Probiotic Consortium Confers Synergistic Anti-Inflammatory Effects in Inflammatory Disorders
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Probiotic Strains
2.3. Colonization of Bacteria in Germ Free Mice
2.4. Isolation of Lymphocytes and Flow Cytometry Analysis
2.5. Flow Cytometry Analysis
2.6. Induction of Experimental Atopic Dermatitis
2.7. Histology
2.8. Induction of Experimental Colitis through Adoptive Transfer of CD4+ T Cells
2.9. Bacterial 16S rRNA Sequencing
2.10. Statistical Analysis
3. Results
3.1. MPRO Consortium Exhibits More Immunoregulatory Potential Compared to Individual Bacterial Species
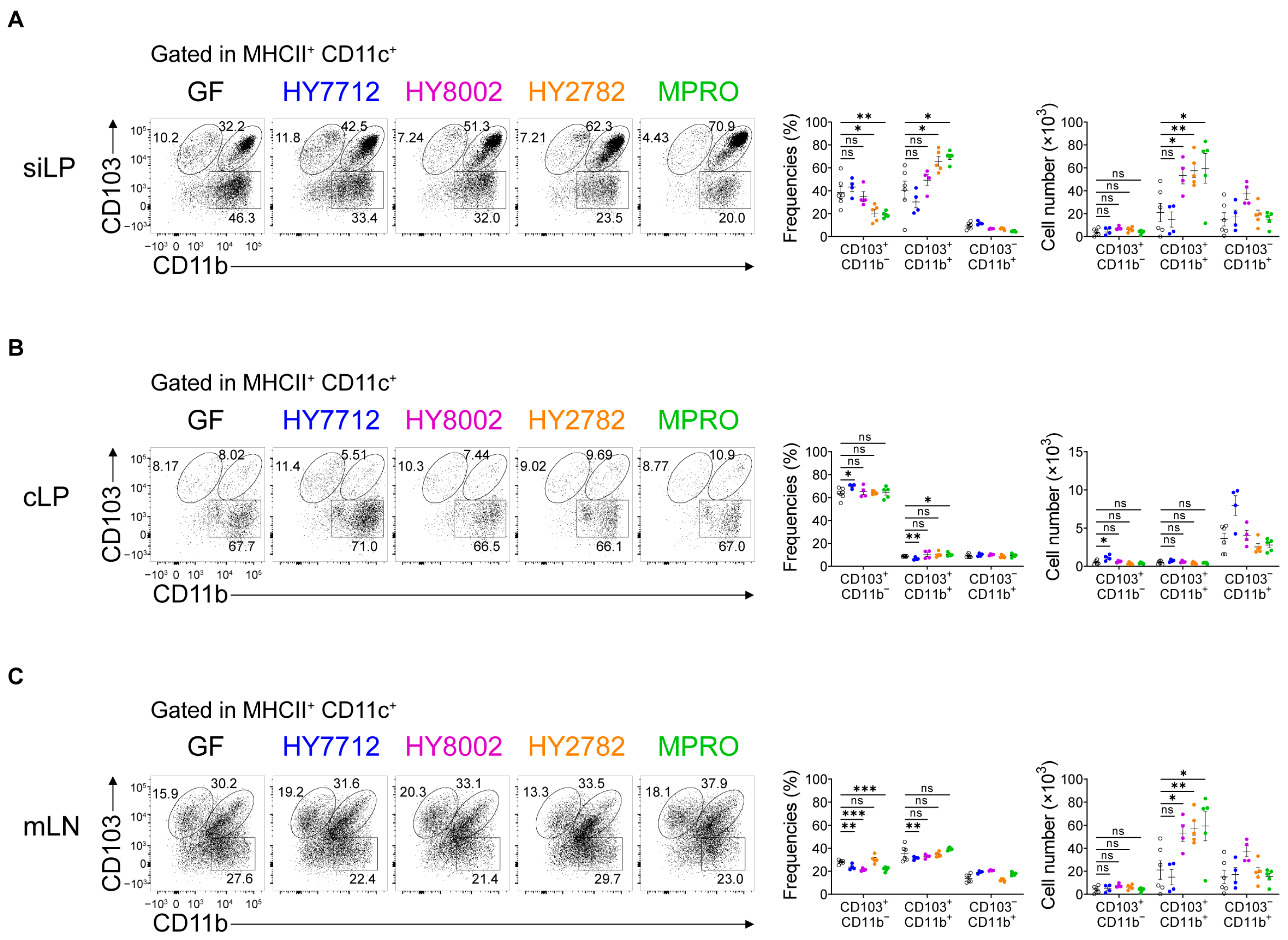
3.2. MPRO Treatment Increases the CD103+CD11b+ Regulatory Dendritic Cells in the Intestines
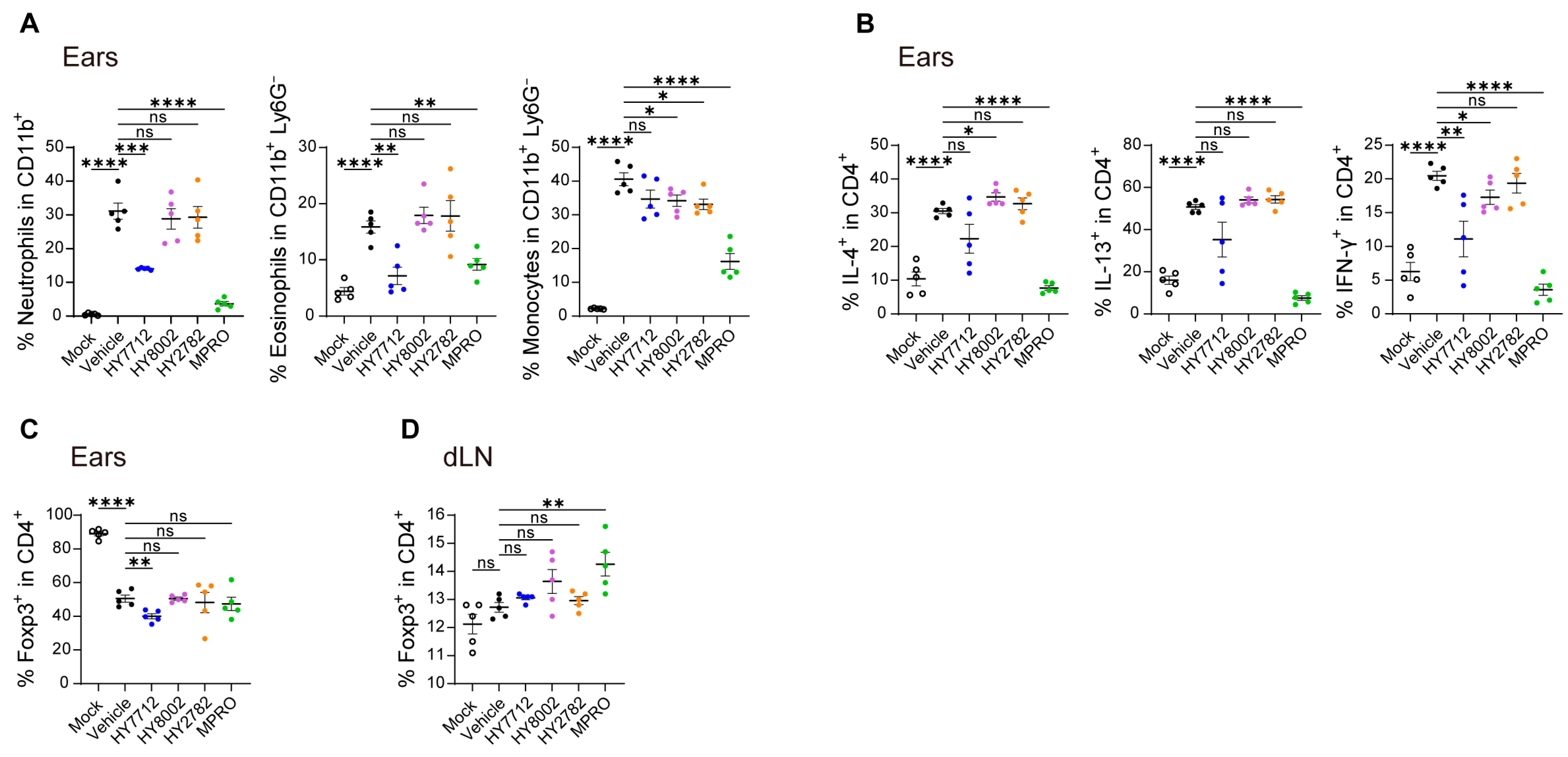
3.3. Administration of MPRO Confers Better Prophylactic Protection against Experimental Atopic Dermatitis Compared to Individual Bacterial Components
3.4. MPRO Demonstrates a Heightened Regulatory Impact on the Immune System in Experimental AD
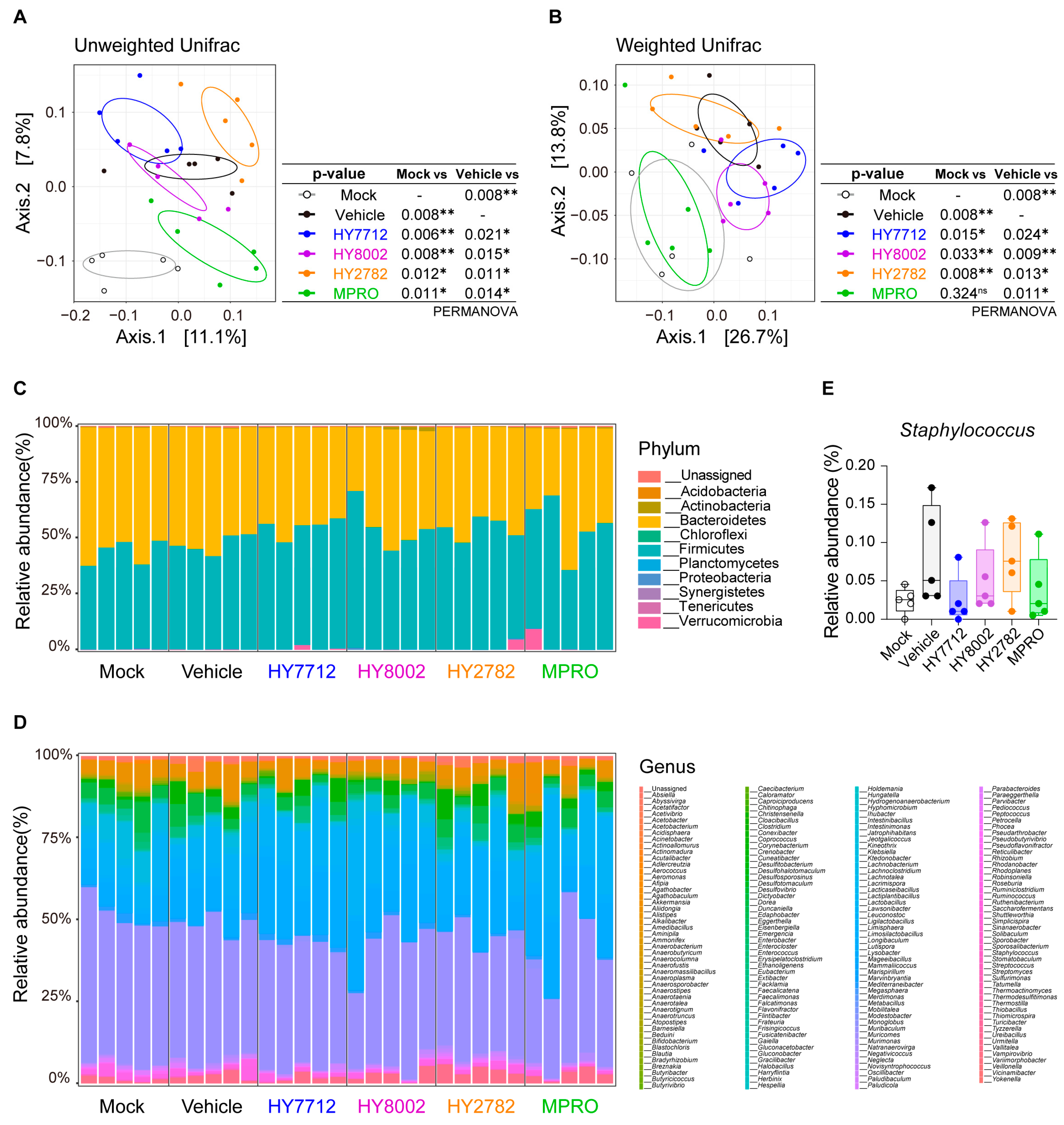
3.5. MPRO Alleviates Experimental Atopic Dermatitis by Reshaping the Microbiome Landscape
3.6. MPRO Administration Ameliorates Experimental Inflammatory Colitis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carabotti, M.; Scirocco, A.; Maselli, M.A.; Severi, C. The gut-brain axis: Interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 2015, 28, 203–209. [Google Scholar] [PubMed]
- Kamada, N.; Seo, S.U.; Chen, G.Y.; Nunez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 2013, 13, 321–335. [Google Scholar] [CrossRef]
- Pascal, M.; Perez-Gordo, M.; Caballero, T.; Escribese, M.M.; Lopez Longo, M.N.; Luengo, O.; Manso, L.; Matheu, V.; Seoane, E.; Zamorano, M.; et al. Microbiome and Allergic Diseases. Front. Immunol. 2018, 9, 1584. [Google Scholar] [CrossRef] [PubMed]
