Abstract
The evidence suggests that diet can modulate endogenous microRNA (miRNA) expression. Changes in miRNA expression may affect metabolic processes and consequently be involved in health status and disease development. The aim of this systematic review was to summarize the evidence of the role of diet and specific food components in the regulation of miRNA expression and discuss its implications for human health and disease development. The PubMed, Embase and Web of Science databases were searched in accordance with the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines for relevant studies. A total of 32 interventional and 5 observational studies performed in adults and evaluating dietary modulation of miRNA expression were included. Energy- and fat-controlled diets along with plant-based foods show substantial evidence of modulating endogenous miRNA levels. Plasma, serum and peripheral blood mononuclear cells (PBMCs) are the main sources used to measure miRNAs. A total of 108 miRNAs modulated by diet were identified. We confirmed that dietary habits are closely associated with the modulation of endogenous miRNAs. Particularly, energy content and fat intake appeared to be key factors influencing miRNA levels. Furthermore, since miRNAs are involved in the regulation of several biological processes, this modulatory process may affect health status and lead to metabolic disorders.
1. Introduction
Noncommunicable diseases (NCDs) such as obesity, type 2 diabetes and cardiovascular disorders have become a health problem of epidemic proportions. According to the World Health Organization (WHO), they are responsible for 71% of all deaths worldwide, corresponding to 41 million people per year, of which 15 million are premature deaths between the ages 30 and 69 [1]. Cardiovascular disease, cancer, respiratory diseases and diabetes are among the NCDs with the highest incidence [1]. Regarding the associated risk factors, along with tobacco, alcohol and physical inactivity, unhealthy dietary habits play a critical role, yet it is worth mentioning that all of these are preventable lifestyle aspects [2]. In recent decades, traditional diets have undergone a westernized shift toward overeating and the abuse of highly processed foods and added sugars, leading consequently to the exacerbation of NCDs [3]. On the other hand, NCD risk can be minimized or prevented by following healthy dietary habits, particularly when they are focused on normocaloric plant-based patterns [4], including a Mediterranean diet [5].
One of the reasons why diet influences the development of diseases is the participation of certain food components in the regulation of the metabolic processes involved [6,7]. Besides providing energy and nutrients, diet also contains bioactive compounds which can modulate biological processes, having an impact on health status. Recent data show that vitamins, polyphenols and fatty acids, as well as specific dietary patterns, regulate metabolism according to mechanisms involving the modulation of endogenous microRNAs (miRNAs) [8] and their expression [9,10,11]. Besides that, the evidence suggests that miRNAs contained in foods can also be absorbed during the digestive process and consequently interact with host gene expression [12,13]. A study assessing the presence of plant miRNAs in the serum of Chinese healthy adults, whose diet primarily consists of rice, has reported the detection of 30 exogenous miRNAs. Among these, significant levels of ath-miR-156a, ath-miR-166a and osa-miR-168a have been found, three miRNAs primarily derived from rice and cruciferous vegetables [12]. In another study that conducted bioinformatics analysis of data from four human small RNA libraries, 35 exogenous miRNAs have been detected in human milk exosomes, with the highest abundance levels observed for the plant-derived miRNAs ath-miR-166a, pab-miR-951, ptc-miR-472a and bdi-miR-168 [13]. Therefore, these studies suggest the potential capability of mature miRNAs to reach human plasma from the gastrointestinal tract and subsequently regulate the expression of human target genes. Due to the potential impact of miRNA modulation on human health and disease development [14,15,16,17], a full understanding of the interplay between diet, miRNAs and their health effects is needed.
miRNAs are endogenous small noncoding RNA sequences of approximately 22 nucleotides in length, which play a key role in the posttranscriptional regulation of gene expression [18]. During their biosynthesis, most miRNAs are first transcribed in the nucleus by RNA polymerase II (Pol II) into large pri-miRNA transcripts, which are cleaved into stem-loops of 70 nucleotides called pre-miRNAs by a complex formed by the RNase III enzyme DROSHA and the double-stranded RNA binding protein DiGeorge syndrome critical region gene 8 (DGCR8). Then, the pre-miRNAs are exported to the cytoplasm by Exportin 5 and cleaved into small double-stranded miRNAs 18–24 nucleotides long by the RNase III enzyme DICER, which is associated with TAR RNA binding protein (TRBP). These miRNA duplexes bind to argonaute proteins, assisted by ATP-dependent chaperone proteins. Subsequently, one of the strands is removed and degraded and the other one is loaded into the RNA-induced silencing complex (RISC), a ribonucleoprotein complex that intervenes in the recognition of the targeted mRNA [9,18]. Finally, this mature form of miRNA is guided to the 3′ UTR of the mRNAs through base pairing, leading to decreased mRNA stability and the repression of mRNA’s translation of target genes [19]. Potentially, each miRNA can modulate the expression of more than one target mRNA, and one mRNA can be modulated by several miRNAs, denoting an intricate miRNA–mRNA interaction network [20]. Additionally, miRNAs can be secreted out of cells and be stably transported into extracellular fluids associated with several carriers, including extracellular vesicles, ribonucleoproteins and lipoprotein complexes [21]. Consequently, miRNAs may have endocrine, paracrine and autocrine regulatory functions, contributing to cell-to-cell communication and participating in essential regulatory pathways involving apoptosis, differentiation, development, proliferation or signal transduction processes [19]. Therefore, disruption of the proper communication carried by miRNAs to cells has been related to the development of chronic disorders [21], including cardiovascular diseases [22,23], type 2 diabetes [24,25] and obesity [26,27].
In this context, animal studies have been successful in demonstrating the capacity of diet to regulate miRNA [28,29,30]. Although the evidence in humans is not as clear as in animal models, some studies have established a clear association between food intake and endogenous miRNAs. The aim of this systematic review is to summarize the evidence of the role of diet and specific food components in the modulation of miRNA expression and to discuss its implications for human health and disease development.
2. Materials and Methods
This systematic review was carried out following the recommendations of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement guidelines [31].
The PubMed, Embase and Web of Science (WOS) databases were manually searched on August 2023 to collect human studies which evaluated the impact of diet on miRNA expression. The search strategy used in PubMed and adapted to Embase and WOS was as follows: (diet OR dietary pattern* OR food OR intake OR dietary OR exogenous) AND (mirna* OR microrna*).
An initial selection of the returned articles was made by checking their suitability by evaluating their titles and keywords and then their abstracts. After this scrutiny, the full texts of the articles were analyzed to select those meeting the eligibility criteria (Table 1).

Table 1.
Eligibility criteria of the systematic review.
All the articles included in this review were summarized. The most relevant information was extracted and sorted according to the following items: (a) first author and publication date, (b) population of the study and (c) health status, (d) study design, (e) dietary strategy, (f) analytical method used to detect/quantify miRNA, (g) type of sample analyzed, (h) outcomes of the study.
Next, a network analysis was performed to characterize the biological functions of the miRNAs identified with further evidence on their association with diet. The miRNet 2.0 software [32] was used for the identification of target genes and, subsequently, for the determination of miRNA–function and miRNA–disease interactions. miRNAs with evidence in at least two studies were included in the network analysis, all of them with the prefix corresponding to the Homo sapiens (hsa-) species: hsa-mir-19b (MI0000074), -mir-19b-3p (MIMAT0000074), -mir-20a-5p (MIMAT0000075), -mir-21-5p (MIMAT0000076), -mir-29a-3p (MIMAT0000086), -mir-29b-3p (MIMAT0000100), -mir-92a (MI0000093), -mir-99a (MI0000101), -mir-99b (MI0000746), -mir-106a (MI0000113), -mir-106b (MI0000734), -mir-122 (MI0000442), -mir-122-5p (MIMAT0000421), -mir-130b (MI0000748), -mir-142-3p (MIMAT0000434), -mir-142-5p (MIMAT0000433), -mir-181a-5p (MIMAT0000256), -mir-181b-5p (MIMAT0000257), -mir-192 (MI0000234), -mir-192-5p (MIMAT0000222), -mir-221 (MI0000298), -mir-223 (MI0000300), -mir-328 (MI0000804), -mir-339-5p (MIMAT0000764), -mir-375 (MI0000783), -mir-411 (MI0003675), -mir-935 (MI0005757), -mir-1260b (MI0014197), -let-7b (MI0000063), -let-7c (MI0000064), -let-7f-5p (MIMAT0000067. The level of significance was set at p < 0.05.
3. Results
3.1. Characterization of the Included Studies
A total of 17,006 records were initially identified through the database search. The removal of records with a poor match with the search strategy turned out 10,148 entries. After the title and abstract screening, 216 manuscripts were considered suitable for eligibility. Finally, 37 articles met the inclusion criteria and were selected for this systematic review (Table 2). The detailed selection strategy is shown in Figure 1.

Table 2.
General characteristics of included studies.
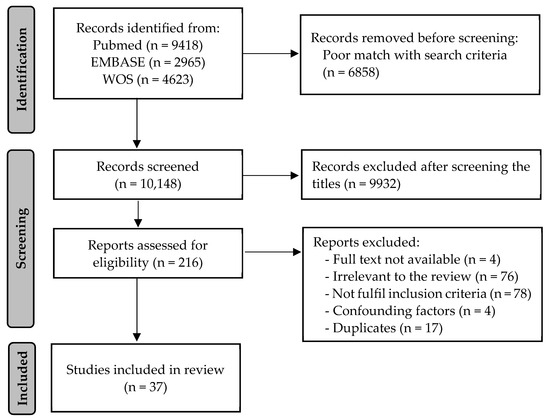
Figure 1.
PRISMA flow diagram of the study selection process.
