| Limosilactobacillus reuteri | ||
| L2 | Wide type | This study |
| Primer | Sequence (5′-3′) | Reference |
|---|---|---|
| FXR-F | GGAACTCCGGACATTCAAC | [24] |
| FXR-R | GTGTCCATCACTGCACATC | |
| SHP-F | TCCTAGCCAAGACAGTAGCCTTCC | [25] |
| SHP-R | TACCGCTGCTGGCTTCCTCTAG | |
| CYP7A1-F | GCTAAGACGCACCTCGTGATCC | |
| CYP7A1-R | CCGCAGAGCCTCCTTGATGATG | |
| FGF15-F | CGGTCGCTCTGAAGACGATTGC | |
| FGF15-R | TACATCCTCCACCATCCTGAACGG | |
| CYP27A1-F | ATTAAGGAGACCCTGCGCCT | |
| CYP27A1-R | AGGCAAGACCGAACCCCATA | |
| CYP8B1-F | AAGGCTGGCTTCCTGAGCTT | [26] |
| CYP8B1-R | AACAGCTCATCGGCCTCATC | |
| IBABP-F | GGCCCGCAACTTCAAGATC | [27] |
| IBABP-R | TAGTGCTGGGACCAAGTGAAGTC | |
| rpL32-F | TCTGGTCCACAACGTCAAGG | [17] |
| rpL32-R | GGATTGGTGACTCTGATGGC |
| Strain | Enzyme Activity (U/mL) | |
|---|---|---|
| GCA | TCA | |
| BPL-1 | 0.84 ± 0.06 *** | 1.83 ± 0.07 *** |
| BPL-2 | 0.44 ± 0.03 *** | 1.37 ± 0.03 *** |
| BPL-4 | 2.11 ± 0.05 *** | 2.53 ± 0.04 *** |
| BPL-5 | 0.84 ± 0.02 *** | 1.93 ± 0.07 *** |
| BPL-8 | 0.95 ± 0.11 *** | 1.90 ± 0.07 *** |
| BPL-9 | 0.93 ± 0.02 *** | 1.60 ± 0.03 *** |
| BPL-10 | 1.24 ± 0.04 *** | 2.96 ± 0.08 *** |
| BPL-11 | 0.60 ± 0.08 *** | 1.71 ± 0.07 *** |
| NB5462 | 0.07 ± 0.01 | 0.07 ± 0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Kuang, W.; Li, M.; Wang, Z.; Liu, Y.; Zhao, M.; Huan, H.; Yang, Y. Cholesterol-Lowering Mechanism of Lactobacillus Bile Salt Hydrolase Through Regulation of Bifidobacterium pseudolongum in the Gut Microbiota. Nutrients 2025, 17, 3019. https://doi.org/10.3390/nu17183019
Liu Y, Kuang W, Li M, Wang Z, Liu Y, Zhao M, Huan H, Yang Y. Cholesterol-Lowering Mechanism of Lactobacillus Bile Salt Hydrolase Through Regulation of Bifidobacterium pseudolongum in the Gut Microbiota. Nutrients. 2025; 17(18):3019. https://doi.org/10.3390/nu17183019
Chicago/Turabian StyleLiu, Yingying, Weijia Kuang, Man Li, Zhihao Wang, Yanrong Liu, Menghuan Zhao, Hailin Huan, and Yao Yang. 2025. "Cholesterol-Lowering Mechanism of Lactobacillus Bile Salt Hydrolase Through Regulation of Bifidobacterium pseudolongum in the Gut Microbiota" Nutrients 17, no. 18: 3019. https://doi.org/10.3390/nu17183019
APA StyleLiu, Y., Kuang, W., Li, M., Wang, Z., Liu, Y., Zhao, M., Huan, H., & Yang, Y. (2025). Cholesterol-Lowering Mechanism of Lactobacillus Bile Salt Hydrolase Through Regulation of Bifidobacterium pseudolongum in the Gut Microbiota. Nutrients, 17(18), 3019. https://doi.org/10.3390/nu17183019