- Pflughoeft, K.J.; Versalovic, J. Human microbiome in health and disease. Annu. Rev. Pathol. 2012, 7, 99–122. [Google Scholar] [CrossRef]
- Cristofori, F.; Dargenio, V.N.; Dargenio, C.; Miniello, V.L.; Barone, M.; Francavilla, R. Anti-Inflammatory and Immunomodulatory Effects of Probiotics in Gut Inflammation: A Door to the Body. Front. Immunol. 2021, 12, 578386. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef]
- Verma, R.; Lee, C.; Jeun, E.J.; Yi, J.; Kim, K.S.; Ghosh, A.; Byun, S.; Lee, C.G.; Kang, H.J.; Kim, G.C.; et al. Cell surface polysaccharides of Bifidobacterium bifidum induce the generation of Foxp3(+) regulatory T cells. Sci. Immunol. 2018, 3, eaat6975. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Liu, B.; Huang, L.; Li, Z.; Cheng, Y.; Tian, Y.; Pan, G.; Li, H.; Xu, Y.; Wu, W.; et al. Probiotic Consortia and Their Metabolites Ameliorate the Symptoms of Inflammatory Bowel Diseases in a Colitis Mouse Model. Microbiol. Spectr. 2022, 10, e0065722. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef]
- Kang, H.J.; Kim, G.C.; Lee, C.G.; Park, S.; Sharma, G.; Verma, R.; Im, S.H.; Kwon, H.K. Probiotics-derived metabolite ameliorates skin allergy by promoting differentiation of FOXP3(+) regulatory T cells. J. Allergy Clin. Immunol. 2021, 147, 1517–1521. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.K.; Lee, C.G.; So, J.S.; Chae, C.S.; Hwang, J.S.; Sahoo, A.; Nam, J.H.; Rhee, J.H.; Hwang, K.C.; Im, S.H. Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc. Natl. Acad. Sci. USA 2010, 107, 2159–2164. [Google Scholar] [CrossRef] [PubMed]
- Chapman, C.M.; Gibson, G.R.; Rowland, I. Health benefits of probiotics: Are mixtures more effective than single strains? Eur. J. Nutr. 2011, 50, 1–17. [Google Scholar] [CrossRef] [PubMed]