Among the 37 studies included in this review, 32 are interventional and 5 observational. Apart from that, 18 evaluate the association between certain dietary strategies and miRNA expression in healthy individuals, whereas the other 19 are focused on subjects with overweight or obesity. Most of the studies quantify the miRNAs in blood components (21 in plasma, 9 in serum and 4 in PBMCs [peripheral blood mononuclear cells]), but, beyond that, other specimens are used (3 studies analyze stool samples, 1 rectal mucosa, 1 subcutaneous adipose tissue, 1 sperm and 1 saliva). Quantitative PCR (qPCR) is the most common technique used to analyze miRNA expression; a total of 15 studies exclusively perform qPCR or a qPCR array, and 13 conduct a first screening followed by validation of the results using qPCR. The screening step is carried out using arrays (11 studies), RNA sequencing (2 studies) and performing qPCR with pooled samples (1 study). Additionally, five studies use only RNA sequencing, two the NanoString nCounter technology and one the miRNA sensor iLluminate.
Concerning the dietary strategies researched, the studies can be classified into four groups—energy-controlled diets [33,34,35,36,37,38,39,40,41,42,43,44,45,46,47], fat-related interventions [48,49,50,51,52,53,54,55,56,57,58,59], observational studies [60,61,62,63,64] and other dietary strategies [17,65,66,67,68]—which are further developed in the next sections. Articles examining the uptake of exogenous miRNAs from dietary sources were deemed insufficient for inclusion in the review. Nonetheless, given the emerging potential of food-derived miRNAs, their implications for health have been discussed below.
Regarding the miRNA nomenclature, some articles refer to the precursor miRNA and others to the mature form. Besides that, some authors do not use a letter after the number of the miRNA, which differentiates members of the same family, or the suffixes -3p and -5p to indicate from which double-stranded RNA the mature sequence comes. For these reasons, the miRNAs included in this review have been analyzed by family, grouping miRNAs with a similar structure and evolutionary origin [69]. Following this criterion, there is evidence that 108 miRNA families are modulated by diet. Despite that, only 37 of them report significant results in more than one study. The most relevant ones are shown in Table 3.

Table 3.
Overview of human endogenous miRNAs modulated by diet in ≥2 studies indicating the source used to measure miRNA expression. The most relevant dietary patterns are included. miRNAs are grouped by family.
3.2. Modulation of miRNA Expression by Energy-Controlled Diets
A total of 15 studies evaluate the impact of energy-restricted diets on miRNA expression [33,34,35,36,37,38,39,40,41,42,43,44,45,46,47], of which 2 are energy-restricted diets without a weight loss purpose [33,34], 10 are weight loss interventions [35,36,37,38,39,40,41,42,43,44] and 3 consider fasting periods [45,46,47]. The characterization of the effects of energy-restricted diets on miRNA expression is mainly performed in healthy adults with overweight or obesity, although three studies also include subjects of a normal weight [35,37,45]. Regarding the experimental design, most of the studies include an intervention period of dietary restriction greater than 8 weeks, except for the three fasting studies [37,45,46], which lasted 5, 10 and 28 days, and two energy restriction studies of 4 and 6 weeks [34,39]. Although there were important differences between the diets implemented and their caloric content, they all agreed on caloric restriction, varying the range of restriction from a reduction of 30% or 500 kcal/day to periods of fasting for several days.
A total of 46 miRNAs have been linked to energy-restricted diets, although only 12 of them have been studied in more than one article (miR-19, miR-22, miR-29, miR-99, miR-122, miR-126, miR-142, miR-221, miR-223, miR-411, miR-935, let-7). In this respect, miR-19 is down-regulated in two fasting studies [46,47] and miR-99 in two energy restriction programs [33,39]. Additionally, miR-142 is down-regulated in responders to a low-fat diet [35] and after a 10-day fasting period [46]; the up-regulation of miR-126 is found after a fasting period [46] and also after a 12-week weight loss diet with a deficit of 500 kcal/day [38]. Interestingly, the subjects who lost less weight after an energy-restricted diet [41,43] were the ones who showed higher levels of miR-935.
Studies involving the remaining miRNAs showed a more complex pattern of regulation. miR-22 is up-regulated in responders (weight loss > 5%) to a 16-week intervention with a low-fat diet [35] but down-regulated after a 10-day fasting period with a daily intake of 250 kcal [46]. miR-122 is down-regulated in subjects who follow a 12-week weight-loss diet with a deficit of 500 kcal/day [38] and after a 12-month weight loss program [36] but also up-regulated in adults who have undergone a 10-week fasting intervention [46]. Concurrently, miR-411 and the let-7 family are down-regulated by energy-restricted diets [33,39] but up-regulated after a fasting period [45,47].
Concerning miR-29 and miR-221, both were down-regulated in the plasma of responders to a weight loss low-fat diet of 16 weeks [35], but other studies show up-regulation related to weight loss diets; miR-29 is up-regulated in the subcutaneous adipose tissue after a 15-week intervention [40], and miR-221 is induced in the plasma of both responders and non-responders after a 16-week intervention [43]. Different outcomes are reported on miR-223 after a weight loss diet depending on sex and the sample analyzed; this miRNA is up-regulated in the plasma after 16 weeks of intervention regardless of the sex of the population [43] but also down-regulated in the serum HDL fraction of men after 12 weeks of intervention [44]. Lower levels of this miRNA have also been seen at baseline in the PBMCs of women who do not respond to 8 weeks of dietary intervention [41].
3.3. Modulation of miRNA Expression by Fat Intake
Twelve studies analyze the impact of different fat-related diets [48,49,50,51,52,53,54,55,56,57,58,59]. Three of them consist of a single high-fat meal [48,49,50], five evaluate nut intake [51,52,53,54,55], two analyze extra virgin olive oil (EVOO) intake [56,57], one a ketogenic diet [58] and one the intake of trans fatty acids [59]. The last report is the only one with no significant results. These studies measure the miRNAs in blood components, except one article about miRNAs in sperm [55]. The method used to analyze miRNAs was qPCR, with an initial screening in some of them [48,51,52,53,55,57], except for one study, which used the NanoString nCounter technology [58].
Two interventional studies evaluate the impact of a single high-fat and high-energy meal on postprandial miRNA expression [48,50], but they analyze different miRNAs. One of them reports nine miRNAs down-regulated (miR-613, miR-629, miR-24-2, miR-555, miR-148a, miR-621, miR-875, miR-513c, miR-1226) and nine up-regulated (miR-653, miR-19b-1, miR-363, miR-885, miR-339, miR-938, miR-148b, miR-593, miR-200b) after a high-fat meal of 800 kcal [48], and the other one finds three miRNAs down-regulated (miR-1260a, miR-92b, miR-205) and six up-regulated (miR-200c, miR-143, miR-200b, miR-143, miR-375, miR-145) after a high-fat meal of 1067 kcal [50]. The miR-200 family is up-regulated after both single-meal studies. Another study using a similar dietary strategy but adding orange juice, a glucose drink or water in a crossover model observes the up-regulation of miR-375 after the high-fat meal with orange juice and down-regulation of miR-205 after the meal with the glucose drink [49].
Regarding the five studies analyzing nut consumption, their interventions lasted at least 8 weeks, and the study population was healthy adults of a normal weight in three of them [51,52,55] and with overweight/obesity in the remaining two [53,54]. Daily intake of 30–60 g of walnuts results in the overexpression of miR-32 [52], miR-29b [52] and miR-551a [51] when compared with the controls who abstained from walnuts [52]. Additionally, the down-regulation of miR-328, miR-330, miR-221 and miR-125a and the up-regulation of miR-192, miR-486, miR-19b, miR-106a, miR-130b, miR-18a and miR-769 are observed after 8 weeks with an almond and walnut intake of 30 g/day [53]. Furthermore, spermatic miR-34b is down-regulated after nut intake when comparing a Western diet avoiding nuts with a Western diet with 60 g/day of nuts [55].
Regarding EVOO intake, both studies evaluate the impact of a single dose of EVOO on the PBMCs [57] and plasma [56] and observe the modulation of several miRNAs. Interestingly, one of them reports the underexpression of miR-192 [57] and the other one overexpression [56]. Beyond that, the ketogenic diet study reports the underexpression of circulating miR-504 and the overexpression of let-7b and miR-143 after 6 weeks following this high-fat low-carbohydrate dietary pattern in a cohort of 12 healthy adults [58].
3.4. Dietary Patterns Related to miRNA Modulation in Observational Studies
Among the observational studies [60,61,62,63,64], four of them evaluate differences between vegan, vegetarian and omnivorous diets [60,61,62,63], and the other one is on the Mediterranean diet [64]. Regarding the differences between vegan, vegetarian and omnivorous diets, one of the studies does not report significant results [60]. The up-regulation of miR-92a in stool and plasma [62] and the down-regulation of miR-636, miR-4488 and 4739 in stool [63] are observed in association with vegan and vegetarian diets [62]. Additionally, in a cohort of 96 healthy adults, miR-3661, miR-320c, miR-29a, miR-320b and miR-204 are overexpressed and miR-132 underexpressed in the plasma of subjects who follow vegan or vegetarian diets [61]. Concerning the impact of the Mediterranean diet on miRNA expression, adherence to this dietary pattern is associated with higher levels of miR-590 in adults with obesity [64].
3.5. Other Dietary Patterns Related to miRNA Modulation
The five remaining studies evaluate the intake of high-red meat [17], protein consumption [65], a Korean diet [66], grape intake [67] and orange juice consumption [68].
A randomized crossover study design, with 23 healthy volunteers, studies the influence of two 4-week dietary interventions: a high red meat (HRM) diet and an HRM diet + supplementation with butyrylated resistant starch on miRNA expression in rectal mucosa. The up-regulation of miR-19a, miR-19b and miR-21 after the HRM diet and down-regulation of miR-17, miR-19a, miR-19b, miR-20a and miR-92a when adding resistant starch are reported [17]. Besides that, the association between protein consumption and circulating miRNA expression is analyzed in a group of three healthy men over the age of 70 [65]. After 2 weeks of study, a high protein intake (1.6 g/kg body weight/day) is associated with the underexpression of miR-125b, miR-100, miR-99a, miR-23b and miR-203 [65].
Apart from that, a 2-week intervention study evaluates the differences between a Korean diet and a westernized Korean diet in circulating and salivary miRNA expression in a group of 10 women with overweight and concludes that the Korean diet down-regulates miR-26a and miR-126 in the plasma and miR-92 and miR-122 in the saliva, while the westernized diet down-regulates miR-25 in the plasma and miR-31 in the saliva [66].
The effects of the intake of 5 g/kg body weight/day of fresh grapes on circulating miRNA expression are analyzed in a cohort of 40 adults with overweight; the up-regulation of two miRNAs (miR-208a and miR-33a) and down-regulation of 18 miRNAs (miR-181a, miR-30e, miR-30d, miR-335, miR-222, miR-15a, miR-421, miR-339, miR-378a, miR-29b, miR-106b, miR-324, miR-1260a, miR-365a, miR-155, miR-335, miR-200c and let-7f) have been observed [67]. let-7f, together with miR-126, is also down-regulated in the PBMCs after 4 weeks of consuming 500 mL of orange juice per day, while miR-144, miR-424 and miR-130b are up-regulated [68].
4. Discussion
Taking together the results of the included articles, the data confirm the involvement of several dietary patterns, as well as specific foods, in the modulation of miRNA expression in human cells, which is mainly reflected in plasma levels. However, the studies collected are quite heterogeneous. The interventions show large differences in terms of their duration and dietary strategy, as well as the characteristics of the selected study population, such as sex and age, with both parameters closely linked to the expression of several miRNAs [70]. The method used to quantify miRNA expression is also worth noting when interpreting the results, as each one provides different information. Three kinds of quantification methods have been identified: targeted analysis, sequencing and a combination of both. Targeted analysis using qPCR is the most used and offers high sensibility and specificity for detecting low levels of miRNAs [71] but has the drawback of quantifying a limited number of miRNAs, which are chosen by researchers following a hypothesis-driven approach. In contrast, miRNA sequencing provides extensive information about the entire miRNAome but has a lower sensibility in detecting those miRNAs expressed in small amounts [72]. Therefore, as some authors have opted for, the development of studies that use a combination of both methods might be advisable. This involves an initial screening using miRNA sequencing, followed by a targeted analysis of the selected miRNAs using qPCR to validate the observations. This approach would bring more reliable results showing not only how specific miRNAs are influenced by diet but also the entire miRNAome. Notwithstanding these limitations, the available data provide enough evidence to point out specific dietary patterns capable of modulating miRNA expression, which are discussed below.
4.1. Influence of Energy Intake on miRNA Regulation
Energy-controlled diets have been widely studied in the context of miRNA regulation, mainly with weight loss purposes. Regarding its consequences, we should differentiate the outputs obtained when considering the duration of the intervention from those based on feeding conditions.
Given the extensive study of hypocaloric diets in miRNA regulation, most of the miRNAs mentioned in this review were associated with energy restriction, either due to long-term daily energy restriction or shorter interventions (Figure 2a). For instance, the up-regulation of miR-126 [38,46] and down-regulation of miR-142 [35,46] have been related to both intervention models [38,46]. Occasionally, the effects observed on the miRNA levels vary depending on the duration of the restriction. This, together with the fact that the experimental designs tend to be quite different, makes it difficult to analyze the outcomes jointly. That is the case with let-7 [33,39,45], miR-22 [35,46], miR-122 [36,38,46] and miR-411 [33,47], whose evidence reported both up- and down-regulations associated with energy restriction. In these cases, some authors reported short-term consequences [46] and others the opposite, long-term outcomes [35,46]. The durations of the interventions prevented us from merging the results.
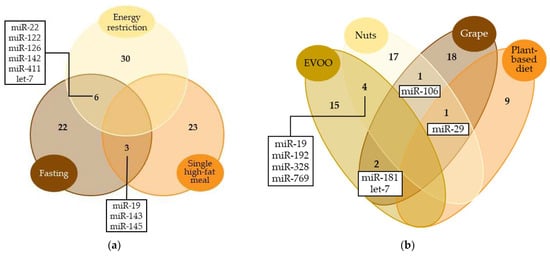
Figure 2.
Venn diagram comparing the most representative miRNAs modulated by (a) energy-controlled dietary patterns and (b) plant-based foods. Only miRNAs with significant results in at least two studies were included. Numbers represent the count of miRNAs modulated by each dietary pattern. miRNAs modulated by ≥2 dietary patterns are specified.
Regarding the feeding conditions, differences in miRNA levels have been observed between fasting and the postprandial state. The up-regulation of circulating miR-19, miR-143, miR-145, miR-200, miR-339 and miR-375 and the down-regulation of miR-92, miR-205 and miR-1260 have been seen in the postprandial state after a single high-energy and high-fat meal when compared with the fasting state [48,49,50]. In line with these results, another study has reported decreased levels of miR-19, miR-143 and miR-145 after a fasting period [46,47]. Apart from that, some of the changes in miRNA expression associated with hypocaloric diets might be influenced by macronutrient content. One example is miR-221, since it is down-regulated with a hypocaloric and low-fat diet [35] and up-regulated with a restricted diet which does not take into account fat content [43]. Fat intake may therefore be implicated in miR-221 modulation and, as the evidence associates miR-221 with insulin levels, it might also be involved in the development of insulin resistance [73,74]. This is the same for miR-223 outputs, as it has shown down-regulation in serum HDL fractions after a weight loss diet high in protein [44] and up-regulation in the plasma after a hypocaloric diet disregarding protein content [43]. The biological sample selected to measure the miRNA levels may also influence the differences between the aforementioned findings. Several miRNAs show tissue specificity, and large differences are found among the miRNA spectrum of several body fluids [75]. This may imply different tissue-dependent effects but with a common purpose [76]. In view of the above, the development of further studies with similar experimental characteristics may help strengthen the evidence available.
Interestingly, miRNA levels have been related to the response to diet-induced weight loss. Non-responders to a weight loss diet presented overexpression of miR-935 and miR-4772 and underexpression of miR-223, miR-224 and miR-376 before the dietary intervention [41,43]. These miRNAs may be considered biomarkers of weight loss susceptibility.
Considering the results of these studies, we could say that energy intake was a major regulatory factor in the human miRNA profile. Changes in the caloric content of the diet lead to the modification of endogenous miRNA levels, and the effects may vary depending on the duration of the intervention and the feeding conditions. Furthermore, the influence of these miRNAs over metabolic pathways and their implication in the development of metabolic diseases has been proposed. For instance, the evidence shows the involvement of miR-22 in the control of metabolic homeostasis [77]. Silencing of this miRNA has been suggested in the treatment of metabolic diseases, including obesity and hepatic steatosis [77,78]. Along with this, miR-122 participates in the hepatic metabolism of lipids, regulating the expression of genes involved in cholesterol and fatty acid synthesis [79]. Additionally, miR-19 and let-7 play a role in insulin signal transduction and have been related to the development of type 2 diabetes and obesity-induced insulin resistance [79]. miR-143 has also been associated with glucose and lipid metabolism [79,80]. These findings reinforce our knowledge about the involvement of energy intake-modulated miRNAs in the regulation of several biological processes [18,81] and their subsequent implications for the development of metabolic diseases [82,83,84,85].
4.2. Influence of a Mediterranean Diet and Plant-Based Foods on miRNA Regulation
The Mediterranean diet is widely known for its health benefits, as most of its characteristic food components have been attributed anti-atherosclerotic, anti-inflammatory or antioxidant effects [86,87,88]. It has been seen that the Mediterranean diet modulates the levels of certain miRNAs that could be involved in the regulation of the aforementioned biological processes. For example, high adherence to the Mediterranean diet has been associated with higher serum levels of miR-590 in adults with morbid obesity [64], a miRNA which has been related to anti-inflammatory effects, a reduction in lipid accumulation and the inhibition of atherosclerotic progression [89,90]. Although this was the only study which evaluated the impact of the Mediterranean diet as a whole, several studies have considered specific foods included in this dietary pattern, such as EVOO [56,57], nuts [51,52,53,54,55], grapes [67] and even plant-based diets [61,62,63]. Those studies showed that the intake of different plant-based foods can modulate the same miRNAs (Figure 2b). For instance, nut and EVOO consumption up-regulate circulating miR-192 [53,56], a miRNA associated with lipid and glucose metabolism [64,91,92], and down-regulate circulating miR-328 [53,56], which, according to the evidence, is related to cardiovascular disease [93,94]. miR-29, miR-106 and miR-181 are also examples of miRNAs modulated by several plant-based foods, and their biological effects are discussed below. Although the way miRNAs impact metabolic control is still not fully understood, their modulation may be one of the mechanisms through which the Mediterranean diet and plant-based foods improve health and reduce the risk of diseases.
4.3. Biological Effects of Diet-Modulated miRNAs
Many studies suggest the modulatory role of miRNAs in key physiological processes and their ensuing impact on health [95]. Particularly, they have been associated with glucose and lipid metabolism [79], which is fundamental to human homeostasis. Here, we have revised the potential of foods and diets to influence miRNA levels and, given the strong involvement of unbalanced diets in the development of metabolic diseases, further investigation into the physiological role of miRNAs could provide new molecular targets that contribute to their prevention.