- McFarland, L.V. Efficacy of Single-Strain Probiotics Versus Multi-Strain Mixtures: Systematic Review of Strain and Disease Specificity. Dig. Dis. Sci. 2021, 66, 694–704. [Google Scholar] [CrossRef] [PubMed]
- van der Lelie, D.; Oka, A.; Taghavi, S.; Umeno, J.; Fan, T.J.; Merrell, K.E.; Watson, S.D.; Ouellette, L.; Liu, B.; Awoniyi, M.; et al. Rationally designed bacterial consortia to treat chronic immune-mediated colitis and restore intestinal homeostasis. Nat. Commun. 2021, 12, 3105. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Xu, T.; Zhang, Y.; Zheng, T.; He, Y.; He, F.; Jiang, Y. Long-term combined administration of Bifidobacterium bifidum TMC3115 and Lactobacillus plantarum 45 alleviates spatial memory impairment and gut dysbiosis in APP/PS1 mice. FEMS Microbiol. Lett. 2020, 367, fnaa048. [Google Scholar] [CrossRef]
- Bieber, T. Atopic dermatitis: An expanding therapeutic pipeline for a complex disease. Nat. Rev. Drug Discov. 2022, 21, 21–40. [Google Scholar] [CrossRef]
- Stander, S. Atopic Dermatitis. N. Engl. J. Med. 2021, 384, 1136–1143. [Google Scholar] [CrossRef]
- Renert-Yuval, Y.; Thyssen, J.P.; Bissonnette, R.; Bieber, T.; Kabashima, K.; Hijnen, D.; Guttman-Yassky, E. Biomarkers in atopic dermatitis-a review on behalf of the International Eczema Council. J. Allergy Clin. Immunol. 2021, 147, 1174–1190. [Google Scholar] [CrossRef]
- Sroka-Tomaszewska, J.; Trzeciak, M. Molecular Mechanisms of Atopic Dermatitis Pathogenesis. Int. J. Mol. Sci. 2021, 22, 4130. [Google Scholar] [CrossRef]
- Lee, S.Y.; Lee, E.; Park, Y.M.; Hong, S.J. Microbiome in the Gut-Skin Axis in Atopic Dermatitis. Allergy Asthma Immunol. Res. 2018, 10, 354–362. [Google Scholar] [CrossRef]