Studies on energy-controlled diets and plant-based foods have substantially shown their relevance in modulating endogenous miRNA levels. However, large differences were found regarding the miRNAs affected by different dietary patterns. This might be attributed to the metabolic effects of dietary nutrients on the body. Each nutrient present in the diet triggers distinct biological processes related to functions essential to our organism. The modulation of endogenous miRNA expression might contribute to the mechanisms underlying the specific functions of each nutrient. Consequently, each dietary pattern might be linked to a specific miRNA expression profile aimed at optimizing nutrient metabolism. Additionally, the evidence suggests that some bioactive compounds in food would be likely to directly change miRNA expression in a positive way, contributing to their health properties. For instance, curcumin, a flavonoid found in turmeric, has been attributed to anti-inflammatory and anti-tumorigenic properties, which are potentially mediated by miRNA activity, including miR-17, miR-20a and miR-27 [96]. The anti-inflammatory properties of resveratrol have also been linked to miRNA regulation. A study assessing resveratrol supplementation in men with type 2 diabetes and hypertension has reported the modulation of a set of miRNAs involved in the inflammatory response [97]. The phenolic compounds from nuts and extra virgin oil are also examples of miRNA activity modulators [96]. Although these findings contribute to our understanding of the role of miRNAs in the health-related properties of diet, the evidence is not sufficient to clearly determine how each dietary pattern influences fundamental biological processes through miRNA modulation. Here, we thoroughly examined the potential physiological functions of these two sets of miRNAs: energy-controlled patterns (Figure 2a) and plant-based foods (Figure 2b). To explore them, first, a miRNA–disease interaction network analysis was performed using the miRNet 2.0 software [32]. Only those miRNAs showing significant results in more than one study were included. The resulting network diagrams are shown in Figure 3. Regarding miRNA–disease interactions, the miRNet software displayed 100 outcomes, of which 57 were related to carcinomas and neoplasms. As miRNAs have been widely studied in relation to cancer and there is strong evidence of this, a large number of results on cancer were expected. Among the 47 remaining outcomes, cardiovascular diseases showed the strongest association with miRNAs modulated by energy-controlled patterns. Obesity, atherosclerosis and stroke also presented a high number of associations (Figure 3a). This highlights the implication of miRNAs in the mechanisms involved in cardiovascular disease development and other risk factors for metabolic syndrome and the need to balance caloric intake to prevent these diseases.
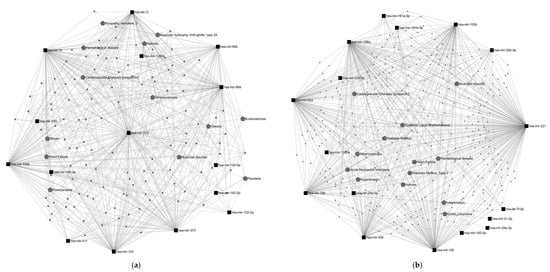
Figure 3.
miRNA–disease interaction network. Network gene analysis was performed using miRNet 2.0 software [32]. Black squares represent the most relevant miRNAs modulated by (a) energy-controlled dietary patterns and (b) plant-based foods; gray pentagons represent diseases related to these miRNAs; bigger pentagons show diseases interacting with ≥3 miRNAs, excluding cancer-related diseases. In addition, a miRNA target network analysis was also performed using miRNet 2.0. A total of 11,928 target genes were found to be associated with miRNAs modulated by energy-controlled patterns, showing the miR-34 and let-7 families had the highest degree of interaction with them. Besides that, this set of miRNAs was significantly involved in 13 metabolic pathways, predominantly in angiogenesis (Figure 4a).
Concerning the set of miRNAs modulated by plant-based foods, the miRNet software also displays 100 diseases interacting with this set of miRNAs, half of them carcinomas and neoplasms. Among the non-cancer outcomes, diabetes, cardiovascular diseases, atherosclerosis and hypertension were some of the most connected in the network (Figure 3b). A total of 9,113 target genes were found. miR-20, miR-15, let-7 and miR-181 were the miRNAs with the strongest interaction. Eight metabolic pathways were significantly influenced by the miRNAs modulated by plant-based foods; among them, adipocyte differentiation and regulation of the AKT pathway appeared to be interesting in the context of diet and health (Figure 4b).
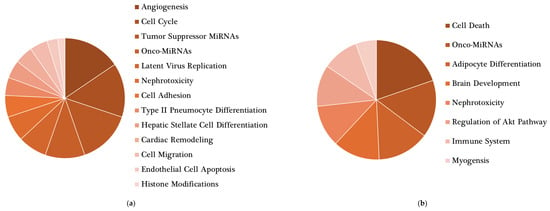
Figure 4.
Main roles of target genes found in the network analysis. miRNet 2.0 software was used to determine relevant functions of miRNAs modulated by (a) energy-controlled dietary patterns and (b) plant-based foods. Only statistically significant (p < 0.05) outcomes are shown.
Of note, a few studies have evaluated the intake of animal-based foods, enabling a comparison with plant-based foods in terms of miRNA modulation. A study assessing the intake of red meat has found an association with miR-19 and miR-21, two miRNAs also modulated by nut [53] and EVOO [56,57] intake. Additionally, one article comparing the effects of plant-based and omnivorous diets reports a trend in miRNA expression, gradually changing its levels from vegan and vegetarian to omnivorous patterns [62]. This suggests that animal and plant origin food may modulate a similar set of miRNAs but in a different way. However, as certain studies evaluating similar dietary patterns obtain opposite results, a clear interpretation cannot be secured.
The network analysis pointed out the involvement of miRNA regulation in metabolic health. Particularly, the two sets of miRNAs analyzed were highly related to cardiovascular health, inflammation and the immune system. Conducting further studies on diet–miRNA and miRNA–health interactions may contribute to the development of nutritional strategies focused on preventing diseases through miRNA modulation.
4.4. Exogenous miRNAs from Dietary Sources
Apart from the regulatory role of diet in miRNA expression, it is also important to consider those miRNAs that can be directly ingested through dietary sources. They are found both in animals and plants, and in both kingdoms, miRNAs can regulate gene expression [98]. As plant miRNAs have been found in mammalian specimens, the evidence suggests that they can regulate cross-kingdom gene expression through dietary intake [98,99,100]. Mature plant-based miRNAs are present not only in raw vegetables but also in their cooked form (as in rice, wheat or potato) and can survive the gastrointestinal tract, even the acidic environment of the stomach [12]. Once exogenous miRNAs enter the intestinal epithelial stem cells, they can be packaged into vesicles and transported through the bloodstream, potentially affecting endogenous processes [101]. Although there is increasing evidence of exogenous miRNAs being acquired through dietary sources, their role as regulators of gene expression is still controversial [102]. Some authors consider that the presence of plant miRNAs in human samples is due to the contamination and oversensitivity of sequencing methods and also that the number of exogenous miRNAs is not enough to affect human gene expression [103,104].
The number of articles was insufficient to include the intake of exogenous miRNAs in the review. Nevertheless, some studies display their absorbability and their potential metabolic role; thus, we should not discard them. Researchers have detected significant levels of ath-miR-156a, osa-miR-168a and ath-miR-166a, three exogenous plant-derived miRNAs, in human serum [12]. Furthermore, in vitro analysis has shown that osa-miR-168a, which is abundant in rice and cruciferous vegetables, has a regulatory role in host gene expression, specifically as a modulator of liver-specific low-density lipoprotein receptor adapter protein 1 (LDLRAP1), which has a role in the removal of LDL from the plasma [12]. Another in vitro model shows that miR-156a, found in rice and green vegetables, can modulate the junction adhesion molecule-A (JAM-A), which is related to the inflammatory recruitment of mononuclear cells in the endothelium of atherosclerotic arteries [14]. Moreover, an in silico analysis which has examined four human datasets has identified 35 exogenous miRNAs in human milk exosomes belonging to 25 plant miRNA families [13].
Exogenous miRNAs acquired through the intake of species in the same kingdom are also noteworthy. Milk is a case in point. This fluid constitutes a relevant source of miRNAs for lactating progeny since they are transported in exosomes or vesicles and protected from degradation and digestion [105]. Humans can absorb biologically effective doses of miRNAs from mammals, as observed with cow milk [8,106]. Considerable amounts of two milk-based miRNAs (miR-29b and miR-200c) have been detected in the postprandial plasma of healthy adults after the consumption of bovine milk [106]. The absorbability of miRNAs from maternal milk has also been proposed [107,108,109] but is still under debate. In this context, the miRNA supply during breastfeeding is of great interest due to the importance of early nutrition to the metabolic programming of babies. Nutrition during the early stages of life has long-term consequences for health, and the metabolic adaptations induced in this critical period of child development can modulate susceptibility to metabolic disorders in adulthood [110,111,112]. Breastfeeding plays an important role in metabolic programming, and the function of miRNAs in metabolic programming should be considered. The evidence in humans is scarce, but animal models have demonstrated its involvement [113,114]. miR-148 is the most abundant miRNA in milk [109,115,116] and has been related to immune regulation, metabolism and development [109,117,118]. Among the articles included in this review, none of them have analyzed the miR-148 levels in milk, but in one of them, the intake of a single hypercaloric high-saturated-fat meal has been related to the PBMC levels of miR-148, that is, the underexpression of miR-148a and the overexpression of miR-148b [48]. Animal models also demonstrate the ability of diet to modulate miRNAs in milk. Higher levels of miR-222 and lower levels of miR-200a and miR-26a have been observed in milk from rats fed with a cafeteria diet (fat-rich hypercaloric diet) compared to controls [28].
In view of the above, further studies are needed to strengthen these data, especially to support the evidence of miRNAs being significantly absorbed from breast milk due to the importance of metabolic programming in early life stages.
5. Conclusions
This review underlines the ability of changes in dietary habits to regulate endogenous miRNA levels. Particularly, energy and fat content appeared to be key factors since they are the nutritional elements with more evidence supporting their role in miRNA modulation. However, the current studies are heterogeneous, which hinders the interpretation of the results jointly. The involvement of miRNAs in the regulation of biological processes and its potential impact on health have also been exposed. Animal models highlight their metabolic repercussions [29,30,119], but the evidence in humans is scarce. In this context, two sets of miRNAs emerged as linked to energy-controlled diets and plant-based foods, and the network analysis revealed its role in cardiometabolic health. Further studies are needed to clarify the consequences of diet on human miRNAs and to reliably identify their relationship with health conditions. Notwithstanding, instead of considering specific miRNAs separately, it would be interesting to identify miRNA profiles related to particular metabolic pathways and determine the dietary patterns that could balance them.
Author Contributions
M.D. performed the data collection and data analysis. M.D., J.S. and F.S. wrote the first draft of the manuscript. All authors worked on the final version of the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the project “PI23/00532”, funded by Instituto de Salud Carlos III and co-funded by the European Union (ERDF/ESF, “Investing in your future”). The Research Group Nutrigenomics, Biomarkers and Risk Evaluation receives financial support from Instituto de Salud Carlos III and Centro de Investigación Biomédica en Red Fisiopatología de la Obesidad y Nutrición (CIBERobn). M.D. has a predoctoral research contract (FI21/00119—Instituto de Salud Carlos III).
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of the data; in the writing of the manuscript or in the decision to publish the results.
References
- WHO. Noncommunicable Diseases. Available online: https://www.who.int/news-room/fact-sheets/detail/noncommunicable-diseases (accessed on 10 June 2023).
- Hunter, D.J.; Reddy, K.S. Noncommunicable Diseases. N. Engl. J. Med. 2013, 369, 1336–1343. [Google Scholar] [CrossRef] [PubMed]
- Willett, W.; Rockström, J.; Loken, B.; Springmann, M.; Lang, T.; Vermeulen, S.; Garnett, T.; Tilman, D.; DeClerck, F.; Wood, A.; et al. Food in the Anthropocene: The EAT-Lancet Commission on Healthy Diets from Sustainable Food Systems. Lancet 2019, 393, 447–492. [Google Scholar] [CrossRef] [PubMed]
- Trautwein, E.A.; McKay, S. The Role of Specific Components of a Plant-Based Diet in Management of Dyslipidemia and the Impact on Cardiovascular Risk. Nutrients 2020, 12, 2671. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, L.J.; Di Bella, G.; Veronese, N.; Barbagallo, M. Impact of Mediterranean Diet on Chronic Non-Communicable Diseases and Longevity. Nutrients 2021, 13, 2028. [Google Scholar] [CrossRef] [PubMed]
- Bruins, M.J.; Van Dael, P.; Eggersdorfer, M. The Role of Nutrients in Reducing the Risk for Noncommunicable Diseases during Aging. Nutrients 2019, 11, 85. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, C.; Gleddie, S.; Xiao, C.W. Soybean Bioactive Peptides and Their Functional Properties. Nutrients 2018, 10, 1211. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Zhou, B.; Ross, S.A.; Zempleni, J. Nutrition, MicroRNAs, and Human Health. Adv. Nutr. 2017, 8, 105–112. [Google Scholar] [CrossRef]
- Milenkovic, D.; Jude, B.; Morand, C. MiRNA as Molecular Target of Polyphenols Underlying Their Biological Effects. Free Radic. Biol. Med. 2013, 64, 40–51. [Google Scholar] [CrossRef]
- Kura, B.; Parikh, M.; Slezak, J.; Pierce, G.N. The Influence of Diet on MicroRNAs That Impact Cardiovascular Disease. Molecules 2019, 24, 1509. [Google Scholar] [CrossRef]
- Otsuka, K.; Yamamoto, Y.; Matsuoka, R.; Ochiya, T. Maintaining Good MiRNAs in the Body Keeps the Doctor Away?: Perspectives on the Relationship between Food-Derived Natural Products and MicroRNAs in Relation to Exosomes/Extracellular Vesicles. Mol. Nutr. Food Res. 2018, 62, 1700080. [Google Scholar] [CrossRef]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous Plant MIR168a Specifically Targets Mammalian LDLRAP1: Evidence of Cross-Kingdom Regulation by MicroRNA. Cell Res. 2012, 22, 107. [Google Scholar] [CrossRef]
- Lukasik, A.; Zielenkiewicz, P. In Silico Identification of Plant MiRNAs in Mammalian Breast Milk Exosomes—A Small Step Forward? PLoS ONE 2014, 9, 99963. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.; He, F.; Ma, L.; Cao, M.; Zhou, Z.; Wei, Z.; Xue, Y.; Sang, X.; Chong, H.; Tian, C.; et al. The Potential Atheroprotective Role of Plant MIR156a as a Repressor of Monocyte Recruitment on Inflamed Human Endothelial Cells. J. Nutr. Biochem. 2018, 57, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Debnath, T.; Deb Nath, N.C.; Kim, E.K.; Lee, K.G. Role of Phytochemicals in the Modulation of MiRNA Expression in Cancer. Food Funct. 2017, 8, 3432–3442. [Google Scholar] [CrossRef] [PubMed]
- Saquib, M.; Agnihotri, P.; Monu, X.; Biswas, S. Exogenous MiRNA: A Perspective Role as Therapeutic in Rheumatoid Arthritis. Curr. Rheumatol. Rep. 2021, 23, 43. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, K.J.; Conlon, M.A.; Young, G.P.; Topping, D.L.; Hu, Y.; Winter, J.M.; Bird, A.R.; Cobiac, L.; Kennedy, N.A.; Michael, M.Z.; et al. Dietary Manipulation of Oncogenic MicroRNA Expression in Human Rectal Mucosa: A Randomized Trial. Cancer Prev. Res. 2014, 7, 786–795. [Google Scholar] [CrossRef] [PubMed]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An Overview of MicroRNAs: Biology, Functions, Therapeutics, and Analysis Methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Bernardo, B.C.; Ooi, J.Y.Y.; Lin, R.C.Y.; Mcmullen, J.R. MiRNA Therapeutics: A New Class of Drugs with Potential Therapeutic Applications in the Heart. Future Med. Chem. 2015, 7, 1771–1792. [Google Scholar] [CrossRef]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A Brief Review on the Mechanisms of MiRNA Regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef]
- Ruiz, G.P.; Camara, H.; Fazolini, N.P.B.; Mori, M.A. Extracellular MiRNAs in Redox Signaling: Health, Disease and Potential Therapies. Free Radic. Biol. Med. 2021, 173, 170–187. [Google Scholar] [CrossRef]
- Kalayinia, S.; Arjmand, F.; Maleki, M.; Malakootian, M.; Singh, C.P. MicroRNAs: Roles in Cardiovascular Development and Disease. Cardiovasc. Pathol. 2021, 50, 107296. [Google Scholar] [CrossRef] [PubMed]
- Çakmak, H.A.; Demir, M. MicroRNA and Cardiovascular Diseases. Balk. Med. J. 2020, 37, 60–71. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Kuang, G.; Wu, Y.; Ou, C. Emerging Roles of Exosomal MiRNAs in Diabetes Mellitus. Clin. Transl. Med. 2021, 11, 11. [Google Scholar] [CrossRef]
- Vasu, S.; Kumano, K.; Darden, C.M.; Rahman, I.; Lawrence, M.C.; Naziruddin, B. MicroRNA Signatures as Future Biomarkers for Diagnosis of Diabetes States. Cells 2019, 8, 1533. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Huang, F.; Gu, X.; Zhang, M.; Wen, J.; Wang, X.; You, L.; Cui, X.; Ji, C.; Guo, X. Adipogenic MiRNA and Meta-Signature MiRNAs Involved in Human Adipocyte Differentiation and Obesity. Oncotarget 2016, 7, 40830–40845. [Google Scholar] [CrossRef]
- Zaiou, M.; El Amri, H.; Bakillah, A. The Clinical Potential of Adipogenesis and Obesity-Related MicroRNAs. Nutr. Metab. Cardiovasc. Dis. 2018, 28, 91–111. [Google Scholar] [CrossRef] [PubMed]
- Pomar, C.A.; Castro, H.; Pico, C.; Serra, F.; Palou, A.; Sanchez, J. Cafeteria Diet Consumption during Lactation in Rats, Rather than Obesity Per Se, Alters MiR-222, MiR-200a, and MiR-26a Levels in Milk. Mol. Nutr. Food Res. 2019, 63, 1800928. [Google Scholar] [CrossRef] [PubMed]
- Guedes, E.C.; da Silva, I.B.; Lima, V.M.; Miranda, J.B.; Albuquerque, R.P.; Ferreira, J.C.B.; Barreto-Chaves, M.L.M.; Diniz, G.P. High Fat Diet Reduces the Expression of MiRNA-29b in Heart and Increases Susceptibility of Myocardium to Ischemia/Reperfusion Injury. J. Cell. Physiol. 2019, 234, 9399–9407. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, B.; Wang, X.; Liu, Z.; Li, J.; Zhang, S.; Gu, X.; Jia, M.; Guo, H.; Feng, N.; et al. P53/PANK1/MiR-107 Signalling Pathway Spans the Gap between Metabolic Reprogramming and Insulin Resistance Induced by High-Fat Diet. J. Cell. Mol. Med. 2020, 24, 3611–3624. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. MiRNet 2.0: Network-Based Visual Analytics for MiRNA Functional Analysis and Systems Biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef]