- Alam, M.J.; Xie, L.; Yap, Y.A.; Marques, F.Z.; Robert, R. Manipulating Microbiota to Treat Atopic Dermatitis: Functions and Therapies. Pathogens 2022, 11, 642. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, Z.; Liu, S.; Zhang, D. Global, regional and national burden of inflammatory bowel disease in 204 countries and territories from 1990 to 2019: A systematic analysis based on the Global Burden of Disease Study 2019. BMJ Open 2023, 13, e065186. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef] [PubMed]
- de Souza, H.S.; Fiocchi, C. Immunopathogenesis of IBD: Current state of the art. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Ng, S.C. The Gut Microbiota in the Pathogenesis and Therapeutics of Inflammatory Bowel Disease. Front. Microbiol. 2018, 9, 2247. [Google Scholar] [CrossRef] [PubMed]
- Coombes, J.L.; Siddiqui, K.R.; Arancibia-Carcamo, C.V.; Hall, J.; Sun, C.M.; Belkaid, Y.; Powrie, F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J. Exp. Med. 2007, 204, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Ruane, D.T.; Lavelle, E.C. The role of CD103(+) dendritic cells in the intestinal mucosal immune system. Front. Immunol. 2011, 2, 25. [Google Scholar] [CrossRef]
- Scott, C.L.; Aumeunier, A.M.; Mowat, A.M. Intestinal CD103+ dendritic cells: Master regulators of tolerance? Trends Immunol. 2011, 32, 412–419. [Google Scholar] [CrossRef]
- Bain, C.C.; Schridde, A. Origin, Differentiation, and Function of Intestinal Macrophages. Front. Immunol. 2018, 9, 2733. [Google Scholar] [CrossRef]
- Na, Y.R.; Stakenborg, M.; Seok, S.H.; Matteoli, G. Macrophages in intestinal inflammation and resolution: A potential therapeutic target in IBD. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 531–543. [Google Scholar] [CrossRef]
- Kim, J.E.; Kim, H.S. Microbiome of the Skin and Gut in Atopic Dermatitis (AD): Understanding the Pathophysiology and Finding Novel Management Strategies. J. Clin. Med. 2019, 8, 444. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Du, X.; Zhai, S.; Tang, X.; Liu, C.; Li, W. Gut microbiota and atopic dermatitis in children: A scoping review. BMC Pediatr. 2022, 22, 323. [Google Scholar] [CrossRef] [PubMed]