- Giardina, S.; Hernández-Alonso, P.; Díaz-López, A.; Salas-Huetos, A.; Salas-Salvadó, J.; Bulló, M. Changes in Circulating MiRNAs in Healthy Overweight and Obese Subjects: Effect of Diet Composition and Weight Loss. Clin. Nutr. 2019, 38, 438–443. [Google Scholar] [CrossRef]
- Margolis, L.M.; Rivas, D.A.; Pasiakos, S.M.; McClung, J.P.; Ceglia, L.; Fielding, R.A. Upregulation of Circulating MyomiR Following Short-Term Energy Restriction Is Inversely Associated with Whole Body Protein Synthesis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2017, 313, R298–R304. [Google Scholar] [CrossRef] [PubMed]
- Assmann, T.S.; Riezu-Boj, J.I.; Milagro, F.I.; Martínez, J.A. Circulating Adiposity-Related MicroRNAs as Predictors of the Response to a Low-Fat Diet in Subjects with Obesity. J. Cell. Mol. Med. 2020, 24, 2956–2967. [Google Scholar] [CrossRef] [PubMed]
- Duggan, C.; Tapsoba, J.d.D.; Scheel, J.; Wang, C.Y.; McTiernan, A. Weight Loss Reduces Circulating Micro-RNA Related to Obesity and Breast Cancer in Postmenopausal Women. Epigenetics 2022, 17, 2082–2095. [Google Scholar] [CrossRef] [PubMed]
- Heianza, Y.; Xue, Q.; Rood, J.; Bray, G.A.; Sacks, F.M.; Qi, L. Circulating Thrifty MicroRNA Is Related to Insulin Sensitivity, Adiposity, and Energy Metabolism in Adults with Overweight and Obesity: The POUNDS Lost Trial. Am. J. Clin. Nutr. 2023, 117, 121–129. [Google Scholar] [CrossRef]
- Hess, A.L.; Larsen, L.H.; Udesen, P.B.; Sanz, Y.; Larsen, T.M.; Dalgaard, L.T. Levels of Circulating MiR-122 Are Associated with Weight Loss and Metabolic Syndrome. Obesity 2020, 28, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Jayasooriya, V.; Johnson, N.; Bradley, A.; Kotarsky, C.; Jepng’etich, L.; Friesner, D.; Stastny, S.; Hackney, K.J.; Nawarathna, D. A Miniaturized MicroRNA Sensor Identifies Targets Associated with Weight Loss in a Diet and Exercise Intervention among Healthy Overweight Individuals. Sensors 2022, 22, 6758. [Google Scholar] [CrossRef]
- Kristensen, M.M.; Davidsen, P.K.; Vigelsø, A.; Hansen, C.N.; Jensen, L.J.; Jessen, N.; Bruun, J.M.; Flemming, D.; Helge, J.W. MiRNAs in Human Subcutaneous Adipose Tissue: Effects of Weight Loss Induced by Hypocaloric Diet and Exercise. Obesity 2017, 25, 572–580. [Google Scholar] [CrossRef]
- Milagro, F.I.; Miranda, J.; Portillo, M.P.; Fernandez-Quintela, A.; Campión, J.; Martínez, J.A. High-Throughput Sequencing of MicroRNAs in Peripheral Blood Mononuclear Cells: Identification of Potential Weight Loss Biomarkers. PLoS ONE 2013, 8, e54319. [Google Scholar] [CrossRef]
- Müller, S.; Wallner, S.; Schmitz, G.; Loew, T.; Stempfl, T.; Möhle, C.; Strack, C.; Sag, S.; Baessler, A.; Fischer, M. SNP Dependent Modulation of Circulating MiRNAs from the MiR25/93/106 Cluster in Patients Undergoing Weight Loss. Gene 2020, 753, 144787. [Google Scholar] [CrossRef] [PubMed]
- Parr, E.B.; Camera, D.M.; Burke, L.M.; Phillips, S.M.; Coffey, V.G.; Hawley, J.A. Circulating MicroRNA Responses between ‘High’ and ‘Low’ Responders to a 16-Wk Diet and Exercise Weight Loss Intervention. PLoS ONE 2016, 11, e0152545. [Google Scholar] [CrossRef] [PubMed]
- Tabet, F.; Torres, L.F.C.; Ong, K.L.; Shrestha, S.; Choteau, S.A.; Barter, P.J.; Clifton, P.; Rye, K.A. High-Density Lipoprotein-Associated MiR-223 Is Altered after Diet-Induced Weight Loss in Overweight and Obese Males. PLoS ONE 2016, 11, e0151061. [Google Scholar] [CrossRef] [PubMed]
- Lilja, S.; Stoll, C.; Krammer, U.; Hippe, B.; Duszka, K.; Debebe, T.; Höfinger, I.; König, J.; Pointner, A.; Haslberger, A. Five Days Periodic Fasting Elevates Levels of Longevity Related Christensenella and Sirtuin Expression in Humans. Int. J. Mol. Sci. 2021, 22, 2331. [Google Scholar] [CrossRef] [PubMed]
- Ravanidis, S.; Grundler, F.; de Toledo, F.W.; Dimitriou, E.; Tekos, F.; Skaperda, Z.; Kouretas, D.; Doxakis, E. Fasting-Mediated Metabolic and Toxicity Reprogramming Impacts Circulating MicroRNA Levels in Humans. Food Chem. Toxicol. 2021, 152, 112187. [Google Scholar] [CrossRef] [PubMed]
- Saini, S.K.; Singh, A.; Saini, M.; Gonzalez-Freire, M.; Leeuwenburgh, C.; Anton, S.D. Time-Restricted Eating Regimen Differentially Affects Circulatory MiRNA Expression in Older Overweight Adults. Nutrients 2022, 14, 1843. [Google Scholar] [CrossRef] [PubMed]
- Lopez, S.; Bermudez, B.; Montserrat-de la Paz, S.; Abia, R.; Muriana, F.J.G. A MicroRNA Expression Signature of the Postprandial State in Response to a High-Saturated-Fat Challenge. J. Nutr. Biochem. 2018, 57, 45–55. [Google Scholar] [CrossRef]
- Quintanilha, B.J.; Chaves, D.F.S.; Brasili, E.; Corrêa, T.A.F.; Capetini, V.C.; Ferreira, F.M.; Castro, I.A.; Hassimotto, N.M.A.; Rogero, M.M.; Lajolo, F.M. Ingestion of Orange Juice Prevents Hyperglycemia and Increases Plasma MiR-375 Expression. Clin. Nutr. ESPEN 2022, 47, 240–245. [Google Scholar] [CrossRef]
- Quintanilha, B.J.; Pinto Ferreira, L.R.; Ferreira, F.M.; Neto, E.C.; Sampaio, G.R.; Rogero, M.M. Circulating Plasma MicroRNAs Dysregulation and Metabolic Endotoxemia Induced by a High-Fat High-Saturated Diet. Clin. Nutr. 2020, 39, 554–562. [Google Scholar] [CrossRef]
- Gil-Zamorano, J.; Cofán, M.; de las Hazas, M.C.L.; García-Blanco, T.; García-Ruiz, A.; Doménech, M.; Serra-Mir, M.; Roth, I.; Valls-Pedret, C.; Rajaram, S.; et al. Interplay of Walnut Consumption, Changes in Circulating MiRNAs and Reduction in LDL-Cholesterol in Elders. Nutrients 2022, 14, 1473. [Google Scholar] [CrossRef]
- López de las Hazas, M.C.; Gil-Zamorano, J.; Cofán, M.; Mantilla-Escalante, D.C.; Garcia-Ruiz, A.; del Pozo-Acebo, L.; Pastor, O.; Yañez-Mo, M.; Mazzeo, C.; Serra-Mir, M.; et al. One-Year Dietary Supplementation with Walnuts Modifies Exosomal MiRNA in Elderly Subjects. Eur. J. Nutr. 2021, 60, 1999–2011. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.J.; Cardona-Alvarado, M.I.; Mercader, J.M.; Moreno-Navarrete, J.M.; Moreno, M.; Sabater, M.; Fuentes-Batllevell, N.; Ramírez-Chávez, E.; Ricart, W.; Molina-Torres, J.; et al. Circulating Profiling Reveals the Effect of a Polyunsaturated Fatty Acid-Enriched Diet on Common MicroRNAs. J. Nutr. Biochem. 2015, 26, 1095–1101. [Google Scholar] [CrossRef]
- Reis, B.Z.; Duarte, G.B.S.; Vargas-Mendez, E.; Ferreira, L.R.P.; Barbosa, F.; Cercato, C.; Rogero, M.M.; Cozzolino, S.M.F. Brazil Nut Intake Increases Circulating MiR-454-3p and MiR-584-5p in Obese Women. Nutr. Res. 2019, 67, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Salas-Huetos, A.; Moraleda, R.; Giardina, S.; Anton, E.; Blanco, J.; Salas-Salvadó, J.; Bulló, M. Effect of Nut Consumption on Semen Quality and Functionality in Healthy Men Consuming a Western-Style Diet: A Randomized Controlled Trial. Am. J. Clin. Nutr. 2018, 108, 953–962. [Google Scholar] [CrossRef] [PubMed]
- Daimiel, L.; Micó, V.; Valls, R.M.; Pedret, A.; Motilva, M.J.; Rubió, L.; Fitó, M.; Farrás, M.; Covas, M.I.; Solá, R.; et al. Impact of Phenol-Enriched Virgin Olive Oils on the Postprandial Levels of Circulating MicroRNAs Related to Cardiovascular Disease. Mol. Nutr. Food Res. 2020, 64, e2000049. [Google Scholar] [CrossRef] [PubMed]
- D’Amore, S.; Vacca, M.; Cariello, M.; Graziano, G.; D’Orazio, A.; Salvia, R.; Sasso, R.C.; Sabbà, C.; Palasciano, G.; Moschetta, A. Genes and MiRNA Expression Signatures in Peripheral Blood Mononuclear Cells in Healthy Subjects and Patients with Metabolic Syndrome after Acute Intake of Extra Virgin Olive Oil. Biochim. Biophys. Acta 2016, 1861, 1671–1680. [Google Scholar] [CrossRef] [PubMed]