- Wagner, C.L.; Taylor, S.N.; Johnson, D. Host factors in amniotic fluid and breast milk that contribute to gut maturation. Clin. Rev. Allergy Immunol. 2008, 34, 191–204. [Google Scholar] [CrossRef]
- Ostanin, D.V.; Pavlick, K.P.; Bharwani, S.; D’Souza, D.; Furr, K.L.; Brown, C.M.; Grisham, M.B. T cell-induced inflammation of the small and large intestine in immunodeficient mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 290, G109–G119. [Google Scholar] [CrossRef] [PubMed]
- Powrie, F.; Leach, M.W.; Mauze, S.; Menon, S.; Caddle, L.B.; Coffman, R.L. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity 1994, 1, 553–562. [Google Scholar] [CrossRef]
- Konieczna, P.; Ferstl, R.; Ziegler, M.; Frei, R.; Nehrbass, D.; Lauener, R.P.; Akdis, C.A.; O’Mahony, L. Immunomodulation by Bifidobacterium infantis 35624 in the murine lamina propria requires retinoic acid-dependent and independent mechanisms. PLoS ONE 2013, 8, e62617. [Google Scholar] [CrossRef]
- Mu, Q.; Tavella, V.J.; Luo, X.M. Role of Lactobacillus reuteri in Human Health and Diseases. Front. Microbiol. 2018, 9, 757. [Google Scholar] [CrossRef]
- Ochoa-Reparaz, J.; Mielcarz, D.W.; Ditrio, L.E.; Burroughs, A.R.; Begum-Haque, S.; Dasgupta, S.; Kasper, D.L.; Kasper, L.H. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J. Immunol. 2010, 185, 4101–4108. [Google Scholar] [CrossRef]
- Nam, W.; Kim, H.; Bae, C.; Kim, J.; Nam, B.; Lee, Y.; Kim, J.; Park, S.; Lee, J.; Sim, J. Lactobacillus HY2782 and Bifidobacterium HY8002 Decrease Airway Hyperresponsiveness Induced by Chronic PM2.5 Inhalation in Mice. J. Med. Food 2020, 23, 575–583. [Google Scholar] [CrossRef]