- Cannataro, R.; Perri, M.; Gallelli, L.; Caroleo, M.C.; De Sarro, G.; Cione, E. Ketogenic Diet Acts on Body Remodeling and MicroRNAs Expression Profile. Microrna 2019, 8, 116–126. [Google Scholar] [CrossRef]
- Desgagné, V.; Guay, S.P.; Guérin, R.; Corbin, F.; Couture, P.; Lamarche, B.; Bouchard, L. Variations in HDL-Carried MiR-223 and MiR-135a Concentrations after Consumption of Dietary Trans Fat Are Associated with Changes in Blood Lipid and Inflammatory Markers in Healthy Men—An Exploratory Study. Epigenetics 2016, 11, 438–448. [Google Scholar] [CrossRef]
- Ferrero, G.; Carpi, S.; Polini, B.; Pardini, B.; Nieri, P.; Impeduglia, A.; Grioni, S.; Tarallo, S.; Naccarati, A. Intake of Natural Compounds and Circulating MicroRNA Expression Levels: Their Relationship Investigated in Healthy Subjects With Different Dietary Habits. Front. Pharmacol. 2021, 11, 619200. [Google Scholar] [CrossRef]
- Liu, T.; Gatto, N.M.; Chen, Z.; Qiu, H.; Lee, G.; Duerksen-Hughes, P.; Fraser, G.; Wang, C. Vegetarian Diets, Circulating MiRNA Expression and Healthspan in Subjects Living in the Blue Zone. Precis. Clin. Med. 2020, 3, 245–259. [Google Scholar] [CrossRef]
- Tarallo, S.; Pardini, B.; Mancuso, G.; Rosa, F.; Di Gaetano, C.; Rosina, F.; Vineis, P.; Naccarati, A. MicroRNA Expression in Relation to Different Dietary Habits: A Comparison in Stool and Plasma Samples. Mutagenesis 2014, 29, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Tarallo, S.; Ferrero, G.; De Filippis, F.; Francavilla, A.; Pasolli, E.; Panero, V.; Cordero, F.; Segata, N.; Grioni, S.; Pensa, R.G.; et al. Stool MicroRNA Profiles Reflect Different Dietary and Gut Microbiome Patterns in Healthy Individuals. Gut 2022, 71, 1302–1314. [Google Scholar] [CrossRef] [PubMed]
- Fontalba-Romero, M.I.; López-Enriquez, S.; Lago-Sampedro, A.; Garcia-Escobar, E.; Pastori, R.L.; Domínguez-Bendala, J.; Alvarez-Cubela, S.; Valdés, S.; Rojo-Martinez, G.; García-Fuentes, E.; et al. Association between the Mediterranean Diet and Metabolic Syndrome with Serum Levels of MiRNA in Morbid Obesity. Nutrients 2021, 13, 436. [Google Scholar] [CrossRef] [PubMed]
- Ramzan, F.; Mitchell, C.J.; Milan, A.M.; Schierding, W.; Zeng, N.; Sharma, P.; Mitchell, S.M.; D’Souza, R.F.; Knowles, S.O.; Roy, N.C.; et al. Comprehensive Profiling of the Circulatory MiRNAome Response to a High Protein Diet in Elderly Men: A Potential Role in Inflammatory Response Modulation. Mol. Nutr. Food Res. 2019, 63, e1800811. [Google Scholar] [CrossRef] [PubMed]
- Shin, P.K.; Kim, M.S.; Park, S.J.; Kwon, D.Y.; Kim, M.J.; Yang, H.J.; Kim, S.H.; Kim, K.; Chun, S.; Lee, H.J.; et al. A Traditional Korean Diet Alters the Expression of Circulating MicroRNAs Linked to Diabetes Mellitus in a Pilot Trial. Nutrients 2020, 12, 2558. [Google Scholar] [CrossRef] [PubMed]
- Tutino, V.; De Nunzio, V.; Milella, R.A.; Gasparro, M.; Cisternino, A.M.; Gigante, I.; Lanzilotta, E.; Iacovazzi, P.A.; Lippolis, A.; Lippolis, T.; et al. Impact of Fresh Table Grape Intake on Circulating MicroRNAs Levels in Healthy Subjects: A Significant Modulation of Gastrointestinal Cancer-Related Pathways. Mol. Nutr. Food Res. 2021, 65, 2100428. [Google Scholar] [CrossRef]
- Capetini, V.C.; Quintanilha, B.J.; de Oliveira, D.C.; Nishioka, A.H.; de Matos, L.A.; Ferreira, L.R.P.; Ferreira, F.M.; Sampaio, G.R.; Hassimotto, N.M.A.; Lajolo, F.M.; et al. Blood Orange Juice Intake Modulates Plasma and PBMC MicroRNA Expression in Overweight and Insulin-Resistant Women: Impact on MAPK and NFκB Signaling Pathways. J. Nutr. Biochem. 2023, 112, 109240. [Google Scholar] [CrossRef]
- Desvignes, T.; Batzel, P.; Berezikov, E.; Eilbeck, K.; Eppig, J.T.; McAndrews, M.S.; Singer, A.; Postlethwait, J.H. MicroRNA Nomenclature: A View Incorporating Genetic Origins, Biosynthetic Pathways, and Sequence Variants. Trends Genet. 2015, 31, 613. [Google Scholar] [CrossRef]
- Ameling, S.; Kacprowski, T.; Chilukoti, R.K.; Malsch, C.; Liebscher, V.; Suhre, K.; Pietzner, M.; Friedrich, N.; Homuth, G.; Hammer, E.; et al. Associations of Circulating Plasma MicroRNAs with Age, Body Mass Index and Sex in a Population-Based Study. BMC Med. Genom. 2015, 8, 61. [Google Scholar] [CrossRef]
- Ban, E.; Song, E.J. Considerations and Suggestions for the Reliable Analysis of MiRNA in Plasma Using QRT-PCR. Genes 2022, 13, 328. [Google Scholar] [CrossRef]
- Wong, R.K.Y.; MacMahon, M.; Woodside, J.V.; Simpson, D.A. A Comparison of RNA Extraction and Sequencing Protocols for Detection of Small RNAs in Plasma. BMC Genom. 2019, 20, 446. [Google Scholar] [CrossRef] [PubMed]
- Meerson, A.; Traurig, M.; Ossowski, V.; Fleming, J.M.; Mullins, M.; Baier, L.J. Human Adipose MicroRNA-221 Is Upregulated in Obesity and Affects Fat Metabolism Downstream of Leptin and TNF-α. Diabetologia 2013, 56, 1971–1979. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Shan, A.; Su, Y.; Cheng, Y.; Ji, H.; Yang, Q.; Lei, Y.; Liu, B.; Wang, W.; Ning, G.; et al. MiR-221/222 Inhibit Insulin Production of Pancreatic β-Cells in Mice. Endocrinology 2020, 161, bqz027. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of Tissue-Specific MicroRNAs from Mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef]
- Hu, Y.; Liu, H.X.; Jena, P.K.; Sheng, L.; Ali, M.R.; Wan, Y.J.Y. MiR-22 Inhibition Reduces Hepatic Steatosis via FGF21 and FGFR1 Induction. JHEP Rep. 2020, 2, 100093. [Google Scholar] [CrossRef]
- Panella, R.; Petri, A.; Desai, B.N.; Fagoonee, S.; Cotton, C.A.; Nguyen, P.K.; Lundin, E.M.; Wagshal, A.; Wang, D.Z.; Näär, A.M.; et al. MicroRNA-22 Is a Key Regulator of Lipid and Metabolic Homeostasis. Int. J. Mol. Sci. 2023, 24, 12870. [Google Scholar] [CrossRef]
- Agbu, P.; Carthew, R.W. MicroRNA-Mediated Regulation of Glucose and Lipid Metabolism. Nat. Rev. Mol. Cell Biol. 2021, 22, 425–438. [Google Scholar] [CrossRef]
- Hartig, S.M.; Hamilton, M.P.; Bader, D.A.; McGuire, S.E. The MicroRNA Interactome in Metabolic Homeostasis. Trends Endocrinol. Metab. 2015, 26, 733. [Google Scholar] [CrossRef]
- Kabekkodu, S.P.; Shukla, V.; Varghese, V.K.; D’Souza, J.; Chakrabarty, S.; Satyamoorthy, K. Clustered MiRNAs and Their Role in Biological Functions and Diseases. Biol. Rev. Camb. Philos. Soc. 2018, 93, 1955–1986. [Google Scholar] [CrossRef]
- Vishnoi, A.; Rani, S. MiRNA Biogenesis and Regulation of Diseases: An Overview. In Methods Mol Biol; Humana Press Inc.: Totowa, NJ, USA, 2017; Volume 1509, pp. 1–10. [Google Scholar]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. MiRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
- Zhou, S.S.; Jin, J.P.; Wang, J.Q.; Zhang, Z.G.; Freedman, J.H.; Zheng, Y.; Cai, L. MiRNAS in Cardiovascular Diseases: Potential Biomarkers, Therapeutic Targets and Challenges. Acta Pharmacol. Sin. 2018, 39, 1073–1084. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Guo, X. The Clinical Potential of Circulating MicroRNAs in Obesity. Nat. Rev. Endocrinol. 2019, 15, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Foscolou, A.; Critselis, E.; Panagiotakos, D. Olive Oil Consumption and Human Health: A Narrative Review. Maturitas 2018, 118, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Nocella, C.; Cammisotto, V.; Fianchini, L.; D’Amico, A.; Novo, M.; Castellani, V.; Stefanini, L.; Violi, F.; Carnevale, R. Extra Virgin Olive Oil and Cardiovascular Diseases: Benefits for Human Health. Endocr. Metab. Immune Disord.-Drug Targets 2018, 18, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Marrone, G.; Guerriero, C.; Palazzetti, D.; Lido, P.; Marolla, A.; Di Daniele, F.; Noce, A. Vegan Diet Health Benefits in Metabolic Syndrome. Nutrients 2021, 13, 817. [Google Scholar] [CrossRef] [PubMed]
- He, P.P.; OuYang, X.P.; Li, Y.; Lv, Y.C.; Wang, Z.B.; Yao, F.; Xie, W.; Tan, Y.L.; Li, L.; Zhang, M.; et al. MicroRNA-590 Inhibits Lipoprotein Lipase Expression and Prevents Atherosclerosis in ApoE Knockout Mice. PLoS ONE 2015, 10, e0138788. [Google Scholar] [CrossRef]
- He, P.P.; Ouyang, X.P.; Tang, Y.Y.; Liao, L.; Wang, Z.B.; Lv, Y.C.; Tian, G.P.; Zhao, G.J.; Huang, L.; Yao, F.; et al. MicroRNA-590 Attenuates Lipid Accumulation and pro-Inflammatory Cytokine Secretion by Targeting Lipoprotein Lipase Gene in Human THP-1 Macrophages. Biochimie 2014, 106, 81–90. [Google Scholar] [CrossRef]
- Liu, X.L.; Cao, H.X.; Wang, B.C.; Xin, F.Z.; Zhang, R.N.; Zhou, D.; Yang, R.X.; Zhao, Z.H.; Pan, Q.; Fan, J.G. MiR-192-5p Regulates Lipid Synthesis in Non-Alcoholic Fatty Liver Disease through SCD-1. World J. Gastroenterol. 2017, 23, 8140–8151. [Google Scholar] [CrossRef]
- Castaño, C.; Kalko, S.; Novials, A.; Párrizas, M. Obesity-Associated Exosomal MiRNAs Modulate Glucose and Lipid Metabolism in Mice. Proc. Natl. Acad. Sci. USA 2018, 115, 12158–12163. [Google Scholar] [CrossRef]
- Li, C.; Li, X.; Gao, X.; Zhang, R.; Zhang, Y.; Liang, H.; Xu, C.; Du, W.; Zhang, Y.; Liu, X.; et al. MicroRNA-328 as a Regulator of Cardiac Hypertrophy. Int. J. Cardiol. 2014, 173, 268–276. [Google Scholar] [CrossRef] [PubMed]
- McManus, D.D.; Rong, J.; Huan, T.; Lacey, S.; Tanriverdi, K.; Munson, P.J.; Larson, M.G.; Joehanes, R.; Murthy, V.; Shah, R.; et al. Messenger RNA and MicroRNA Transcriptomic Signatures of Cardiometabolic Risk Factors. BMC Genom. 2017, 18, 139. [Google Scholar] [CrossRef]
- Vienberg, S.; Geiger, J.; Madsen, S.; Dalgaard, L.T. MicroRNAs in Metabolism. Acta Physiol. 2017, 219, 346–361. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy, A.; Ezekiel, U. Phytochemical Modulation of MiRNAs in Colorectal Cancer. Medicines 2019, 6, 48. [Google Scholar] [CrossRef] [PubMed]
- Tomé-Carneiro, J.; Larrosa, M.; Yáñez-Gascón, M.J.; Dávalos, A.; Gil-Zamorano, J.; Gonzálvez, M.; García-Almagro, F.J.; Ruiz Ros, J.A.; Tomás-Barberán, F.A.; Espín, J.C.; et al. One-Year Supplementation with a Grape Extract Containing Resveratrol Modulates Inflammatory-Related MicroRNAs and Cytokines Expression in Peripheral Blood Mononuclear Cells of Type 2 Diabetes and Hypertensive Patients with Coronary Artery Disease. Pharmacol. Res. 2013, 72, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Sanchita; Trivedi, R.; Asif, M.H.; Trivedi, P.K. Dietary Plant MiRNAs as an Augmented Therapy: Cross-Kingdom Gene Regulation. RNA Biol. 2018, 15, 1433–1439. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W. XenomiRs and MiRNA Homeostasis in Health and Disease: Evidence That Diet and Dietary MiRNAs Directly and Indirectly Influence Circulating MiRNA Profiles. RNA Biol. 2012, 9, 1147. [Google Scholar] [CrossRef]
- Jia, M.; He, J.; Bai, W.; Lin, Q.; Deng, J.; Li, W.; Bai, J.; Fu, D.; Ma, Y.; Ren, J.; et al. Cross-Kingdom Regulation by Dietary Plant MiRNAs: An Evidence-Based Review with Recent Updates. Food Funct. 2021, 12, 9549–9562. [Google Scholar] [CrossRef]
- Díez-Sainz, E.; Lorente-Cebrián, S.; Aranaz, P.; Riezu-Boj, J.I.; Martínez, J.A.; Milagro, F.I. Potential Mechanisms Linking Food-Derived MicroRNAs, Gut Microbiota and Intestinal Barrier Functions in the Context of Nutrition and Human Health. Front. Nutr. 2021, 8, 586564. [Google Scholar] [CrossRef]
- Li, J.; Lei, L.; Ye, F.; Zhou, Y.; Chang, H.; Zhao, G. Nutritive Implications of Dietary MicroRNAs: Facts, Controversies, and Perspectives. Food Funct. 2019, 10, 3044–3056. [Google Scholar] [CrossRef]
- Title, A.; Denzler, R.; Stoffel, M. Reply to Diet-Responsive MicroRNAs Are Likely Exogenous. J. Biol. Chem. 2015, 290, 25198. [Google Scholar] [CrossRef] [PubMed]
- Snow, J.W.; Hale, A.E.; Isaacs, S.K.; Baggish, A.L.; Chan, S.Y. Ineffective Delivery of Diet-Derived MicroRNAs to Recipient Animal Organisms. RNA Biol. 2013, 10, 1107–1116. [Google Scholar] [CrossRef]
- Cintio, M.; Polacchini, G.; Scarsella, E.; Montanari, T.; Stefanon, B.; Colitti, M. MicroRNA Milk Exosomes: From Cellular Regulator to Genomic Marker. Animals 2020, 10, 1126. [Google Scholar] [CrossRef]
- Baier, S.R.; Nguyen, C.; Xie, F.; Wood, J.R.; Zempleni, J. MicroRNAs Are Absorbed in Biologically Meaningful Amounts from Nutritionally Relevant Doses of Cow Milk and Affect Gene Expression in Peripheral Blood Mononuclear Cells, HEK-293 Kidney Cell Cultures, and Mouse Livers. J. Nutr. 2014, 144, 1495–1500. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Izumi, H.; Sekine, K.; Ochiya, T. MicroRNA as a New Immune-Regulatory Agent in Breast Milk. Silence 2010, 1, 7. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Du, X.; Li, J.; Lönnerdal, B. Human Milk Exosomes and Their MicroRNAs Survive Digestion in Vitro and Are Taken up by Human Intestinal Cells. Mol. Nutr. Food Res. 2017, 61, 1700082. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Lozano, E.; Sebastián-Valles, F.; Knott-Torcal, C. Circulating MicroRNAs in Breast Milk and Their Potential Impact on the Infant. Nutrients 2020, 12, 66. [Google Scholar] [CrossRef]
- Langley-Evans, S.C. Nutrition in Early Life and the Programming of Adult Disease: A Review. J. Hum. Nutr. Diet. 2015, 28, 1–14. [Google Scholar] [CrossRef]
- Carolan-Olah, M.; Duarte-Gardea, M.; Lechuga, J. A Critical Review: Early Life Nutrition and Prenatal Programming for Adult Disease. J. Clin. Nurs. 2015, 24, 3716–3729. [Google Scholar] [CrossRef]
- Picó, C.; Palou, M.; Priego, T.; Sánchez, J.; Palou, A.; Sanchez, J.; Palou, A. Metabolic Programming of Obesity by Energy Restriction during the Perinatal Period: Different Outcomes Depending on Gender and Period, Type and Severity of Restriction. Front. Physiol. 2012, 3, 436. [Google Scholar] [CrossRef]
- Alonso-Bernáldez, M.; Asensio, A.; Palou-March, A.; Sánchez, J.; Palou, A.; Serra, F.; Palou, M. Breast Milk MicroRNAs Related to Leptin and Adiponectin Function Can Be Modulated by Maternal Diet and Influence Offspring Phenotype in Rats. Int. J. Mol. Sci. 2022, 23, 7237. [Google Scholar] [CrossRef] [PubMed]
- Pomar, C.A.; Serra, F.; Palou, A.; Sánchez, J. Lower MiR-26a Levels in Breastmilk Affect Gene Expression in Adipose Tissue of Offspring. FASEB J. 2021, 35, e21924. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; Stremmel, W.; Weiskirchen, R.; John, S.M.; Schmitz, G. Exosome-Derived MicroRNAs of Human Milk and Their Effects on Infant Health and Development. Biomolecules 2021, 11, 851. [Google Scholar] [CrossRef] [PubMed]
- Golan-Gerstl, R.; Elbaum Shiff, Y.; Moshayoff, V.; Schecter, D.; Leshkowitz, D.; Reif, S. Characterization and Biological Function of Milk-Derived MiRNAs. Mol. Nutr. Food Res. 2017, 61, 1700009. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Li, M.; Wang, X.; Li, Q.; Wang, T.; Zhu, Q.; Zhou, X.; Wang, X.; Gao, X.; Li, X. Immune-Related MicroRNAs Are Abundant in Breast Milk Exosomes. Int. J. Biol. Sci. 2012, 8, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.B.; Chernausek, S.D.; Garman, L.D.; Pezant, N.P.; Plows, J.F.; Kharoud, H.K.; Demerath, E.W.; Fields, D.A. Human Milk Exosomal MicroRNA: Associations with Maternal Overweight/Obesity and Infant Body Composition at 1 Month of Life. Nutrients 2021, 13, 1091. [Google Scholar] [CrossRef] [PubMed]
- Torres, L.F.; Cogliati, B.; Otton, R. Green Tea Prevents NAFLD by Modulation of MiR-34a and MiR-194 Expression in a High-Fat Diet Mouse Model. Oxid. Med. Cell. Longev. 2019, 2019, 4168380. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).