- Park, I.J.; Lee, J.H.; Kye, B.H.; Oh, H.K.; Cho, Y.B.; Kim, Y.T.; Kim, J.Y.; Sung, N.Y.; Kang, S.B.; Seo, J.M.; et al. Effects of PrObiotics on the Symptoms and Surgical ouTComes after Anterior REsection of Colon Cancer (POSTCARE): A Randomized, Double-Blind, Placebo-Controlled Trial. J. Clin. Med. 2020, 9, 2181. [Google Scholar] [CrossRef] [PubMed]
- Stagg, A.J. Intestinal Dendritic Cells in Health and Gut Inflammation. Front. Immunol. 2018, 9, 2883. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Nguyen, A.; Gommerman, J.L. Dendritic Cell Subsets in Intestinal Immunity and Inflammation. J. Immunol. 2020, 204, 1075–1083. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, C.; Kim, S.W.; Verma, R.; Noh, J.; Park, J.C.; Park, S.; Lee, H.; Park, H.E.; Kim, C.J.; Byun, S.; et al. Probiotic Consortium Confers Synergistic Anti-Inflammatory Effects in Inflammatory Disorders. Nutrients 2024, 16, 790. https://doi.org/10.3390/nu16060790
Lee C, Kim SW, Verma R, Noh J, Park JC, Park S, Lee H, Park HE, Kim CJ, Byun S, et al. Probiotic Consortium Confers Synergistic Anti-Inflammatory Effects in Inflammatory Disorders. Nutrients. 2024; 16(6):790. https://doi.org/10.3390/nu16060790
Chicago/Turabian StyleLee, Changhon, Seung Won Kim, Ravi Verma, Jaegyun Noh, John Chulhoon Park, Sunhee Park, Haena Lee, Hye Eun Park, Chan Johng Kim, Seohyun Byun, and et al. 2024. "Probiotic Consortium Confers Synergistic Anti-Inflammatory Effects in Inflammatory Disorders" Nutrients 16, no. 6: 790. https://doi.org/10.3390/nu16060790
APA StyleLee, C., Kim, S. W., Verma, R., Noh, J., Park, J. C., Park, S., Lee, H., Park, H. E., Kim, C. J., Byun, S., Ko, H., Choi, S., Kim, I., Jeon, S., Lee, J., & Im, S.-H. (2024). Probiotic Consortium Confers Synergistic Anti-Inflammatory Effects in Inflammatory Disorders. Nutrients, 16(6), 790. https://doi.org/10.3390/nu16060790





